Abstract
Diverse ascomycete fungi oxidize manganese(II) [Mn(II)] and produce Mn(III, IV) oxides in terrestrial and freshwater environments. Although multicopper oxidase (MCO) is considered to be a key catalyst in mediating Mn(II) oxidation in ascomycetes, the responsible gene and its product have not been identified. In this study, a gene, named mco1, encoding Mn(II)-oxidizing MCO from Acremonium strictum strain KR21-2 was cloned and heterologously expressed in the methylotrophic yeast Pichia pastoris. Based on the phylogenetic relationship, similarity of putative copper-binding motifs, and homology modeling, the gene product Mco1 was assigned to a bilirubin oxidase. Mature Mco1 was predicted to be composed of 565 amino acids with a molecular mass of 64.0 kDa. The recombinant enzyme oxidized Mn(II) to yield spherical Mn oxides, several micrometers in diameter. Zinc(II) ions added to the reaction mixture were incorporated by the Mn oxides at a Zn/Mn molar ratio of 0.36. The results suggested that Mco1 facilitates the growth of the micrometer-sized Mn oxides and affects metal sequestration through Mn(II) oxidation. This is the first report on heterologous expression and identification of the Mn(II) oxidase enzyme in Mn(II)-oxidizing ascomycetes. The cell-free, homogenous catalytic system with recombinant Mco1 could be useful for understanding Mn biomineralization by ascomycetes and the sequestration of metal ions in the environment
1. Introduction
Several microorganisms can convert manganese(II) [Mn(II)] to insoluble Mn(III, IV) oxides and are responsible for Mn deposition in natural environments [1]. Abiotic oxidation of Mn(II) at neutral pH proceeds only at a limited rate. Biogenic Mn oxides serve as fine adsorbents for cationic species and as oxidants for redox-sensitive elements, implying their important role in the geochemical cycles of numerous elements [1,2]. Based on these functions, microbial Mn(II) oxidation has attracted attention in environmental applications related to the removal of Mn and other toxic elements [3,4,5]. Since Mn oxides serve as an oxidant of organic compounds, recent studies have also focused on using biogenic Mn oxides for the treatment of wastewater contaminated with organic pollutants [6]. Studies on catalytic oxidation of Mn(II) by microorganisms are of growing significance, not only because of such geochemical implications of Mn minerals but also due to its biotechnological applicability.
In bacterial Mn(II) oxidation, multicopper oxidase (MCO) is considered a key enzyme that catalyzes the conversion of Mn(II) to Mn oxides [2,7]. Another enzyme that oxidizes Mn(II) directly is the animal heme peroxidase [7]. Mn(II)-oxidizing MCOs were identified in many Mn(II)-oxidizing bacteria including, Bacillus sp. strains SG-1 and PL-12, Leptothrix discophora, Pseudomonas putida, and Pedomicrobium sp. ACM 3067 [7]. Several genes encoding Mn(II)-oxidizing MCO have been cloned and heterologously expressed [8,9,10,11,12]. Expression of an MCO protein, MnxG, along with other Mnx subunit proteins from Bacillus sp. strain PL-12 yielded active Mn(II) oxidase enzyme [8]. The microbial cell-free Mn-oxidizing system with Mnx protein complex provides an insight into the enzymatic Mn(II) oxidation reaction and Mn(IV) oxide mineralization at the atomic and molecular levels [13,14].
MCOs constitute an enzyme superfamily, including laccases (EC 1.10.3.2), ascorbate oxidases (EC 1.10.3.3), ferroxidases (EC 1.16.3.1), and bilirubin oxidases (EC 1.3.3.5); they mediate one-electron oxidation of a broad range of substrates by O2 as an electron acceptor. Diverse fungi belonging to the phyla Basidiomycota and Ascomycota also use MCO for Mn(II) oxidation. In the basidiomycetes, laccase is known to catalyze one-electron oxidation of Mn(II) in the presence of a suitable Mn chelator [15,16,17] and accumulate soluble Mn(III) chelate, as well as manganese peroxidase [18]. These enzymes are involved in lignin decomposition. Laccase or laccase-like MCOs appear to mediate the Mn(II) oxidation in ascomycetes [3,19,20,21] in addition to an indirect system with biotic superoxide (O2−) [22]. The ascomycete MCOs catalyze Mn(II) oxidation without a Mn chelator to yield insoluble Mn oxide. However, catalytic Mn(II) oxidation and the resulting Mn precipitation has not been substantiated in ascomycetes. Since the gene responsible for Mn(II) oxidation has not been identified in the ascomycetes yet, overexpression of the enzyme cannot be used for unveiling the reaction mechanism.
The ascomycete enzyme that converts Mn(II) to Mn oxides was first purified from the spent culture medium of Acremonium strictum strain KR21-2 [23]. The enzyme reaction yielded micrometer-sized particles consisting mainly of Mn(IV). Based on the biochemical traits and N-terminal amino acid sequence similarity, the Mn(II)-oxidizing enzyme was identified as a laccase-like MCO [23]. A. strictum KR21-2 cultures have been used as a fungal model system for understanding Mn oxide formation, and sorption, redox reactions, and sequestrations of various elements on the Mn oxides. Like the bacterial Mn oxides, the strain KR21-2 Mn oxides also resemble vernadite (δ–MnO2), a natural nanostructured and turbostratic variety of birnessite, and the crystallites have domain dimensions of ~10 nm in the layer plane [24]. Based on the small size of the crystallites and the layer structures with 22% to 30% vacant octahedral sites, the Mn oxides of strain KR21-2 show high capacity for sorption of heavy metal cations. The molar ratios of zinc(II) [Zn(II)], cobalt(II) [Co(II)], and nickel(II) loadings to solid Mn have been observed to be as much as 0.3, 0.23, and 0.12, respectively [25]. Recent research with strain KR21-2 has demonstrated that newly formed Mn oxides possess Mn(II) oxidase activity, which strongly affects the sorption and redox reactions of metals on the solid Mn phase. The enzyme activity enhances the sorption of metal cations [26,27,28] and oxidation of arsenic(III) [29,30], Co(II) [31], and chromium (III) [32] by the Mn oxides. These observations suggest the crucial role of Mn(II)-oxidizing enzymes associated with solid Mn phase in the fate of diverse elements.
Since A. strictum KR21-2 secretes only a small amount of enzyme into the culture medium [23], an enhanced production system is required for large-scale production of the enzyme. The aim of this study was to identify the Mn(II)-oxidizing enzyme of A. strictum KR21-2 by gene cloning and heterologous expression in the methylotrophic yeast Pichia pastoris. Although laccase or laccase-like MCOs are considered to be Mn oxidases in ascomycetes, the present study indicated that the enzyme from strain KR21-2 belonged to a different class of MCOs. The recombinant enzyme obtained in this study can serve as a useful tool for investigating the Mn biomineralization driven by ascomycetes and the Mn oxide–metal interactions.
2. Results
2.1. Identification of Mco1 Gene
The genome of A. strictum strain KR21-2 was analyzed by whole-genome shotgun sequencing. The genome sequence assembly consisted of 298 scaffolds with an N50 of 237.2 kb with the genome size of approximately 35.25 Mb. The genome contained one putative gene, 2021 bp in length; its sequences corresponded to the N-terminus of the KR21-2 Mn(II) oxidase [23] and type-1 (T1) to -3 (T3) copper-binding sites conserved in the related MCOs (Figure S1) [33]. No other putative gene encoding MCO was found in the genome. The MCO-encoding region appeared to be interrupted by three introns, with splice junctions that followed the GT-AG rule (Figure S1).
Mn(II) oxidation in the liquid culture of strain KR21-2 started at 40 h (Figure 1). The Mn(II) oxidase activity was monitored in cultures without Mn(II) and was found between 34 to 58 h. The cDNA of the targeted protein, named Mco1 as the product of gene mco1, was obtained from the total RNA collected from the culture at 42 h and was subjected to cloning and sequencing. It was predicted that Mco1 consisted of 614 amino acids, where the first 49 residues could be the signal peptide for extracellular secretion because the N-terminus of the mature polypeptide [23] started from the 50th position (Figure S1). The mature Mco1 was predicted to have a molecular weight of 64.02 kDa. The cDNA sequence contained the copper (Cu)-binding regions that are highly conserved among fungal bilirubin oxidases, a class of MCOs (Figure 2). The sequence contained four conserved histidine motifs for Cu ligands: HXH, HXH, HXXHXH, and HXHXXXH (Figure 2). The first and second motifs of Mco1 were consistent with fungal bilirubin oxidases (HLH and HDH) and bacterial CotA, an endospore coat protein of Bacillus spp., but tryptophan (W) and serine (S) that are found caught between two histidine (H) residues in laccases were replaced with leucine (L) and aspartic acid (D), respectively, in Mco1. The third and fourth motifs also showed high similarities with bilirubin oxidases and CotA.
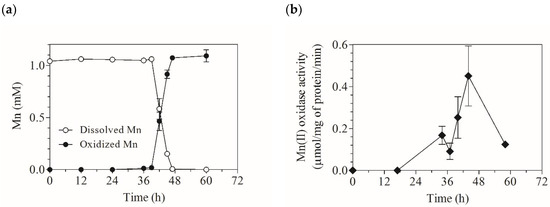
Figure 1.
Mn(II) oxidation by A. strictum KR21-2 and Mn(II) oxidase activity in spent culture medium. (a) Formation of Mn oxides in the culture; (b) Extracellular Mn(II) oxidase activity of the culture supernatant. Data represent mean ± SD from duplicate experiments.
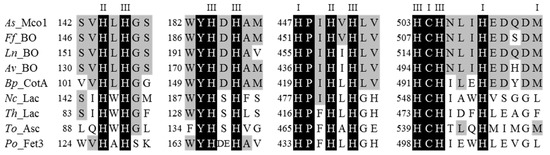
Figure 2.
Amino acid sequence alignment of the four conserved Cu-binding regions in A. strictum KR21-2 (As) Mco1 and related MCO proteins including fungal bilirubin oxidases (BO), laccases (Lac), L-ascorbate oxidase (Asc), Fet3 ferroxidase, and bacterial CotA protein. Ff, Fusarium floridanum; Ln, Lophiotrema nucula; Av, Albifimbria verrucaria; Bp, Bacillus pumilus; Nc, Neurospora crassa; Th, Trametes hirsuta; To, Tolypocladium ophioglossoides; Po, Pleurotus ostreatus. The Cu-binding T1, T2, and T3 residues are represented by I, II, and III, respectively. For accession number of each sequence, see Figure S2. In fungal proteins, the positions of amino acid residues were counted for the polypeptide accompanied by N-terminal signal sequence.
Phylogenetic analysis of the deduced amino acid sequence clearly indicated that Mco1 could be assigned to a bilirubin oxidase instead of laccase (Figure S2). Crystal structure of bilirubin oxidase from Albifimbria verrucaria (AvBOx) has been solved [34]. Homology model of Mco1 was generated and was compared with the structure of AvBOx (Protein Data Bank number, 2XLL). Overall structure of the Mco1 model and AvBOx was almost identical (RMSD = 0.245 Å, 463 Cα atoms) (Figure S3). Moreover, structures of Cu-binding sites were highly conserved (Figure S3), further supporting that Mco1 is bilirubin oxidase. The KR21-2 enzyme was also related to the bacterial MCOs including CotA and CueO, a periplasmic Cu efflux system-associated protein (Figure S2).
2.2. Heterologous Expression and Purification
Pichia pastoris transformed with pPICZα-mco1 deposited Mn oxides, indicating the expression of Mco1 with Mn(II) oxidase activity (Figure 3). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of undenatured protein followed by activity staining demonstrated the secretion of an active protein in liquid cultures, induced by methanol (Figure 3). The culture supernatant was collected at 36 h and Mco1 was purified with Superdex 200 and Mono Q columns. The purified fraction showed a blue color, consistent with the nature of MCO. A total of 4.5 mg of recombinant protein was produced when 1 L of culture was subjected to methanol induction. SDS-PAGE of the denatured protein represented a minor band with an apparent molecular weight of 46 kDa, along with a major band of 59 kDa (Figure 3).
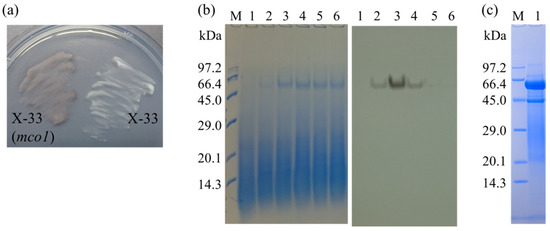
Figure 3.
Expression of A. strictum KR21-2 mco1 gene in P. pastoris. (a) Insoluble Mn oxide production by P. pastoris X-33 transformed with pPCIZα-mco1 (left); (b) SDS-PAGE of undenatured recombinant Mco1 followed by in-gel assay of Mn(II) oxidase activity, wherein the protein bands were visualized by Coomassie brilliant blue staining (left panel) and by formation of insoluble Mn oxides (right panel); (c) SDS-PAGE of denatured recombinant Mco1 (purified by FPLC) (lane 1). Lane M was loaded with protein markers (sizes are shown on the left). Lanes 1–6 in (b) were loaded with the supernatant fluid from the transformant cultures collected at 0, 12, 36, 48, 60, and 72 h after induction by methanol.
2.3. Mn(II) Oxidation by Recomninant Mco1
We observed that the recombinant Mco1 completed the oxidation of 0.6 mM Mn(II) within 3 h (Figure 4). The majority of solid Mn (96.1%) was present in the oxidized form, while only a portion (3.9%) was in the sorbed form that is exchangeable with excess Cu (II) ions. Scanning electron microscopy (SEM) and energy-dispersive X-ray (EDX) analysis demonstrated that the enzyme reaction yielded spherical Mn oxides with fine-crumpled surfaces (Figure 4 and Figure S3). The diameter of the particles was 2.8 ± 0.6 μm (mean ± standard deviation, n = 60). To examine the capacity for metal sorption, the produced oxides were washed and then soaked in a Zn(II)-containing solution. The Zn(II) loaded to Mn oxides and reached a solid Zn/solid Mn molar ratio of 0.25 (Table 1). At this time, no detectable amount of Mn(II) was released from the Mn oxides despite the high Zn(II) loading. Mn(II) oxidation or Zn(II) precipitation was not observed for the reaction mixtures without enzyme, indicating that the enzyme activity contributed thoroughly to the metal sequestrations.
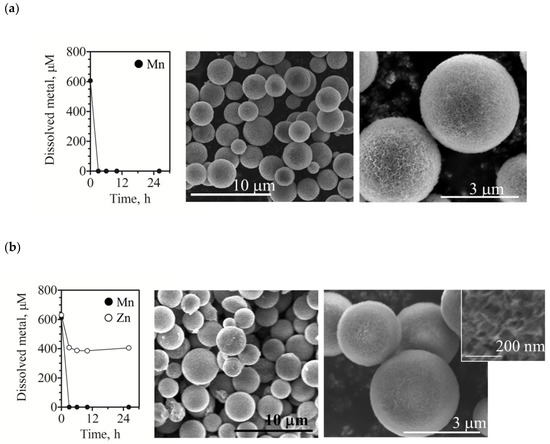
Figure 4.
Mn(II) oxidation by recombinant A. strictum KR21-2 Mco1 and SEM images of solid-phase Mn produced by the enzyme reaction. (a) Precipitation of dissolved Mn(II) during the enzyme reaction without ZnSO4; (b) Precipitation of dissolved Zn(II) along with Mn(II) during the reaction in the presence of ZnSO4. The data of metal concentrations represent mean ± SD from duplicate runs; error bars are smaller than the symbols. The SEM images display the formation of spherical Mn oxide particles with fine-crumpled surfaces, which were collected after 26 h of incubation.

Table 1.
Sorption of Zn(II) on Mn oxides produced by recombinant Mco1 1.
The enzyme was able to complete Mn(II) oxidation within 3 h even in the presence of a similar concentration of Zn(II) (~0.65 mM; Figure 4 and Figure S3 and Table 1). The diameter of the particles was 3.1 ± 0.8 μm, which was not significantly different from that of the particles obtained in the absence of Zn(II) (p = 0.0583). Concentration of aqueous-phase Mn(II) and Zn(II) did not decrease when the enzyme was omitted from the reaction mixture (data not shown). The result indicated that the incorporation of Zn(II) was concurrent with Mn oxide formation. In this case, the molar ratio of solid Zn/solid Mn was ~0.4 (Figure 4 and Table 1), which was 1.6 times higher than that when Zn(II) was added later. Solid-phase Mn was mostly present in an oxidized form (Cu-exchangeable fraction, <3%).
2.4. Sequence Data Deposition
The cDNA sequence of mco1 has been deposited in DNA Data Bank of Japan (DDBJ) under the accession number LC547411.
3. Discussion
In A. strictum KR21-2, the Mn(II)-oxidizing enzyme was previously identified as a laccase-like MCO due to its biochemical nature, including the presence of laccase substrate oxidizing activity, Cu content, and N-terminus similarity [19,23]. In the present study, the gene encoding the Mn(II) oxidase enzyme from the fungus was successfully cloned and overexpressed in P. pastoris. We concluded that enzyme Mco1 belongs to a class of bilirubin oxidases based on the phylogenetic relationship among MCOs, the similarity between the conserved Cu-binding histidine motifs, and the result of homology modeling. The presence of extracellular activity and the additional signal sequence in mco1 gene indicated that the enzyme was secreted to the culture medium. The Mco1 sequence also showed a high similarity with the bacterial CotA, which possesses bilirubin [35,36] and Mn(II) [9] oxidation activities. Based on Mco1 analysis in this study, Mn(II) oxidizing activity may be widely distributed in fungal bilirubin oxidases and related enzymes.
Mco1 was produced in active form by the recombinant P. pastoris expression system. SDS-PAGE of the denatured protein resulted in the appearance of two protein bands. This is consistent with a previous study on the enzyme purified from A. strictum KR21-2 culture [23]. Thus, the 46-kDa band was identified as the secondary form of Mco1 rather than a second protein [23]. Due to the fact that the mco1 gene encoded a single polypeptide of 64.02 kDa, we concluded that the recombinant Mco1 was successfully produced by the P. pastoris transformant and was purified to homogeneity via column chromatography. Mco1 was most likely monomeric because the recombinant protein single-handedly oxidized Mn(II) in the homogenous reaction mixture, as reported for bacterial MCOs such as CotA of B. pumilus [9] and CueO of Escherichia coli [10]. Though Mco1 showed similarity with CotA, yet these two enzymes differ in the kinetic properties for the Mn(II) oxidation. The kinetic properties of Mco1 enzyme purified from strain KR21-2 have been described previously [23]. The pH optimum for Mn(II) oxidation was 7.0, and no activity was detected at pH 8.1. The enzyme did not require additional Cu (II) for the oxidation, and the apparent half-saturation constant was 0.20 mM. CotA oxidizes Mn(II) faster when pH exceeds 8.0 and the reaction is enhanced largely by addition of 0.8 mM Cu (II) [9]. Furthermore, CotA shows the apparent half-saturation constant of ~15 mM [9], much higher than that of Mco1. Unlike these enzymes, the Mn(II)-oxidizing Bacillus sp. strain PL-12 produces a multimeric enzyme, including MnxG (an MCO) and two accessory proteins MnxE and MnxF with molecular mass of ~200 kDa [8]. MnxE and MnxF are suggested to be redox active and involved in a series of Mn mineralization processes, along with MnxG. The Mnx complex facilitates two one-electron transfers from Mn(II) to Mn(III) and from Mn(III) to Mn(IV). Although Mn species in the oxides were not investigated in this study, X-ray absorption near-edge structure spectroscopic data from a previous study revealed that 30-min reaction with the purified enzyme yielded mainly Mn(IV) oxide, possibly with a minor amount of Mn(III) (~10%) [23]. Further study is required to elucidate whether (or how) monomeric Mco1 mediates similar electron transfers during Mn(II) oxidation to Mn(IV).
Interestingly, the Mn(II) oxidation by Mco1 resulted in the formation of well-proportioned, spherical Mn oxide phase with diameters up to several micrometers, somewhat larger than the Mn oxides (0.7–1 μm) obtained with the purified enzyme from the A. strictum cultures in a previous study [23]. The surface of the particles showed fine-crumpled structures, which are consistent with the formation of vernadite (δ–MnO2) in fungal cultures [23,24]. The formation of spherical shapes has not been observed in the fungal cultures; it is likely due to the use of a simple, homogenous solution consisting of recombinant Mco1, Mn(II), and a buffering salt. Furthermore, such spherical forms may be characteristic of Mco1. In the bacterial CueO, Mn(II) is shown to be converted to a granular phase including a Mn(II, III) oxide, γ–Mn3O4, with crystals sized ~350 nm within 24 h [9]. This enzyme reaction was conducted in mixtures with high concentrations of Mn(II) (5 mM) at pH 8.0. Such low valency of Mn might represent abiotic secondary product from reaction of Mn(II) with biotic Mn(IV) oxide [37,38,39]. In a previous study, it was observed that recombinant Mnx complex converted Mn(II) to nanoparticulate crystallites with dimensions of 1–10.5 nm during the 30-min-enzyme reaction [40]. Research has unveiled the bacterial Mn biomineralization process, wherein the nanoparticulate crystallites are released from the Mnx complex and then aggregated into larger particles [13,40,41]. The nanoparticle formation and subsequent aggregation process was also demonstrated in Mn mineralization by Roseobacter sp., wherein enzymatically generated superoxide (O2−) induced formation of colloidal Mn oxides with dimensions of 20–100 nm [42,43]. Aggregation of colloidal Mn oxides requires unknown organic reactants (e.g., proteins, organic metabolites, and exopolymers) present in the culture broth. If a similar process is involved in Mn(II) oxidation by Mco1, Mco1 may also participate in the growth of Mn oxide particles. Given that the shape of the particles is almost spherical; aggregation of the particles should not be a disordered process. Otherwise, it may result from continuous accumulation of Mn that is sorbed, oxidized (enzymatically or nonenzymatically), and then fixed as oxide on the surfaces of the initial particulate product. Although the mechanism is unknown, this hypothesis appears inconsistent with a previous result that the enzyme of A. strictum KR21-2 immobilized on the formed Mn oxides and participated in Mn oxidation [44].
The Mn oxides produced by recombinant Mco1 showed high sorption capacity for Zn(II). The Mn oxides formed during the 3-h incubation period were immediately collected and subjected to the sorption experiments. Given the relatively short time taken for these experimental procedures, it is likely that the enzyme remains active during the sorption experiment, as also shown previously [27,44]. This is supported by the fact that no Mn(II) ion was released from the Mn oxides despite the high Zn(II) loading (Zn/Mn molar ratio = 0.25, Table 1). In the previous study [27], newly formed Mn oxides of A. strictum KR21-2 released Mn(II) at a molar concentration corresponding to approximately 20% of sorbed Zn(II) through an ion-exchange reaction only when the enzyme was inactivated under anaerobic conditions. The significance of Mn(II)-oxidizing activity during Zn(II) sorption was also described for Paraconiothyrium sp. [45]. Similarly, Mn(II) could be oxidized by existing recombinant Mco1 even if released.
The present study also demonstrated that the enzyme worked in the presence of relatively high concentration of Zn(II). Zn(II) sorption is concurrent with the Mn(II) oxidation that resulted in the increase in Zn(II) loading (1.6 times in this study), which is consistent with results from previous studies on fungal cultures [27,45]. Metal cations compete with Mn(II) for binding to the enzyme, inducing the inhibition of Mn(II) oxidation [46,47]. In L. discophora SS-1, the Mn(II)-oxidizing activity in the culture broth was inhibited by the addition of 10 μM Zn(II) [46]. A. strictum Mco1 appeared to be much less susceptible because ~0.7 mM Mn(II) was oxidized within 3 h even in the presence of a similar concentration of Zn(II). Previous results showed that the Mn(II) oxidation ability in the cultures decreased with increasing Zn(II) concentration and was completely lost at 0.5 mM [27]. If the fungal cultures preformed Mn oxides, then the cultures were able to oxidize Mn(II) rapidly even with 0.5 mM Zn(II). These observations suggest that the presence of Mn oxides and the remaining enzyme activity can lower the inhibitory effect of Zn(II) and enhance metal sequestration on Mn oxides.
Although Mn(II) oxidation by fungi is less studied in comparison with that of bacteria, studies have documented the contribution of fungi to Mn immobilization in various environments [48,49,50]. A recent work demonstrated selenium cycle driven by Mn(II)-oxidizing ascomycetes, further reinforcing their significant role in biogeochemical cycles [51]. In biotreatment of mine drainage containing Mn(II), increasing attention has been paid to Mn(II)-oxidizing fungi and their participation in the Mn removal, along with bacterial Mn oxidizers [52,53,54]. Enzymatic Mn(II) oxidation followed by formation of Mn oxide phases are considered the key reactions in such biogeochemical cycles and wastewater treatment. Future work employing Mco1 enzyme produced by P. pastoris would shed light on these processes driven by ascomycetes.
4. Materials and Methods
4.1. Organisms and Culture Conditions
A. strictum strain KR21-2 was grown at 25 °C in HEPES [4-(2-hydroxyethyl)-1-piperazinyl-ethane-2-sulfonic acid]-buffered AY (acetate-yeast extract) medium [19] supplemented with 1.0 μM CuSO4. HEPES-buffered AY medium contained 410 mg of sodium acetate trihydrate, 150 mg of yeast extract (Difco, Detroit, MI, USA), 50 mg of MgSO4 7H2O, 5.0 mg of K2HPO4, and 2.0 mL of trace mineral salts solution [19] per liter of 20 mM HEPES buffer (pH 7.0).
For gene analyses, E. coli strain DH5-T1R and P. pastoris strain X-33 were purchased from Invitrogen (Carlsbad, CA, USA). Culture media preparation and other culture procedures for these microorganisms were conducted according to the manufacturer’s instructions unless stated otherwise. The E. coli transformants were grown on Luria–Bertani (LB) medium, and P. pastoris was grown on YPD (yeast extract-peptone-dextrose) medium, BMGY (buffered minimal glycerol complex) medium, and BMMY (buffered methanol-complex) medium, which were prepared according to the EASYSelect Pichia Expression Kit user manual (Invitrogen). Pichia transformants were also grown on modified HEPES-buffered AY, which contained 240 mg of glycerol, 150 mg of yeast extract, 50 mg of (NH4)2SO4, 50 mg of MgSO4 7H2O, 5.0 mg of K2HPO4, 2.0 mL of trace mineral salts solution, 0.4 mg of biotin, 1 mmol of MnSO4, and 0.01 mmol of CuSO4 per liter of 20 mM HEPES buffer (pH 7.0).
4.2. Culture Experiments
To examine Mn(II) oxidation by the strain KR21-2, MnSO4 was added to HEPES-buffered AY at 1 mM. Duplicate cultures (50 mL per flask) were set and incubated at 25 °C for 60 h after each medium was inoculated with the fungal conidiospore suspension, as described previously [19]. The entire volume of the culture was centrifuged at 10,000× g for 10 min (10 °C). The obtained precipitate including mycelium and Mn solids were washed with Milli-Q water and sequentially treated with 10 mM CuSO4 and 50 mM hydroxylamine hydrochloride, to extract adsorbed Mn(II) and oxidized Mn, respectively [19]. Concentrations of Mn in the culture supernatants and extracts were determined by inductively coupled plasma optical emission spectroscopy (ICP-OES; iCAP 6000, Thermo Fisher Scientific, San Jose, CA, USA).
For monitoring the Mn(II) oxidase activity, the strain KR21-2 was grown in HEPES-buffered AY without 1 mM MnSO4. The culture supernatants collected by centrifugation were concentrated to approximately 80 folds by ultrafiltration (4500× g, 10 °C) with a polyethersulfone membrane (cut-off molecular mass, 10 kDa; Sartorius, Göttingen, Germany) and stored at −20 °C until use.
4.3. Genome Extraction and Sequencing
The mycelium of strain KR21-2 was harvested from cultures at 63 h and washed once in sterile deionized water. The genomic DNA was extracted from the mycelium with a commercial DNA extraction kit (ISOIL for Beads Beating, Nippon Gene, Tokyo, Japan) and then purified with QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the instructions. The purified DNA was stored at −20 °C. Two micrograms of genomic DNA was used for constructing the Illumina sequence library. The high molecular DNA was sheared by sonication using a Covaris M220 apparatus (Woburn, MA, USA) to an average size of 550 bp, and then TruSeq DNA PCR Free Library Prep Kit (Illumina, San Diego, CA, USA) was used according to the manufacturer’s instructions. The paired-end (2 × 300-bp) reads were obtained with the MiSeq system (Illumina). The raw reads were filtered and assembled by the CLC Genomic Workbench (Qiagen).
4.4. RNA Extraction and Gene Cloning and Heterologous Expression
The fungus was grown for 42 h in the culture medium without 1 mM Mn(II) and the mycelia were collected and immediately stored at −80 °C. Total RNA was extracted using the RNA PowerSoil Total RNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, USA) and reverse-transcribed into cDNA with the PrimeScript II High Fidelity RT-PCR Kit (Takara, Shiga, Japan). Oligonucleotide primers, named AKM-P2-F and AKM-P1-R (Figure S1 and Table S1), were designed for amplifying the full length of the MCO gene based on whole-genome sequencing data. The amplified cDNA fragments were TA-cloned and the constructs were transformed into E. coli competent cells, using TOPO TA Cloning Kit (Invitrogen). The cDNA fragments were analyzed by conventional Sanger sequencing. For phylogenetic analysis, the alignments of the deduced amino acid sequence and the related sequences from GenBank were generated using Clustal W, and neighbor-joining phylogenies were constructed by MEGA X [55], based on Kimura’s two-parameter distance. Bootstrap analysis (1000 replicates) was performed to evaluate the reliability of the phylogenetic reconstructions.
Homology model of Mco1 was created using Phyre2 web server [56]. Figure was drawn with PyMOL software (Schrödinger, LLC, New York, NY, USA).
Mco1-coding gene sequence, excluding the predicted signal sequence, was amplified with primers MCO-comp-F and MCO-comp-R (Figure S1 and Table S1). The Pichia expression vector pPICZαA with the α-factor secretion signal was digested with EcoR I and Xba I, and the mco1 gene was inserted into the predigested vector to construct pPICZα-mco1, which was sequenced to confirm the accuracy of the open reading frame. The plasmid pPICZα-mco1 was transformed into E. coli cells for maintenance.
This plasmid was linearized with Pme I and then transformed into the competent cells of P. pastoris strain X-33, which were prepared according to the EasySelect Pichia Expression Kit manual. The transformants were cultured at 30 °C on YPD plates containing 0.1 mg/mL Zeocin (Invitrogen). Genomic DNA was extracted from the positive transformants with the ISOIL for Beads Beating kit to confirm the maintenance of mco1 gene, which was amplified with primers MCO-comp-F and MCO-comp-R. Thus-obtained transformants were selected for Mn(II) oxidation activity on the modified HEPES-buffered AY plates with 1 mM MnSO4. After the colonies grew, a small volume of methanol was placed on the lid of each Petri dish to induce the gene expression. The Petri dishes were then sealed with parafilm and further incubated overnight at 25 °C. The transformants with Mn(II) oxidase activity deposited dark-brown Mn oxides positive for reaction with leucoberbelin blue I (Sigma-Aldrich, St. Louis, MS, USA).
Production of Mco1 enzyme by the Pichia transformant was conducted at 28 °C with 1-L culture of BMMY medium with 0.05 mM CuSO4 according to the manufacturer’s instructions. To maintain enzyme expression, the cultures were shaken vigorously and supplied with methanol (final concentration, 0.5% (v/v)) once every day. A small portion of the culture broth (2 mL) was sampled during the process, and the supernatant was obtained by centrifugation at 15,000× g for 10 min at 4 °C and stored at −20 °C. The optimal cultivation time was determined by the enzyme production in the supernatants, which was assayed by SDS–PAGE. In a separate experiment, the culture supernatant was collected at the optimal time (36 h) for recovery of the recombinant protein.
4.5. Purification of Recombinant Enzyme
The supernatant of the 1-L culture of P. pastoris transformant grown for 36 h was obtained by centrifugation (4000× g, 20 min, 4 °C) and then concentrated to ~80 folds by ultrafiltration described above. The concentrated supernatant was subjected to enzyme purification using Äkta FPLC purifier system with a Superdex 200 Increase 10/300 GL column (GE Healthcare, Uppsala, Sweden). The proteins were eluted with 20 mM HEPES (pH 7.0) containing 0.15 M NaCl and monitored by absorbance at 280 nm. Fractions with Mn(II)-oxidizing activity were combined, concentrated by ultrafiltration, and applied to the Superdex 200 column again. The active fractions were concentrated and dialyzed against 5 mM HEPES (pH 7.0) at 8 °C. The dialyzed sample was further applied to a Mono Q 5/50 GL column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 7.6). The proteins were eluted with a linear gradient of NaCl from 0 to 0.5 M, and the active fractions were concentrated by ultrafiltration and then dialyzed against 20 mM HEPES (pH 7.0). The enzyme solution was stored at –80 °C.
4.6. Enzyme Assays
The Mn(II) oxidase activity in A. strictum KR21-2 culture broth was assayed as described previously [57]. The enzyme reaction was conducted at 30 °C with 1 mM MnSO4 in 20 mM HEPES (pH 7.0) and the Mn oxides produced were determined with leucoberbelin blue. No oxidation of Mn(II) was detected in the reaction mixture without enzyme, so that abiotic Mn(II) oxidation was able to be neglected under the reaction conditions used in this study. Protein concentration was determined with bicinchoninic acid protein assay kit (Pierce, Rockford, IL, USA).
SDS-PAGE in-gel assay [57] was also used for monitoring the enzyme activity in the culture broth and the eluate from the purification column. The samples were mixed with Laemmli buffer (without heating) and run in a 12.5% polyacrylamide gel. After electrophoresis, the gel was washed twice in 10 mM HEPES (pH 7.0) for 20 min and incubated in the same buffer containing 1 mM MnSO4. The protein band was visualized by Coomassie brilliant blue staining.
When the molecular mass of the purified enzyme was measured by SDS-PAGE, it was denatured by heating after being mixed with the Laemmli buffer and run in the gel along with a set of marker proteins (14.3–97.2 kDa; Atto, Tokyo, Japan).
4.7. Mn Oxide Production by the Recombinant Enzyme and Its Characterization
Duplicate runs were conducted with a reaction mixture (1 or 3 mL) containing 12 μg/mL (0.19 μM) purified protein in 20 mM HEPES (pH 7.0) and 0.6–0.7 mM MnSO4 at 30 °C. Effect of Zn(II) ion on the enzymatic activity was examined by addition of 0.65 mM ZnSO4 to the reaction mixture. Abiotic oxidation of Mn(II) or precipitation of Zn(II) was examined with the reaction mixtures without enzyme. For electron microscopy, suspended Mn solids in the reaction mixture were collected by centrifugation (20,000× g, 10 min, 10 °C), washed with Milli-Q water, and freeze-dried. For the metal sorption experiment, the Mn solids collected by centrifugation were washed with 20 mM HEPES buffer (pH 7.0) and used immediately without drying. The sorbed Mn(II) and oxidized Mn in the solids were determined by the sequential extraction described above. Dissolved Mn and Zn in the Cu (II)-exchangeable and reducible fractions were determined by ICP-OES.
The Mn solids were analyzed with a field emission scanning electron microscope (FE-SEM; SU 8010, Hitachi, Tokyo, Japan) at 6 kV, equipped with an energy-dispersive X-ray spectrometer (EDX; EMAX X-act, Horiba, Kyoto, Japan). Non-coated samples were used for SEM observation.
In Zn(II) sorption experiment, the Mn solids were washed and resuspended in equal volumes of 20 mM HEPES (pH 7.0) with 0.65 mM ZnSO4 and incubated for 26 h at 30 °C. The supernatant was collected by centrifugation (20,000× g, 10 min, 10 °C) and the concentration of remaining dissolved Mn(II) and Zn(II) was determined.
5. Conclusions
In this study, the gene encoding Mn(II) oxidase in A. strictum strain KR21-2 was cloned and successfully expressed in P. pastoris. Laccase-like MCO has been identified as a Mn(II) oxidase in ascomycete fungi previously, but the present study demonstrated that the Mco1 of strain KR21-2 was a bilirubin oxidase instead of being true laccase. In the reaction mixture with the recombinant enzyme, the Mn oxide phase grew into well-proportioned spherical forms of several micrometers in diameter. This may suggest the progress of Mn mineralization in a manner other than simple aggregation of smaller, nano-sized particles. Besides high Zn(II) sorption capacity, the Mn(II) oxidation occurred in the presence of Zn(II), possibly leading to the incorporation of more Zn(II). The results were consistent with the hypothesis that the enzyme activity enhances the sequestration of both Mn(II) and Zn(II) on the Mn oxides. The role of enzyme activity in Mn biomineralization has not been elucidated in many ascomycetes yet. The results of this study suggest that the homogenous Mn(II) oxidation system with recombinant Mco1 serves as a simple but useful model for understanding its role in ascomycetes.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4344/10/6/686/s1, Figure S1: Nucleotide and deduced amino acid sequences of mco1 gene from A. strictum KR21-2, Figure S2: Neighbor-joining phylogenetic tree showing the relationship of A. strictum KR21-2 Mco1 with related MCOs, Figure S3: Homology model of A. strictum KR21-2 Mco1 and crystal structure of bilirubin oxidase from Albifimbria verrucaria (AvBOx), Figure S4: SEM images of solid-phase Mn produced by the enzyme reaction, Table S1: Primers used in this study.
Author Contributions
Conceptualization, N.M. and Y.T.; methodology, F.T., N.M., J.F. and Y.T.; investigation, F.T., A.K., N.M., K.O. and J.F.; Formal analysis, R.S.; writing—original draft preparation, N.M. and J.F.; writing—review and editing, F.T., K.O., J.F. and Y.T.; supervision, N.M.; funding acquisition, N.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Japan Society for Promotion of Science, JSPS KAKENHI Grant Number JP16K12619.
Acknowledgments
The authors thank Yoichiro Shimura, Ryoki Asano, and Yayoi Kobayashi for their technical support in the protein purification and analysis of genomic data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tebo, B.M.; Bargar, J.R.; Clement, B.G.; Dick, G.J.; Murray, K.J.; Parker, D.; Verity, R.; Webb, S.M. Biogenic manganese oxides: Properties and mechanisms of formation. Annu. Rev. Earth Planet. Sci. 2004, 32, 287–328. [Google Scholar] [CrossRef]
- Tebo, B.M.; Johnson, H.A.; McCarthy, J.K.; Templeton, A.S. Geomicrobiology of manganese(II) oxidation. Trend Microbiol. 2005, 13, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Miyata, N.; Tani, Y.; Sakata, M.; Iwahori, K. Microbial manganese oxide formation and interaction with toxic metal ions. J. Biosci. Bioeng. 2007, 104, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hennebel, T.; De Gusseme, B.; Boon, N.; Verstraete, W. Biogenic metals in advanced water treatment. Trend Biotechnol. 2009, 27, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, Z.; Ma, H.; Hursthouse, A.S. Removal of manganese(II) from acid mine wastewater: A review of the challenges and opportunities with special emphasis on Mn-oxidizing bacteria and microalgae. Water 2019, 11, 2493. [Google Scholar] [CrossRef]
- Remucal, C.K.; Ginder-Vogel, M. A critical review of the reactivity of manganese oxides with organic contaminants. Environ. Sci. Process. Impacts 2014, 16, 1247–1266. [Google Scholar] [CrossRef]
- Geszvain, K.; Butterfield, C.; Davis, R.E.; Madison, A.S.; Lee, S.-W.; Parker, D.L.; Soldatova, A.; Spiro, T.G.; Luther III, G.W.; Tebo, B.M. The molecular biogeochemistry of manganese(II) oxidation. Biochem. Soc. Trans. 2012, 40, 1244–1248. [Google Scholar] [CrossRef]
- Butterfield, C.N.; Soldatova, A.V.; Lee, S.-W.; Spiro, T.G.; Tebo, B.M. Mn(II,III) oxidation and MnO2 mineralization by an expressed bacterial multicopper oxidase. Proc. Natl. Acad. Sci. USA 2013, 110, 11731–11735. [Google Scholar] [CrossRef]
- Su, J.; Bao, P.; Bai, T.; Deng, L.; Wu, H.; Liu, F.; He, J. CotA, a multicopper oxidase from Bacillus pumilus WH4, exhibits manganese-oxidase activity. PLoS ONE 2013, 8, e60573. [Google Scholar] [CrossRef]
- Su, J.; Deng, L.; Huang, L.; Guo, S.; Liu, F.; He, J. Catalytic oxidation of manganese(II) by multicopper oxidase CueO and characterization of the biogenic Mn oxide. Water Res. 2014, 56, 304–313. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Z.; Chen, H.; Liu, J.; Liu, C.; Ni, H.; Zhao, C.; Ali, M.; Liu, F.; Li, L. Surface Mn(II) oxidation actuated by a multicopper oxidase in a soil bacterium leads to the formation of manganese oxide minerals. Sci. Rep. 2015, 5, 10895. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zhang, M.; Liu, Y.; Tang, W. Manganese(II) oxidation by the multi-copper oxidase CopA from Brevibacillus panacihumi MK-8. Enzyme Microb. Technol. 2018, 117, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Romano, C.A.; Zhou, M.; Song, Y.; Wysocki, V.H.; Dohnalkova, A.C.; Kovarik, L.; Paša-Tolić, L.; Tebo, B.M. Biogenic manganese oxide nanoparticle formation by a multimeric multicopper oxidase Mnx. Nat. Commun. 2017, 8, 746. [Google Scholar] [CrossRef] [PubMed]
- Soldatova, A.V.; Romano, C.A.; Tao, L.; Stich, T.A.; Casey, W.H.; Britt, R.D.; Tebo, B.M.; Spiro, T.G. Mn(II) oxidation by the multicopper oxidase complex Mnx: A coordinated two-stage Mn(II)/(III) and Mn(III)/(IV) mechanism. J. Am. Chem. Soc. 2017, 139, 11381–11391. [Google Scholar] [CrossRef]
- Höfer, C.; Schlosser, D. Novel enzymatic oxidation of Mn2+ to Mn3+ catalyzed by a fungal laccase. FEBS Lett. 1999, 51, 186–190. [Google Scholar] [CrossRef]
- Schlosser, D.; Höfer, C. Laccase-catalyzed oxidation of Mn2+ in the presence of natural Mn3+ chelators as a novel source of extracellular H2O2 production and its impact on manganese peroxidase. Appl. Environ. Microbiol. 2002, 68, 3514–3521. [Google Scholar] [CrossRef]
- Gorbacheva, M.; Morozova, O.; Shumakovich, G.; Streltsov, A.; Shleev, S.; Yaropolov, A. Enzymatic oxidation of manganese ions catalysed by laccase. Bioorg. Chem. 2009, 37, 1–5. [Google Scholar] [CrossRef]
- Glenn, J.K.; Akileswaran, L.; Gold, M.H. Mn(II) oxidation is the principal function of the extracellular Mn-peroxidase from Phanerochaete chrysosporium. Arch. Biochem. Biophys. 1986, 251, 688–696. [Google Scholar] [CrossRef]
- Miyata, N.; Tani, Y.; Iwahori, K.; Soma, M. Enzymatic formation of manganese oxides by an Acremonium-like hyphomycete fungus, strain KR21-2. FEMS Microbiol. Ecol. 2004, 47, 101–109. [Google Scholar] [CrossRef]
- Miyata, N.; Maruo, K.; Tani, Y.; Tsuno, H.; Seyama, H.; Soma, M.; Iwahori, K. Production of biogenic manganese oxides by anamorphic ascomycete fungi isolated from streambed pebbles. Geomicrobiol. J. 2006, 23, 63–73. [Google Scholar] [CrossRef]
- Thompson, I.A.; Huber, D.M.; Schulze, D.G. Evidence of a multicopper oxidase in Mn oxidation by Gaeumannomyces graminis var. tritici. Phytopathology 2006, 96, 130–136. [Google Scholar] [CrossRef]
- Hansel, C.M.; Zeiner, C.A.; Santelli, C.M.; Webb, S.M. Mn(II) oxidation by an ascomycete fungus is linked to superoxide production during asexual reproduction. Proc. Natl. Acad. Sci. USA 2012, 109, 12621–12625. [Google Scholar] [CrossRef] [PubMed]
- Miyata, N.; Tani, Y.; Maruo, K.; Tsuno, H.; Sakata, M.; Iwahori, K. Manganese(IV) oxide production by Acremonium sp. strain KR21-2 and extracellular Mn(II) oxidase activity. Appl. Environ. Microbiol. 2006, 72, 6467–6673. [Google Scholar] [CrossRef] [PubMed]
- Grangeon, S.; Lanson, B.; Miyata, N.; Tani, Y.; Manceau, A. Structure of nanocrystalline phyllomanganates produced by freshwater fungi. Am. Mineral. 2010, 95, 1608–1616. [Google Scholar] [CrossRef]
- Tani, Y.; Ohashi, M.; Miyata, N.; Seyama, H.; Iwahori, K.; Soma, M. Sorption of Co(II), Ni(II), and Zn(II) on biogenic manganese oxides produced by a Mn-oxidizing fungus, strain KR21-2. J. Environ. Sci. Health A 2004, 39, 2641–2660. [Google Scholar] [CrossRef]
- Chang, J.; Tani, Y.; Naitou, H.; Miyata, N.; Seyama, H. Sequestration of Cd(II) and Ni(II) ions on fungal manganese oxides associated with Mn(II) oxidase activity. Appl. Geochem. 2014, 47, 198–208. [Google Scholar] [CrossRef]
- Chang, J.; Tani, Y.; Naitou, H.; Miyata, N.; Tojo, F.; Seyama, H. Zn(II) sequestration by fungal biogenic manganese oxide through enzymatic and abiotic processes. Chem. Geol. 2014, 383, 155–163. [Google Scholar] [CrossRef]
- Zheng, H.; Tani, Y.; Naitou, H.; Miyata, N.; Tojo, F.; Seyama, H. Sequestration of La3+ by fungal manganese oxides and the effect of Mn(II) oxidase activity. J. Environ. Chem. Eng. 2017, 5, 735–743. [Google Scholar] [CrossRef]
- Tani, Y.; Miyata, N.; Ohashi, M.; Ohnuki, T.; Seyama, H.; Iwahori, K.; Soma, M. Interaction of inorganic arsenic with biogenic manganese oxide produced by a Mn-oxidizing fungus, strain KR21-2. Environ. Sci. Technol. 2004, 38, 6618–6624. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, J.; Tani, Y.; Chang, J.; Miyata, N.; Naitou, H.; Seyama, H. As(III) oxidation kinetics of biogenic manganese oxides formed by Acremonium strictum strain KR21-2. Chem. Geol. 2013, 347, 227–232. [Google Scholar] [CrossRef]
- Chang, J.; Tani, Y.; Naitou, H.; Miyata, N.; Seyama, H.; Tanaka, K. Cobalt(II) sequestration on fungal biogenic manganese oxide enhanced by manganese(II) oxidase activity. Appl. Geochem. 2013, 37, 170–178. [Google Scholar] [CrossRef]
- Suzuki, R.; Tani, Y.; Naitou, H.; Miyata, N.; Tanaka, K. Sequestration and oxidation of Cr(III) by fungal Mn oxides with Mn(II) oxidizing activity. Catalysts 2020, 10, 44. [Google Scholar] [CrossRef]
- Solomon, E.I.; Sundaram, U.M.; Machonkin, T.E. Multicopper oxidases and oxygenases. Chem. Rev. 1996, 96, 2563–2605. [Google Scholar] [CrossRef] [PubMed]
- Cracknell, J.A.; McNamara, T.P.; Lowe, E.D.; Blanford, C.F. Bilirubin oxidase from Myrothecium verrucaria: X-ray determination of the complete crystal structure and a rational surface modification for enhanced electrocatalytic O2 reduction. Dalton Trans. 2011, 40, 6668–6675. [Google Scholar] [CrossRef]
- Sakasegawa, S.-I.; Ishikawa, H.; Imamura, S.; Sakuraba, H.; Goda, S.; Ohshima, T. Bilirubin oxidase activity of Bacillus subtilis CotA. Appl. Environ. Microbiol. 2006, 72, 972–975. [Google Scholar] [CrossRef]
- Durand, F.; Kjaergaard, C.H.; Suraniti, E.; Gounel, S.; Hadt, R.G.; Solomon, E.I.; Mano, N. Bilirubin oxidase from Bacillus pumilus: A promising enzyme for the elaboration of efficient cathodes in biofuel cells. Biosens. Bioelectron. 2012, 35, 140–146. [Google Scholar] [CrossRef]
- Mandernack, K.W.; Post, J.; Tebo, B.M. Manganese mineral formation by bacterial spores of the marine Bacillus, strain SG-1: Evidence for the direct oxidation of Mn(II) to Mn(IV). Geochim. Cosmochim. Acta 1995, 59, 4393–4408. [Google Scholar] [CrossRef]
- Bargar, J.R.; Tebo, B.M.; Villinsky, J.E. In situ characterization of Mn(II) oxidation by spores of the marine Bacillus sp. strain SG-1. Geochim. Cosmochim. Acta 2000, 64, 2775–2778. [Google Scholar] [CrossRef]
- Bargar, J.R.; Tebo, B.M.; Bergmann, U.; Webb, S.M.; Glatzel, P.; Chiu, V.Q.; Villalobos, M. Biotic and abiotic products of Mn(II) oxidation by spores of the marine Bacillus sp. strain SG-1. Am. Mineral. 2005, 90, 143–154. [Google Scholar] [CrossRef]
- Soldatova, A.V.; Butterfield, C.; Oyerinde, O.F.; Tebo, B.M.; Spiro, T.G. Multicopper oxidase involvement in both Mn(II) and Mn(III) oxidation during bacterial formation of MnO2. J. Biol. Inorg. Chem. 2012, 17, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Soldatova, A.V.; Balakrishnan, G.; Oyerinde, O.F.; Romano, C.A.; Tebo, B.M.; Spiro, T.G. Biogenic and synthetic MnO2 nanoparticles: Size and growth probed with absorption and Raman spectroscopies and dynamic light scattering. Environ. Sci. Technol. 2019, 53, 4185–4197. [Google Scholar] [CrossRef] [PubMed]
- Learman, D.R.; Wankel, S.D.; Webb, S.M.; Martinez, N.; Madden, A.S.; Hansel, C.M. Coupled biotic-abiotic Mn(II) oxidation pathway mediates the formation and structural evolution of biogenic Mn oxides. Geochim. Cosmochim. Acta 2011, 75, 6048–6063. [Google Scholar] [CrossRef]
- Learman, D.R.; Voelker, B.M.; Madden, A.S.; Hansel, C.M. Constraints on superoxide mediated formation of manganese oxides. Front. Microbiol. 2013, 4, 262. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Tani, Y.; Naitou, H.; Miyata, N.; Seyama, H. Fungal Mn oxides supporting Mn(II) oxidase activity as effective Mn(II) sequestering materials. Environ. Technol. 2013, 34, 2781–2787. [Google Scholar] [CrossRef]
- Yu, Q.; Sasaki, K.; Tanaka, K.; Ohnuki, T.; Hirajima, T. Zinc sorption during bio-oxidation and precipitation of manganese modifies the layer stacking of biogenic birnessite. Geomicrobiol. J. 2013, 30, 829–839. [Google Scholar] [CrossRef]
- Adams, L.F.; Ghiorse, W.C. Characterization of extracellular Mn2+-oxidizing activity and isolation of an Mn2+-oxidizing protein from Leptothrix discophora SS-1. J. Bacteriol. 1987, 169, 1279–1285. [Google Scholar] [CrossRef]
- Murray, K.J.; Webb, S.M.; Bargar, J.R.; Tebo, B.M. Indirect oxidation of Co(II) in the presence of the marine Mn(II)-oxidizing bacterium Bacillus sp. strain SG-1. Appl. Environ. Microbiol. 2007, 73, 6905–6909. [Google Scholar] [CrossRef]
- Sterflinger, K. Fungi as geologic agents. Geomicrobiol. J. 2000, 17, 97–124. [Google Scholar] [CrossRef]
- Carmichael, S.K.; Zorn, B.T.; Santelli, C.M.; Roble, L.A.; Carmichael, M.J.; Bräuer, S.L. Nutrient input influences fungal community composition and size and can stimulate manganese (II) oxidation in caves. Environ. Microbiol. Rep. 2015, 7, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Gadd, G.M. The geomycology of elemental cycling and transformations in the environment. Microbiol. Spectr. 2017, 5, FUNK-0010-2016. [Google Scholar]
- Rosenfeld, C.E.; Sabuda, M.C.; Hinkle, M.A.G.; James, B.R.; Santelli, C.M. A fungal-mediated cryptic selenium cycle linked to manganese biogeochemistry. Environ. Sci. Technol. 2020, 54, 3570–3580. [Google Scholar] [CrossRef] [PubMed]
- Mariner, R.; Johnson, D.B.; Hallberg, K.B. Characterisation of an attenuation system for the remediation of Mn(II) contaminated waters. Hydrometallurgy 2008, 94, 100–104. [Google Scholar] [CrossRef]
- Santelli, C.M.; Pfister, D.H.; Lazarus, D.; Sun, L.; Burgos, W.D.; Hansel, C.M. Promotion of Mn(II) oxidation and remediation of coal mine drainage in passive treatment systems by diverse fungal and bacterial communities. Appl. Environ. Microbiol. 2010, 76, 4871–4875. [Google Scholar] [CrossRef] [PubMed]
- Santelli, C.M.; Webb, S.M.; Dohnalkova, A.C.; Hansel, C.M. Diversity of Mn oxides produced by Mn(II)-oxidizing fungi. Geochim. Cosmochim. Acta 2011, 75, 2762–2776. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Boogerd, F.C.; de Vrind, J.P.M. Manganese oxidation by Leptothrix discophora. J. Bacteriol. 1987, 169, 489–494. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).