Aspergillus: A Powerful Protein Production Platform
Abstract
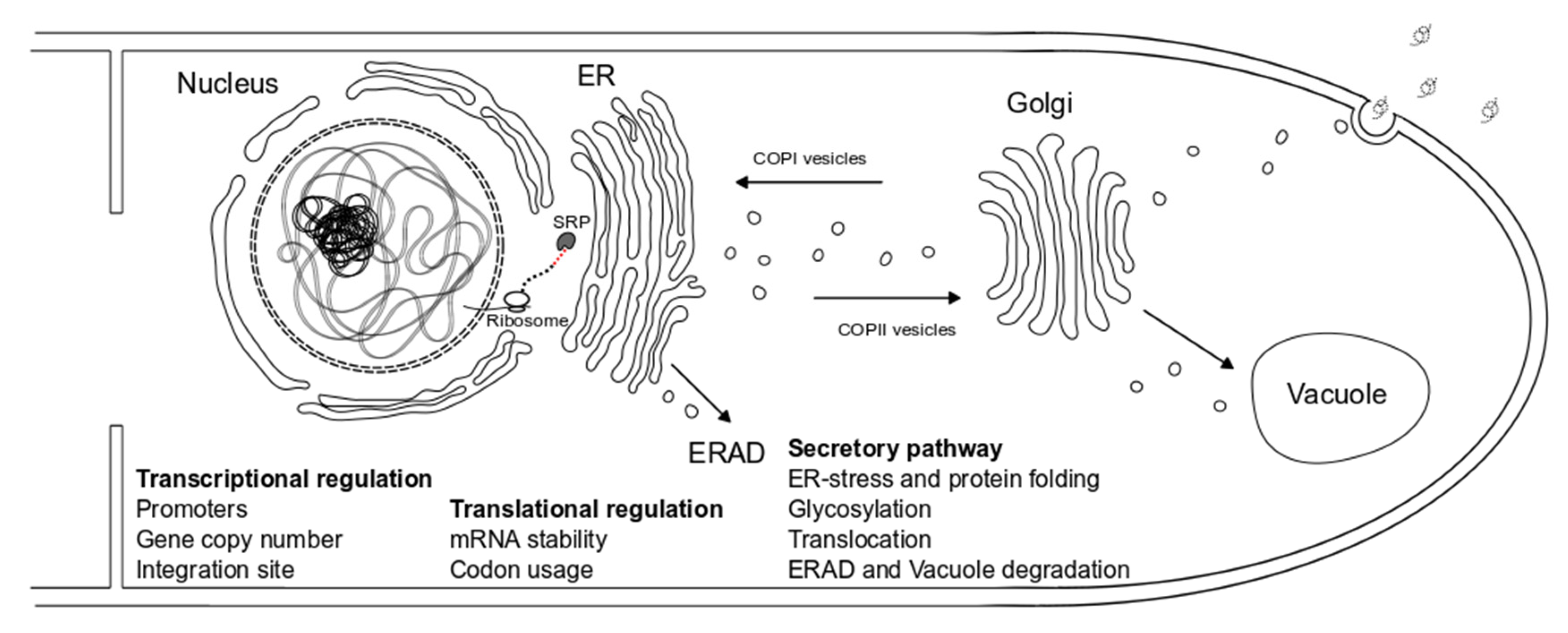
:1. Introduction
2. Industrial Application of Aspergilli
2.1. Traditional Uses of Aspergillus Species
2.2. The Use of Aspergillus Species in Heterologous Protein Production
3. Genetic Engineering Approaches for Aspergillus Strain Improvement
3.1. Transcriptional Regulation
3.1.1. Promoters
3.1.2. Gene Copy Number and Integration Site
3.2. Translational Regulation
Codon Usage and mRNA Stability
3.3. Glycosylation
3.4. Secretion
3.4.1. The fungal Secretory Pathway
- ER-stress and protein folding
- Glycosylation
- Protein translocation
- Protein degradation pathways—ERAD and Vacuole
3.4.2. Carrier Proteins
3.5. Proteases
3.6. Altering Fungal Morphology Using Genetic Engineering
4. Fermentation Conditions for Improved Heterologous Production in Aspergillus
4.1. Fermentation Conditions
4.2. Altering Fungal Morphology Using Bioprocessing
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Puetz, J.; Wurm, F.M. Recombinant proteins for industrial versus pharmaceutical purposes: A review of process and pricing. Processes 2019, 7, 476. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.-M.; Han, S.-S.; Kim, H.-S. Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnol. Adv. 2015, 33, 1443–1454. [Google Scholar] [CrossRef] [PubMed]
- Baeshen, N.A.; Baeshen, M.N.; Sheikh, A.; Bora, R.S.; Ahmed, M.M.M.; Ramadan, H.A.I.; Saini, K.S.; Redwan, E.M. Cell factories for insulin production. Microb. Cell Fact. 2014, 13, 141. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Ullah, M.W.; Siddique, R.; Nabi, G.; Manan, S.; Yousaf, M.; Hou, H. Role of recombinant DNA technology to improve life. Int. J. Genomics 2016, 2016, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Walsh, G. Biopharmaceutical benchmarks 2018. Nat. Biotechnol. 2018, 36, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.d.G.; Azzoni, A.R.; Freitas, S. Techno-economic analysis of the industrial production of a low-cost enzyme using E. coli: The case of recombinant β-glucosidase. Biotechnol. Biofuels 2018, 11, 81. [Google Scholar] [CrossRef]
- Nevalainen, H.; Peterson, R. Making recombinant proteins in filamentous fungi—Are we expecting too much? Front. Microbiol. 2014, 5, 1–10. [Google Scholar] [CrossRef]
- Demain, A.L.; Vaishnav, P. Production of recombinant proteins by microbes and higher organisms. Biotechnol. Adv. 2009, 27, 297–306. [Google Scholar] [CrossRef]
- Baghban, R.; Farajnia, S.; Rajabibazl, M.; Ghasemi, Y.; Mafi, A.; Hoseinpoor, R.; Rahbarnia, L.; Aria, M. Yeast expression systems: Overview and recent advances. Mol. Biotechnol. 2019, 61, 365–384. [Google Scholar] [CrossRef]
- Xie, Y.; Han, X.; Miao, Y. An effective recombinant protein expression and purification system in Saccharomyces cerevisiae. Curr. Protoc. Mol. Biol. 2018, 123, e62. [Google Scholar] [CrossRef]
- Meyer, V.; Basenko, E.Y.; Benz, J.P.; Braus, G.H.; Caddick, M.X.; Csukai, M.; de Vries, R.P.; Endy, D.; Frisvad, J.C.; Gunde-Cimerman, N.; et al. Growing a circular economy with fungal biotechnology: A white paper. Fungal Biol. Biotechnol. 2020, 7, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Havlik, D.; Brandt, U.; Bohle, K.; Fleißner, A. Establishment of Neurospora crassa as a host for heterologous protein production using a human antibody fragment as a model product. Microb. Cell Fact. 2017, 16, 128. [Google Scholar] [CrossRef] [PubMed]
- Landowski, C.P.; Mustalahti, E.; Wahl, R.; Croute, L.; Sivasiddarthan, D.; Westerholm-Parvinen, A.; Sommer, B.; Ostermeier, C.; Helk, B.; Saarinen, J.; et al. Enabling low cost biopharmaceuticals: High level interferon alpha-2b production in Trichoderma reesei. Microb. Cell Fact. 2016, 15, 104. [Google Scholar] [CrossRef] [Green Version]
- Magaña-Ortíz, D.; Fernández, F.; Loske, A.M.; Gómez-Lim, M.A. Extracellular expression in Aspergillus niger of an antibody fused to Leishmania sp. antigens. Curr. Microbiol. 2018, 75, 40–48. [Google Scholar] [CrossRef]
- Arnau, J.; Yaver, D.; Hjort, C.M. Strategies and challenges for the development of industrial enzymes using fungal cell factories. In Grand Challenges in Fungal Biotechnology; Springer: Cham, Switzerland, 2020; pp. 179–210. ISBN 9783030295417. [Google Scholar]
- Sun, X.; Su, X. Harnessing the knowledge of protein secretion for enhanced protein production in filamentous fungi. World J. Microbiol. Biotechnol. 2019, 35, 54. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V. Genetic engineering of filamentous fungi—Progress, obstacles and future trends. Biotechnol. Adv. 2008, 26, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Shukla, P. Advanced technologies for improved expression of recombinant proteins in bacteria: Perspectives and applications. Crit. Rev. Biotechnol. 2016, 36, 1089–1098. [Google Scholar] [CrossRef]
- Baeshen, M.N.; Al-Hejin, A.M.; Bora, R.S.; Ahmed, M.M.M.; Ramadan, H.A.I.; Saini, K.S.; Baeshen, N.A.; Redwan, E.M. Production of biopharmaceuticals in E. coli: Current scenario and future perspectives. J. Microbiol. Biotechnol. 2015, 25, 953–962. [Google Scholar] [CrossRef]
- Contreras-Gómez, A.; Sánchez-Mirón, A.; García-Camacho, F.; Molina-Grima, E.; Chisti, Y. Protein production using the baculovirus-insect cell expression system. Biotechnol. Prog. 2014, 30, 1–18. [Google Scholar] [CrossRef]
- Hunter, M.; Yuan, P.; Vavilala, D.; Fox, M. Optimization of protein expression in mammalian cells. Curr. Protoc. Protein Sci. 2019, 95, e77. [Google Scholar] [CrossRef] [Green Version]
- Houdebine, L.-M. Production of pharmaceutical proteins by transgenic animals. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 107–121. [Google Scholar] [CrossRef]
- Łojewska, E.; Kowalczyk, T.; Olejniczak, S.; Sakowicz, T. Extraction and purification methods in downstream processing of plant-based recombinant proteins. Protein Expr. Purif. 2016, 120, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Weng, Y.; Dickey, A.; Wang, K. Plants as factories for human pharmaceuticals: Applications and challenges. Int. J. Mol. Sci. 2015, 16, 28549–28565. [Google Scholar] [CrossRef]
- Currie, J.N. The citric acid fermentation of Aspergillus niger. J. Biol. Chem. 1917, 31, 15–37. [Google Scholar]
- Cairns, T.C.; Nai, C.; Meyer, V. How a fungus shapes biotechnology: 100 years of Aspergillus niger research. Fungal Biol. Biotechnol. 2018, 5, 13. [Google Scholar] [CrossRef] [Green Version]
- Dai, Z.; Zhou, H.; Zhang, S.; Gu, H.; Yang, Q.; Zhang, W.; Dong, W.; Ma, J.; Fang, Y.; Jiang, M.; et al. Current advance in biological production of malic acid using wild type and metabolic engineered strains. Bioresour. Technol. 2018, 258, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Barrios-González, J.; Miranda, R.U. Biotechnological production and applications of statins. Appl. Microbiol. Biotechnol. 2010, 85, 869–883. [Google Scholar] [CrossRef]
- Meyer, V.; Andersen, M.R.; Brakhage, A.A.; Braus, G.H.; Caddick, M.X.; Cairns, T.C.; de Vries, R.P.; Haarmann, T.; Hansen, K.; Hertz-Fowler, C.; et al. Current challenges of research on filamentous fungi in relation to human welfare and a sustainable bio-economy: A white paper. Fungal Biol. Biotechnol. 2016, 3, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins-Santana, L.; Nora, L.C.; Sanches-Medeiros, A.; Lovate, G.L.; Cassiano, M.H.A.; Silva-Rocha, R. Systems and synthetic biology approaches to engineer fungi for fine chemical production. Front. Bioeng. Biotechnol. 2018, 6, 117. [Google Scholar] [CrossRef] [Green Version]
- Meyer, V.; Wu, B.; Ram, A.F.J. Aspergillus as a multi-purpose cell factory: Current status and perspectives. Biotechnol. Lett. 2011, 33, 469–476. [Google Scholar] [CrossRef] [Green Version]
- Leynaud-Kieffer, L.M.C.; Curran, S.C.; Kim, I.; Magnuson, J.K.; Gladden, J.M.; Baker, S.E.; Simmons, B.A. A new approach to Cas9-based genome editing in Aspergillus niger that is precise, efficient and selectable. PLoS ONE 2019, 14, e0210243. [Google Scholar] [CrossRef] [PubMed]
- Sarkari, P.; Marx, H.; Blumhoff, M.L.; Mattanovich, D.; Sauer, M.; Steiger, M.G. An efficient tool for metabolic pathway construction and gene integration for Aspergillus niger. Bioresour. Technol. 2017, 245, 1327–1333. [Google Scholar] [CrossRef] [PubMed]
- Cairns, T.C.; Feurstein, C.; Zheng, X.; Zhang, L.H.; Zheng, P.; Sun, J.; Meyer, V. Functional exploration of co-expression networks identifies a nexus for modulating protein and citric acid titres in Aspergillus niger submerged culture. Fungal Biol. Biotechnol. 2019, 6, 18. [Google Scholar] [CrossRef] [Green Version]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katayama, T.; Tanaka, Y.; Okabe, T.; Nakamura, H.; Fujii, W.; Kitamoto, K.; Maruyama, J.I. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol. Lett. 2016, 38, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Fuller, K.K.; Chen, S.; Loros, J.J.; Dunlap, J.C. Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot. Cell 2015, 14, 1073–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunn-Coleman, N.S.; Bloebaum, P.; Berka, R.M.; Bodie, E.; Robinson, N.; Armstrong, G.; Ward, M.; Przetak, M.; Carter, G.L.; LaCost, R.; et al. Commercial levels of chymosin production by Aspergillus. Bio/Technology 1991, 9, 976–981. [Google Scholar] [CrossRef]
- Ward, P.P.; Lo, J.-Y.; Duke, M.; May, G.S.; Headon, D.R.; Conneely, O.M. Production of biologically active recombinant human lactoferrin in Aspergillus Oryzae. Nat. Biotechnol. 1992, 10, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.I.; Asakura, T.; Maruyama, J.I.; Morita, Y.; Oike, H.; Shimizu-Ibuka, A.; Misaka, T.; Sorimachi, H.; Arai, S.; Kitamoto, K.; et al. Extracellular production of neoculin, a sweet-tasting heterodimeric protein with taste-modifying activity, by Aspergillus oryzae. Appl. Environ. Microbiol. 2006, 72, 3716–3723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Den Hondel, C.A.M.J.J.; Punt, P.J.; Van Gorcom, R.F.M. Heterologous Gene Expression in Filamentous Fungi; Academic Press, Inc.: Cambridge, MA, USA, 1991. [Google Scholar]
- Fleißner, A.; Dersch, P. Expression and export: Recombinant protein production systems for Aspergillus. Appl. Microbiol. Biotechnol. 2010, 87, 1255–1270. [Google Scholar] [CrossRef] [PubMed]
- Rendsvig, J.K.H.; Workman, C.T.; Hoof, J.B. Bidirectional histone-gene promoters in Aspergillus: Characterization and application for multi-gene expression. Fungal Biol. Biotechnol. 2019, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Qiu, R.; Zhu, X.; Liu, L.; Tang, G. Detection of a protein, AngCP, which binds specifically to the three upstream regions of glaA gene in A. niger T21. Sci. China Ser. C 2002, 45, 527. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, J.; Qiu, R.X.; Zhu, X.G.; Dong, Z.Y.; Tang, G.M. Improving heterologous gene expression in Aspergillus niger by introducing multiple copies of protein-binding sequence containing CCAAT to the promoter. Lett. Appl. Microbiol. 2003, 36, 358–361. [Google Scholar] [CrossRef]
- Moralejo, F.-J.; Cardoza, R.-E.; Gutierrez, S.; Martin, J.F. Thaumatin production in Aspergillus awamori by use of expression cassettes with strong fungal promoters and high gene dosage. Appl. Environ. Microbiol. 1999, 65, 1168–1174. [Google Scholar] [CrossRef] [Green Version]
- Pachlinger, R.; Mitterbauer, R.; Adam, G.; Strauss, J. Metabolically independent and accurately adjustable Aspergillus sp. expression system. Appl. Environ. Microbiol. 2005, 71, 672–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bando, H.; Hisada, H.; Ishida, H.; Hata, Y.; Katakura, Y.; Kondo, A. Isolation of a novel promoter for efficient protein expression by Aspergillus oryzae in solid-state culture. Appl. Microbiol. Biotechnol. 2011, 92, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Gressler, M.; Hortschansky, P.; Geib, E.; Brock, M. A new high-performance heterologous fungal expression system based on regulatory elements from the Aspergillus terreus terrein gene cluster. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Jun, W.; Yuk, S.L. Polynucleotide Fragment, Expression Vector Comprising Same, Aspergillus niger Genetic Engineering Strain and Application of Aspergillus niger Genetic Engineering. CN Patent No. CN107304431A, 31 October 2017. [Google Scholar]
- Li, W.; Yu, J.; Li, Z.; Yin, W.B. Rational design for fungal laccase production in the model host Aspergillus nidulans. Sci. China Life Sci. 2019, 62, 84–94. [Google Scholar] [CrossRef]
- Yin, W.; Li, W.; Ma, Z. Construction and Application of Heterologous Expression System of Aspergillus nidulans. CN Patent No. CN108795970A, 13 November 2018. [Google Scholar]
- Li, M.; Lu, F.; Chen, Y. Fungus Promoter and Application Thereof 2019. CN Patent No. CN110331144A, 15 October 2019. [Google Scholar]
- Gladden, J.M.; Campen, S.A.; Zhang, J.; Magnuson, J.K.; Baker, S.E.; Simmons, B.A. Promoter Useful for High Expression of a Heterologous Gene of Interest in Aspergillus niger. U.S. Patent No. US20,190,169,584A1, 6 June 2019. [Google Scholar]
- Meyer, V.; Wanka, F.; van Gent, J.; Arentshorst, M.; van den Hondel, C.A.M.J.J.; Ram, A.F.J. Fungal gene expression on demand: An inducible, tunable, and metabolism-independent expression system for Aspergillus niger. Appl. Environ. Microbiol. 2011, 77, 2975–2983. [Google Scholar] [CrossRef] [Green Version]
- Vogt, K.; Bhabhra, R.; Rhodes, J.C.; Askew, D.S. Doxycycline-regulated gene expression in the opportunistic fungal pathogen Aspergillus fumigatus. BMC Microbiol. 2005, 5, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rantasalo, A.; Landowski, C.P.; Kuivanen, J.; Korppoo, A.; Reuter, L.; Koivistoinen, O.; Valkonen, M.; Penttilä, M.; Jäntti, J.; Mojzita, D. A universal gene expression system for fungi. Nucleic Acids Res. 2018, 46, e111. [Google Scholar] [CrossRef] [PubMed]
- Verdoes, J.C.; Punt, P.J.; Schrickx, J.M.; van Verseveld, H.W.; Stouthamer, A.H.; van den Hondel, C.A.M.J.J. Glucoamylase overexpression in Aspergillus niger: Molecular genetic analysis of strains containing multiple copies of the glaA gene. Transgenic Res. 1993, 2, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Wallis, G.L.F.; Swift, R.J.; Hemming, F.W.; Trinci, A.P.J.; Peberdy, J.F. Glucoamylase overexpression and secretion in Aspergillus niger: Analysis of glycosylation. Biochim. Biophys. Acta 1999, 1472, 576–586. [Google Scholar] [CrossRef]
- Tada, S.; Iimura, Y.; Gomi, K.; Takahashi, K.; Hara, S.; Yoshizawa, K. Cloning and nucleotide sequence of the genomic taka-amylase a gene of Aspergillus oryzae. Agric. Biol. Chem. 1989, 53, 593–599. [Google Scholar] [CrossRef] [Green Version]
- Schalén, M.; Anyaogu, D.C.; Hoof, J.B.; Workman, M. Effect of secretory pathway gene overexpression on secretion of a fluorescent reporter protein in Aspergillus nidulans. Fungal Biol. Biotechnol. 2016, 3, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Lubertozzi, D.; Keasling, J.D. Marker and promoter effects on heterologous expression in Aspergillus nidulans. Appl. Microbiol. Biotechnol. 2006, 72, 1014–1023. [Google Scholar] [CrossRef]
- Verdoes, J.C.; Punt, P.J.; van den Hondel, C.A.M.J.J. Molecular genetic strain improvement for the overproduction of fungal proteins by filamentous fungi. Appl. Microbiol. Biotechnol. 1995, 43, 195–205. [Google Scholar] [CrossRef]
- Gouka, R.J.; Hessing, J.G.M.; Stam, H.; Musters, W.; van den Hondel, C.A.M.J.J. A novel strategy for the isolation of defined pyrG mutants and the development of a site-specific integration system for Aspergillus awamori. Curr. Genet. 1995, 27, 536–540. [Google Scholar] [CrossRef]
- Qin, L.; Jiang, X.; Dong, Z.; Huang, J.; Chen, X. Identification of two integration sites in favor of transgene expression in Trichoderma reesei. Biotechnol. Biofuels 2018, 11, 142. [Google Scholar] [CrossRef]
- Hansen, B.G.; Salomonsen, B.; Nielsen, M.T.; Nielsen, J.B.; Hansen, N.B.; Nielsen, K.F.; Regueira, T.B.; Nielsen, J.; Patil, K.R.; Mortensen, U.H. Versatile enzyme expression and characterization system for Aspergillus nidulans, with the Penicillium brevicompactum polyketide synthase gene from the mycophenolic acid gene cluster as a test case. Appl. Environ. Microbiol. 2011, 77, 3044–3051. [Google Scholar] [CrossRef] [Green Version]
- Mauro, V.P. Codon optimization in the production of recombinant biotherapeutics: Potential risks and considerations. BioDrugs 2018, 32, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, C.; Minshull, J.; Govindarajan, S.; Ness, J.; Villalobos, A.; Welch, M. Engineering genes for predictable protein expression. Protein Expr. Purif. 2012, 83, 37–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, M.; Tokuoka, M.; Gomi, K. Effects of codon optimization on the mRNA levels of heterologous genes in filamentous fungi. Appl. Microbiol. Biotechnol. 2014, 98, 3859–3867. [Google Scholar] [CrossRef] [PubMed]
- Tokuoka, M.; Tanaka, M.; Ono, K.; Takagi, S.; Shintani, T.; Gomi, K. Codon optimization increases steady-state mRNA levels in Aspergillus oryzae heterologous gene expression. Appl. Environ. Microbiol. 2008, 74, 6538–6546. [Google Scholar] [CrossRef] [Green Version]
- Koda, A.; Bogaki, T.; Minetoki, T.; Hirotsune, M. High expression of a synthetic gene encoding potato α-glucan phosphorylase in Aspergillus niger. J. Biosci. Bioeng. 2005, 100, 531–537. [Google Scholar] [CrossRef]
- Gouka, R.J.; Punt, P.J.; Hessing, J.G.M.; Van Den Hondel, C.A.M.J.J. Analysis of heterologous protein production in defined recombinant Aspergillus awamori strains. Appl. Environ. Microbiol. 1996, 62, 1951–1957. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, M.; Tokuoka, M.; Shintani, T.; Gomi, K. Transcripts of a heterologous gene encoding mite allergen Der f 7 are stabilized by codon optimization in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2012, 96, 1275–1282. [Google Scholar] [CrossRef]
- Yu, C.-H.; Dang, Y.; Zhou, Z.; Wu, C.; Zhao, F.; Sachs, M.S.; Liu, Y. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell 2015, 59, 744–754. [Google Scholar] [CrossRef] [Green Version]
- Gouka, R.J.; Punt, P.J.; Van Den Hondel, C.A.M.J.J. Glucoamylase gene fusions alleviate limitations for protein production in Aspergillus awamori at the transcriptional and (post)translational levels. Appl. Environ. Microbiol. 1997, 63, 488–497. [Google Scholar] [CrossRef] [Green Version]
- Carraway, K.L.; Hull, S.R. O-glycosylation pathway for mucin-type glycoproteins. BioEssays 1989, 10, 117–121. [Google Scholar] [CrossRef]
- Bause, E. Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes. Biochem. J. 1983, 209, 331–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshpande, N.; Wilkins, M.R.; Packer, N.; Nevalainen, H. Protein glycosylation pathways in filamentous fungi. Glycobiology 2008, 18, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, S.H.; Jensen, B.; Olsen, J. Effect of N-linked glycosylation on secretion, activity, and stability of α-amylase from Aspergillus oryzae. Curr. Microbiol. 1998, 37, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Maras, M.; van Die, I.; Contreras, R.; van den Hondel, C.A. Filamentous fungi as production organisms for glycoproteins of bio-medical interest. Glycoconj. J. 1999, 16, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Kainz, E.; Gallmetzer, A.; Hatzl, C.; Nett, J.H.; Li, H.; Schinko, T.; Pachlinger, R.; Berger, H.; Reyes-Dominguez, Y.; Bernreiter, A.; et al. N-Glycan modification in Aspergillus species. Appl. Environ. Microbiol. 2008, 74, 1076–1086. [Google Scholar] [CrossRef] [Green Version]
- Kasajima, Y.; Yamaguchi, M.; Hirai, N.; Ohmachi, T.; Yoshida, T. In vivo expression of UDP-N-acetylglucosamine: Alpha-3-D-mannoside beta-1,2-N-acetylglucosaminyltransferase I (GnT-1) in Aspergillus oryzae and effects on the sugar chain of alpha-amylase. Biosci. Biotechnol. Biochem. 2006, 70, 2662–2668. [Google Scholar] [CrossRef] [Green Version]
- Ichishima, E.; Taya, N.; Ikeguchi, M.; Chiba, Y.; Nakamura, M.; Kawabata, C.; Inoue, T.; Takahashi, K.; Minetoki, T.; Ozeki, K.; et al. Molecular and enzymic properties of recombinant 1, 2-alpha-mannosidase from Aspergillus saitoi overexpressed in Aspergillus oryzae cells. Biochem. J. 1999, 339, 589–597. [Google Scholar] [CrossRef]
- Kalsner, I.; Hintz, W.; Reid, L.S.; Schachter, H. Insertion into Aspergillus nidulans of functional UDP-GlcNAc: Alpha 3-D- mannoside beta-1,2-N-acetylglucosaminyl-transferase I, the enzyme catalysing the first committed step from oligomannose to hybrid and complex N-glycans. Glycoconj. J. 1995, 12, 360–370. [Google Scholar] [CrossRef]
- Guillemette, T.; van Peij, N.N.M.E.; Goosen, T.; Lanthaler, K.; Robson, G.D.; van den Hondel, C.A.M.J.J.; Stam, H.; Archer, D.B. Genomic analysis of the secretion stress response in the enzyme-producing cell factory Aspergillus niger. BMC Genom. 2007, 8, 158. [Google Scholar] [CrossRef] [Green Version]
- Kwon, M.J.; Jørgensen, T.R.; Nitsche, B.M.; Arentshorst, M.; Park, J.; Ram, A.F.; Meyer, V. The transcriptomic fingerprint of glucoamylase over-expression in Aspergillus niger. BMC Genom. 2012, 13, 701. [Google Scholar] [CrossRef] [Green Version]
- Geysens, S.; Whyteside, G.; Archer, D.B. Genomics of protein folding in the endoplasmic reticulum, secretion stress and glycosylation in the aspergilli. Fungal Genet. Biol. 2009, 46, S121–S140. [Google Scholar] [CrossRef]
- Valkonen, M.; Ward, M.; Wang, H.; Penttilä, M.; Saloheimo, M. Improvement of foreign-protein production in Aspergillus niger var. awamori by constitutive induction of the unfolded-protein response. Appl. Environ. Microbiol. 2003, 69, 6979–6986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derkx, P.M.F.; Madrid, S.M. Peptidyl prolyl cis-trans isomerases 2000. World Patent WO0018934, 6 April 2000. [Google Scholar]
- Wang, H.; Ward, M. Molecular characterization of a PDI-related gene prpA in Aspergillus niger var. awamori. Curr. Genet. 2000, 37, 57–64. [Google Scholar] [CrossRef]
- Moralejo, F.; Watson, A.; Jeenes, D.; Archer, D.; Martín, J. A defined level of protein disulfide isomerase expression is required for optimal secretion of thaumatin by Aspergillus awamori. Mol. Genet. Genom. 2001, 266, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Jeenes, D.; Archer, D.B.; van den Hondel, C.A.; Punt, P.J. Calnexin overexpression increases manganese peroxidase production in Aspergillus niger. Appl. Environ. Microbiol. 2002, 68, 846–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombraña, M.; Moralejo, F.J.; Pinto, R.; Martín, J.F. Modulation of Aspergillus awamori thaumatin secretion by modification of bipA gene expression. Appl. Environ. Microbiol. 2004, 70, 5145–5152. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Vishwanath, P.; Cui, J.; Kelleher, D.J.; Gilmore, R.; Robbins, P.W.; Samuelson, J. The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc. Natl. Acad. Sci. USA 2007, 104, 11676–11681. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Feng, D.; Fang, W.; Ouyang, H.; Luo, Y.; Du, T.; Jin, C. Comparative proteomic analysis of an Aspergillus fumigatus mutant deficient in glucosidase I (AfCwh41). Microbiology 2009, 155, 2157–2167. [Google Scholar] [CrossRef] [Green Version]
- Ríos, S.; Fernández-Monistrol, I.; Laborda, F. Effect of tunicamycin on α-galactosidase secretion by Aspergillus nidulans and the importance of N-glycosylation. FEMS Microbiol. Lett. 1994, 120, 169–175. [Google Scholar] [CrossRef]
- Perlińska-lenart, U.; Kurzątkowski, W.; Janas, P.; Palamarczyk, G.; Kruszewska, J.S. Protein production and secretion in an Aspergillus nidulans mutant impaired in glycosylation. Acta Biochim. Pol. 2005, 52, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Van den Brink, H.J.M.; Petersen, S.G.; Rahbek-Nielsen, H.; Hellmuth, K.; Harboe, M. Increased production of chymosin by glycosylation. J. Biotechnol. 2006, 125, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. Gene Inactivated Mutants with Altered Protein Production. World Patent No. WO2006110677, 19 October 2006. [Google Scholar]
- Dai, Z.; Aryal, U.K.; Shukla, A.; Qian, W.J.; Smith, R.D.; Magnuson, J.K.; Adney, W.S.; Beckham, G.T.; Brunecky, R.; Himmel, M.E.; et al. Impact of alg3 gene deletion on growth, development, pigment production, protein secretion, and functions of recombinant Trichoderma reesei cellobiohydrolases in Aspergillus niger. Fungal Genet. Biol. 2013, 61, 120–132. [Google Scholar] [CrossRef] [PubMed]
- Pantazopoulou, A. The Golgi apparatus: Insights from filamentous fungi. Mycologia 2016, 108, 603–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiedler, M.R.M.; Barthel, L.; Kubisch, C.; Nai, C.; Meyer, V. Construction of an improved Aspergillus niger platform for enhanced glucoamylase secretion. Microb. Cell Fact. 2018, 17, 1–12. [Google Scholar] [CrossRef]
- Fiedler, M.R.M.; Cairns, T.C.; Koch, O.; Kubisch, C.; Meyer, V. Conditional expression of the small GTPase ArfA impacts secretion, morphology, growth, and actin ring position in Aspergillus niger. Front. Microbiol. 2018, 9, 1–17. [Google Scholar] [CrossRef]
- Hoang, H.-D.; Maruyama, J.; Kitamoto, K. Modulating endoplasmic reticulum-golgi cargo receptors for improving secretion of carrier-fused heterologous proteins in the filamentous fungus Aspergillus oryzae. Appl. Environ. Microbiol. 2015, 81, 533–543. [Google Scholar] [CrossRef] [Green Version]
- Bonifacino, J.S.; Weissman, A.M. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol. 1998, 14, 19–57. [Google Scholar] [CrossRef]
- Jacobs, D.I.; Olsthoorn, M.M.A.; Maillet, I.; Akeroyd, M.; Breestraat, S.; Donkers, S.; van der Hoeven, R.A.M.; van den Hondel, C.A.M.J.J.; Kooistra, R.; Lapointe, T. Effective lead selection for improved protein production in Aspergillus niger based on integrated genomics. Fungal Genet. Biol. 2009, 46, S141–S152. [Google Scholar] [CrossRef]
- Carvalho, N.D.S.P.; Arentshorst, M.; Kooistra, R.; Stam, H.; Sagt, C.M.; Van Den Hondel, C.A.M.J.J.; Ram, A.F.J. Effects of a defective ERAD pathway on growth and heterologous protein production in Aspergillus niger. Appl. Microbiol. Biotechnol. 2011, 89, 357–373. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.; Aishan, T.; Maruyama, J.; Kitamoto, K. Enhanced production and secretion of heterologous proteins by the filamentous fungus Aspergillus oryzae via disruption of vacuolar protein sorting receptor gene Aovps10. Appl. Environ. Microbiol. 2010, 76, 5718–5727. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.; Kikuma, T.; Maruyama, J.; Kitamoto, K. Enhanced production of bovine chymosin by autophagy deficiency in the filamentous fungus Aspergillus oryzae. PLoS ONE 2013, 8, e62512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; An, Z. Heterologous protein expression in yeasts and filamentous fungi. In Manual of Industrial Microbiology and Biotechnology; Baltz, R.H., Demain, A.L., Davies, J.E., Bull, A.T., Junker, B., Katz, L., Lynd, L.R., Masurekar, P., Reeves, C.D., Zhao, H., Eds.; American Society of Microbiology Press: Washington, DC, USA, 2010; pp. 145–156. ISBN 9781555815127. [Google Scholar]
- Lawlis, V.B. DNA Sequences, Vectors, and Fusion Polypeptides to Increase Secretion of Desired Polypeptides from Filamentous Fungi. World Patent No. WO9015860, 27 December 1990. [Google Scholar]
- Ward, M.; Wilson, L.J.; Kodama, K.H.; Rey, M.W.; Berka, R.M. Improved production of chymosin in Aspergillus by expression as a glucoamylase-chymosin fusion. Nat. Biotechnol. 1990, 8, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Cullen, D.; Gray, G.L.; Wilson, L.J.; Hayenga, K.J.; Lamsa, M.H.; Rey, M.W.; Norton, S.; Berka, R.M. Controlled expression and secretion of bovine chymosin in Aspergillus nidulans. Nat. Biotechnol. 1987, 5, 369–376. [Google Scholar] [CrossRef]
- Broekhuijsen, M.P.; Mattern, I.E.; Contreras, R.; Kinghorn, J.R.; van den Hondel, C.A.M.J.J. Secretion of heterologous proteins by Aspergillus niger: Production of active human interleukin-6 in a protease-deficient mutant by KEX2-like processing of a glucoamylase-hIL6 fusion protein. J. Biotechnol. 1993, 31, 135–145. [Google Scholar] [CrossRef]
- Frenken, L.G.J.; Van Gorcom, R.F.M.; Hessing, J.G.M.; Van Den Hondel, C.A.; Musters, W.; Verbakel, J.M.A.; Verrips, C.T. Process for Producing Fusion Proteins Comprising Scfv Fragments by a Transformed Mould. World Patent No. WO9429457, 22 December 1994. [Google Scholar]
- Ward, M.; Lin, C.; Victoria, D.C.; Fox, B.P.; Fox, J.A.; Wong, D.L.; Meerman, H.J.; Pucci, J.P.; Fong, R.B.; Heng, M.H.; et al. Characterization of humanized antibodies secreted by Aspergillus niger. Appl. Environ. Microbiol. 2004, 70, 2567–2576. [Google Scholar] [CrossRef] [Green Version]
- Segato, F.; Damásio, A.R.L.; Gonçalves, T.A.; de Lucas, R.C.; Squina, F.M.; Decker, S.R.; Prade, R.A. High-yield secretion of multiple client proteins in Aspergillus. Enzyme Microb. Technol. 2012, 51, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.J.; Watanabe, T.; Juvvadi, P.R.; Maruyama, J.; Arioka, M.; Kitamoto, K. Double disruption of the proteinase genes, tppA and pepE, increases the production level of human lysozyme by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2007, 76, 1059–1068. [Google Scholar] [CrossRef]
- Ohno, A.; Maruyama, J.; Nemoto, T.; Arioka, M.; Kitamoto, K. A carrier fusion significantly induces unfolded protein response in heterologous protein production by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2011, 92, 1197–1206. [Google Scholar] [CrossRef]
- Gouka, R.J.; Van Den Hondel, C.A.; Musters, W.; Stam, H.; Verbakel, J.M.A. Process for Producing/Secreting a Protein by a Transformed Mould Using Expression/Secretion Regulating Regions Derived from an Aspergillus Endoxylanase II Gene. World Patent No. WO9312237, 24 June 1993. [Google Scholar]
- Jalving, R.; Van De Vondervoort, P.J.I.; Visser, J.; Schaap, P.J. Characterization of the Kexin-Like Maturase of Aspergillus niger. Appl. Environ. Microbiol. 2000, 66, 363–368. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Ward, M. Kex2 Cleavage Regions of Recombinant Fusion Proteins. U.S. Patent No. US8,936,917B2, 20 January 2015. [Google Scholar]
- O’Donnell, D.; Wang, L.; Xu, J.; Ridgway, D.; Gu, T.; Moo-Young, M. Enhanced heterologous protein production in Aspergillus niger through pH control of extracellular protease activity. Biochem. Eng. J. 2001, 8, 187–193. [Google Scholar] [CrossRef]
- Van Den Hombergh, J.P.T.W.; Van De Vondervoort, P.J.I.; Fraissinet-Tachet, L.; Visser, J. Aspergillus as a host for heterologous protein production: The problem of proteases. Trends Biotechnol. 1997, 15, 256–263. [Google Scholar] [CrossRef]
- Berka, R.M.; Ward, M.; Wilson, L.J.; Hayenga, K.J.; Kodama, K.H.; Carlomagne, L.P.; Thompson, S.A. Molecular cloning and deletion of the gene encoding aspergillopepsin A from Aspergillus awamori. Gene 1990, 86, 153–162. [Google Scholar] [CrossRef]
- Mattern, I.E.; van Noort, J.M.; van den Berg, P.; Archer, D.B.; Roberts, I.N.; van den Hondep, C.A. Isolation and characterization of mutants of Aspergillus niger deficient in extracellular proteases. Mol. Gen. Genet. 1992, 332–336. [Google Scholar] [CrossRef]
- Kamaruddin, N.; Storms, R.; Mahadi, N.M.; Illias, R.M.; Bakar, F.D.A.; Murad, A.M.A. Reduction of extracellular proteases increased activity and stability of heterologous protein in Aspergillus niger. Arab. J. Sci. Eng. 2018, 43, 3327–3338. [Google Scholar] [CrossRef]
- Berka, R.M.; Hayenga, K.; Lawlis, V.B.; Ward, M. Aspartic Proteinase Deficient Filamentous Fungi. World Patent No. WO199000192, 11 January 1990. [Google Scholar]
- Punt, P.J.; Schuren, F.H.J.; Lehmbeck, J.; Christensen, T.; Hjort, C.; van den Hondel, C.A.M.J.J. Characterization of the Aspergillus niger prtT, a unique regulator of extracellular protease encoding genes. Fungal Genet. Biol. 2008, 45, 1591–1599. [Google Scholar] [CrossRef]
- Yoon, J.; Kimura, S.; Maruyama, J.; Kitamoto, K. Construction of quintuple protease gene disruptant for heterologous protein production in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2009, 82, 691–701. [Google Scholar] [CrossRef]
- Yoon, J.; Maruyama, J.; Kitamoto, K. Disruption of ten protease genes in the filamentous fungus Aspergillus oryzae highly improves production of heterologous proteins. Appl. Microbiol. Biotechnol. 2011, 89, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Lehmbeck, J. Novel Host Cells and Methods of Producing Proteins. World Patent No. WO9812300, 26 March 1998. [Google Scholar]
- Shinkawa, S.; Mitsuzawa, S.; Tanaka, M.; Imai, T. Aspergillus Mutant Strain and Transformant Thereof. U.S. Patent No. US9,567,563, 11 August 2016. [Google Scholar]
- Taheri-Talesh, N.; Horio, T.; Araujo-Bazán, L.; Dou, X.; Espeso, E.A.; Peñalva, M.A.; Osmani, S.A.; Oakley, B.R. The tip growth apparatus of Aspergillus nidulans. Mol. Biol. Cell 2008, 19, 1439–1449. [Google Scholar] [CrossRef] [Green Version]
- Grimm, L.H.; Kelly, S.; Krull, R.; Hempel, D.C. Morphology and productivity of filamentous fungi. Appl. Microbiol. Biotechnol. 2005, 69, 375–384. [Google Scholar] [CrossRef]
- Sisniega, H.; Río, J.-L.; Amaya, M.-J.; Faus, I. Strategies for large-scale production of recombinant proteins in filamentous fungi. In Microbial Processes and Products; Humana Press: Totowa, NJ, USA, 2005; Volume 18, pp. 225–237. [Google Scholar]
- El-Enshasy, H.A. Filamentous fungal cultures—Process characteristics, products, and applications. In Bioprocessing for Value-Added Products from Renewable Resources; Elsevier: Amsterdam, The Netherlands, 2007; pp. 225–261. [Google Scholar]
- Workman, M.; Andersen, M.R.; Thykaer, J. Integrated approaches for assessment of cellular performance in industrially relevant filamentous fungi. Ind. Biotechnol. 2013, 9, 337–344. [Google Scholar] [CrossRef]
- Wang, L.; Ridgway, D.; Gu, T.; Moo-Young, M. Bioprocessing strategies to improve heterologous protein production in filamentous fungal fermentations. Biotechnol. Adv. 2005, 23, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Gyamerah, M.; Merichetti, G.; Adedayo, O.; Scharer, J.; Moo-Young, M. Bioprocessing strategies for improving hen egg-white lysozyme (HEWL) production by recombinant Aspergillus niger HEWL WT-13-16. Appl. Microbiol. Biotechnol. 2002, 60, 403–407. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, D.A.; Gendron, L.C.G.; Jeenes, D.J.; Archer, D.B. Physiological optimization of secreted protein production by Aspergillus niger. Enzyme Microb. Technol. 1994, 16, 276–280. [Google Scholar] [CrossRef]
- Wang, L.; Ridgway, D.; Gu, T.; Moo-Young, M. Effects of process parameters on heterologous protein production in Aspergillus niger fermentation. J. Chem. Technol. Biotechnol. 2003, 78, 1259–1266. [Google Scholar] [CrossRef]
- Swift, R.J.; Karandikar, A.; Griffen, A.M.; Punt, P.J.; van den Hondel, C.A.M.J.J.; Robson, G.D.; Trinci, A.P.J.; Wiebe, M.G. The Effect of organic nitrogen sources on recombinant glucoamylase production by Aspergillus niger in chemostat culture. Fungal Genet. Biol. 2000, 31, 125–133. [Google Scholar] [CrossRef]
- El-Enshasy, H.; Kleine, J.; Rinas, U. Agitation effects on morphology and protein productive fractions of filamentous and pelleted growth forms of recombinant Aspergillus niger. Process Biochem. 2006, 41, 2103–2112. [Google Scholar] [CrossRef]
- Eibes, G.M.; Lú-Chau, T.A.; Ruiz-Dueñas, F.J.; Feijoo, G.; Martínez, M.J.; Martínez, A.T.; Lema, J.M. Effect of culture temperature on the heterologous expression of Pleurotus eryngii versatile peroxidase in Aspergillus hosts. Bioprocess Biosyst. Eng. 2009, 32, 129–134. [Google Scholar] [CrossRef] [Green Version]
- Amanullah, A.; Christensen, L.H.; Hansen, K.; Nienow, A.W.; Thomas, C.R. Dependence of morphology on agitation intensity in fed-batch cultures of Aspergillus oryzae and its implications for recombinant protein production. Biotechnol. Bioeng. 2002, 77, 815–826. [Google Scholar] [CrossRef]
- Archer, D.B.; MacKenzie, D.A.; Ridout, M.J. Heterologous protein secretion by Aspergillus niger growing in submerged culture as dispersed or aggregated mycelia. Appl. Microbiol. Biotechnol. 1995, 44, 157–160. [Google Scholar] [CrossRef]
- Driouch, H.; Sommer, B.; Wittmann, C. Morphology engineering of Aspergillus niger for improved enzyme production. Biotechnol. Bioeng. 2010, 105, 1058–1068. [Google Scholar] [CrossRef]
- Driouch, H.; Hänsch, R.; Wucherpfennig, T.; Krull, R.; Wittmann, C. Improved enzyme production by bio-pellets of Aspergillus niger: Targeted morphology engineering using titanate microparticles. Biotechnol. Bioeng. 2012, 109, 462–471. [Google Scholar] [CrossRef] [PubMed]
| Expression Platform | Genetic Manipulation | Growth Rate | Product Titers | Product Quality | Product Purification | Contamination Risk | Production Cost | Relevant Literature |
|---|---|---|---|---|---|---|---|---|
| Bacteria (Escherichia coli) | Simple | Fast | High | Products can be non-functional (codon bias, no adequate post-translation modifications) | Can be problematic (e.g., inclusion bodies) | Medium (endotoxins) | Low | [18,19] |
| Yeasts (Saccharomyces cerevisiae, Pichia pastoris, etc.) | Simple | Fast | S. cerevisiae limited P. pastoris higher | Hypermannosylation of glycoproteins often occurs (shortens half-life of the protein in vivo, leads to immunogenic reactions) | Feasible | Low | Low | [9,10] |
| Filamentous fungi (Aspergillus niger, Trichoderma reesei, Neurospora crassa) | Feasible | Medium | High | Less hypermannosylation compared to yeasts, but still differences from mammalian glycosylation patterns | Simple | Medium (mycotoxins) | Low | [7] |
| Insect cells (Spodoptera frugiperda, Drosophila melanogaster) | Laborious | Fast | High | Not able to carry out N-glycosylation | Feasible | Very low | High | [20] |
| Mammalian cells (CHO cells, Human cell lines) | Laborious | Slow | Low | High quality therapeutic proteins, human-like glycosylation pattern | Simple | High (viruses and prions) | High | [21] |
| Transgenic animals (goats, chickens) | Laborious | Very slow | High | High quality therapeutic proteins | Simple | High (viruses and prions) | High, ethically questionable | [22] |
| Transgenic plants (rice, bananas, carrots, potatoes) | Feasible | Slow | High | Some differences in glycan structures from human-like pattern | Complex and expensive downstream processing | Very low | Medium | [23,24] |
| Process | Modification | Performance | Improvement Factor | Reference | |
|---|---|---|---|---|---|
| Promoters | Use of several promoters (P) in A. awamori | PB2 from Acremonium chrysogenum: 0.25–2 mg/L thaumatin | - | [46] | |
| PpcbC from Penicillium chrysogenum: 0.25–2 mg/L thaumatin | |||||
| PgdhA from A. awamori: 1–9 mg/L thaumatin | |||||
| PgpdA from A. nidulans: 0.75–11 mg/L thaumatin | |||||
| Insertion of multiple copies of an activator protein-binding site from the cis-regulatory region of A. niger glaA to the new promoter in A. niger | 396.0 ± 51.5 mg/L of Vitreoscilla hemoglobin compared to 19.7 ± 4.8 mg/L from the strain with 1 copy | 20 | [45] | ||
| Use of hybrid promoters (combination of a human hERa-activated promoter (pERE), S. cerevisiae URA3 promoter and A. nidulans nirA promoter) in A. nidulans | pERE-RS-nirA + lacZ: 25 U of β-galactosidase activity/mg of protein | - | [47] | ||
| pERE-URA-nirA + lacZ: 100 U of β-galactosidase activity/mg of protein | 4 | ||||
| pERE-URA-RS + lacZ: 1400 U of β-galactosidase activity/mg of protein [1 pM inducer (DES)] | 56 | ||||
| Use of a hemolysin-like protein promoter (Phyl) for heterologous production in A. oryzae | Reporter gene: Endoglucanase Cel B Pamy: 24.1 ± 5.5 U/mL, Phyl: 57.9 ± 17.4 U/mL | 2.4 | [48] | ||
| Reporter gene: Trichoderma endoglucanase I Pamy: 7.7 ± 3.9 U/mL, Phyl: 27.8 ± 1.3 U/mL | 3.6 | ||||
| Reporter gene: Trichoderma endoglucanase III Pamy:4.0 ± 0.6 U/mL, hyl:31.7 ± 3.3 U/mL | 7.9 | ||||
| Regulatory elements (TerR and PterA) from A. terreus terrain gene cluster for E. coli lacZ expression in A. niger | Promoter activity ~5000 mU/mg when TerR under PgpdA (No activity when TerR under the native promoter) | - | [49] | ||
| Promoter activity ~10,000 mU/mg (when TerR under PgpdA in 2 copies) | 2 | ||||
| Promoter activity ~15,000 mU/mg (when TerR under PamyB) | 3 | ||||
| A. niger α-glucosyltransferase produced under the A. niger pyruvate kinase promoter | 2000 U/mL total activity of α-glucosyltransferase compared to 600 U/mL in the wild type | 3.3 | [50] | ||
| Overexpression of the transcription factor RsmA, while the aflR promoter was inserted in front of the pslcc in A. nidulans | 0.06 U/mL of Pycnoporus sanguineus laccase compared to 0.004 U/mL in the control strain | 15 | [51,52] | ||
| A novel promoter from Talaromyces emersonii (Pglucan1200) for expressing glaA in A. niger | 6000 U/mL of GlaA, enzyme activity increased by about 25% compared to 5000 U/mL in the strain with the PglaA | 1.2 | [53] | ||
| The constitutive promoter of ecm33 (Pecm33) from A. niger in A. niger | Maltose: | Pecm33 activity induced by 1.7 compared to PglaA activity that induced by 2.7 | - | [54] | |
| Glucose: | Pecm33 activity induced by 1.1 compared to PglaA activity that induced by 1.8 | ||||
| Xylose: | Pecm33 activity induced by 2 compared to PglaA activity that induced by 1.3 Increased Pecm33 activity at 37 °C | ||||
| Process | Modification | Performance | Improvement Factor | Reference |
|---|---|---|---|---|
| Copy number | Integration of up to 200 additional copies of the glaA in A. niger | 355 mg/L of glucoamylase compared to parental strain (50 mg/L) | 7.1 | [58] |
| Integration of 80 additional copies of the glaA in A. niger | 1268 mU of glucoamylase/mL culture filtrate compared to 280 mU/mL in the parental strain | 4.5 | [59] | |
| Integration of multiple copies of cassettes with thaumatin gene in A. awamori (two types of cassettes, 1 with PB2 and 1 with PgdhA) | 8 copies: 10 ± 0.4 mg/L thaumatin | - | [46] | |
| 11 copies: 14 ± 1.1 mg/L thaumatin | 1.4 | |||
| 14 copies: 11 ± 0.8 mg/L thaumatin | 1.1 | |||
| 10 copies: 14 ± 1.3 mg/L thaumatin | 1.4 | |||
| Insertion of multiple copies of the cassette pERE-URA-RS + lacZ in A. nidulans | 1 copy of pERE-URA-RS + lacZ: 9500 U of β-galactosidase/mg of protein | 5.4 | [47] | |
| Multiple copies of pERE-URA-RS + lacZ: 51,000U of β-galactosidase/mg of protein [1 nM inducer (DES)] | ||||
| Integration of an additional copy of the glaA- RFP (2 in total) in A. nidulans | A 70% increase in maximum fluorescence level (quantification data not available) | 1.7 | [61] |
| Process | Modification | Performance | Improvement Factor | Reference |
|---|---|---|---|---|
| Codon usage | A Cyamopsis tetragonoloba α-galactosidase gene optimized based on Saccharomyces cerevisiae codon usage and expressed in A. awamori | Synthetic gene: 0.4 mg/L α-galactosidase Wild type gene: Undetectable levels | - | [72] |
| A Solanum tuberosum α-glucan phosphorylase synthetic gene optimized based on A. niger-preferred codon usage for production in A. niger | Synthetic gene: 39.6–94.6 mg/L α-glucan phosphorylase Wild type gene: <0.1 mg/L | - | [71] | |
| A codon optimized Dermatophagoides farina der f7 gene based on A. oryzae codon usage and expressed in A. oryzae | Non-fused: undetectable level to a detectable level Fused to GlaA: 3 to 5 fold increase (Signal intensity quantification of the bands from the SDS-PAGE) | 3–5 | [70] |
| Process | Modification | Performance | Improvement Factor | Reference |
|---|---|---|---|---|
| ER-stress and protein folding | Overexpression of prpA (multicopy integrated vector) in A. niger var. awamori | The level of chymosin by the control transformants was similar to the transformants with overexpression of the prpA | - | [90] |
| Deletion of prpA in A. niger var. awamori | The production level of bovine prochymosin was lower than expected for randomly isolated transformants (3/19 transformants) | - | [90] | |
| Overexpression of cypB in A. niger | Twofold increase in glucoamylase production | 2 | [89] | |
| Insertion of multiple copies of pdiA in A. awamori | 19 mg/L thaumatin compared to 5 mg/L in the parental strain | 3.8 | [91] | |
| Optimal bioreactor conditions: 150 mg/L thaumatin compared to 40 mg/L in the parental strain | 3.8 | |||
| Overexpression of clxA in A.niger | 60–73 mg/L P. chrysosporium manganese peroxidase compared to 14 mg/L in the parental strain | 4–5 | [92] | |
| Overexpression of bipA in A.niger | P. chrysosporium manganese peroxidase was severely reduced-almost undetectable levels | - | [92] | |
| Overexpression of hacA in A. niger var. awamori | 13–34 mg/L chymosin compared to 12.5 mg/L in parental strain | 1.3–2.8 | [88] | |
| 3.9–8.5 nkat/mL laccase compared to 0.9 nkat/mL in parental strain | 3–7.6 | |||
| Overexpression of bipA in A. awamori | 20 mg/L thaumatin compared to 9 mg/L in the parental strain | 2–2.5 | [93] | |
| Constitutive expression of the active form of hacA cDNA in A. oryzae | 2 mg/L neoculin compared to 1.5 mg/L in the control strain | 1.5 | [40] |
| Process | Modification | Performance | Improvement Factor | Reference |
|---|---|---|---|---|
| Glycosylation | Overexpression of S. cerevisiae DPM1 in A. nidulans strains impaired in DPMS activity | No significant increase in protein secretion observed Production of invertase and glucoamylase was higher but the proteins were trapped in the periplasmic space | 28 | [97] |
| Bovine prochymosin synthetic gene with a single mutation (S335T)—This mutation resulted in a potentially better N-glycosylation site (NHT) in A. niger | 207 IMCU/mL (0.9 g/L) chymosin compared to 90 IMCU/mL in the parental strain 90% of the chymosin molecules were glycosylated compared to 10% in the parental strain | 3 | [98] | |
| Bovine prochymosin synthetic gene with a N–S–T glycosylation site (TDNST) in the short peptide linker in A. niger | 141 International Milk Clotting Unit/mL (0.6 g/L) chymosin compared to 90 IMCU/mL in the parental strain Same glycosylation pattern | 1.5 | [98] | |
| Deletion of mnn9, mnn10, ochA in A. niger | Δmnn9: 14.6% increase in Tramete laccase production | 1.1 | [99] | |
| Δmnn10: 12.7% increase | 1.1 | |||
| ΔochA: 7.2% increase | 1.1 | |||
| Δmnn9/ochA: 16.8% increase | 1.2 |
| Process | Modification | Performance | Improvement Factor | Reference |
|---|---|---|---|---|
| Protein translocation | Deletion of Aovip36 in A. oryzae | 50 mg/L chymosin compared to 27 mg/L in the parental strain | 1.9 | [104] |
| 300% EGFP in culture supernatant compared to 100% in the parental strain The α-amylase activity (native protein) was reduced by approximately 30% compared with the activity in the control strain | 3 | |||
| Deletion of AoEmp47 in A. oryzae | 50 mg/L chymosin compared to 27 mg/L in the parental strain | 1.9 | [104] | |
| 210% EGFP in culture supernatant compared to 100% in the parental strain No difference in the α-amylase activity (native protein) | 2.1 | |||
| Overexpression of rabD in A. nidulans | 25% increase in mRFP secretion in submerged cultivations in shake flasks | 1.3 | [61] | |
| 40% increase in RFP secretion in 2l bioreactor | 1.4 | |||
| Deletion of racA in A. niger | Native GlaA secreted into the culture medium is four times more compared to its parental strain, when ensuring continuous high-level expression of glaA. Quantification was done by dot blot analysis using a monoclonal antibody (Arbitrary units) | 4 | [102] | |
| Overexpression of arfA in A. niger | Quantitative abundance of GlaA-dtomato reporter protein was 397.4 absolute fluorescence compared to 298.7 in control strain | 1.3 | [103] |
| Process | Modification | Performance | Improvement Factor | Reference |
|---|---|---|---|---|
| Protein degradation pathways—ERAD and Vacuole | Deletion of derA and derB in A. niger | ΔderA: 80% decrease in Tramete laccase production | 0.2 | [99] |
| - | ΔderB: 15.7% increase in Tramete laccase | 1.15 | ||
| Deletion of doaA and overexpression of sttC in A. niger | Higher GUS activity compared to parental strain (no quantitative data available) | - | [106] | |
| Disruption of Aovps10 in A. oryzae | 83.1 and 70.3 mg/L chymosin compared to 28.7 mg/L in parental strain | 3–2.5 | [108] | |
| 22.6 and 24.6 mg/L human lysozyme compared to 11.1 mg/L in parental strain | 2–2.2 | |||
| Deletion of ERAD key genes (derA, doaA, hrdC, mifA and mnsA) in A. niger | ΔderA and ΔhrdC: 2-fold increase compared to parental strain (single-copy) | 2 | [107] | |
| ΔderA: 6-fold increase compared to parental strain (multi-copy) Relative amount of intracellular GlaGus (β-glucuronidase levels) fusion protein detected in total protein extracts of strains with impaired ERAD and respective parental strain | 6 | |||
| Disruption of genes involved in autophagy in A. oryzae | ΔAoatg1: 60 mg/L chymosin | 2.3 | [109] | |
| ΔAoatg13: 37 mg/L chymosin | 1.4 | |||
| ΔAoatg4: 80 mg/L chymosin | 3.1 | |||
| ΔAoatg8: 66 mg/L chymosin | 2.5 | |||
| ΔAoatg15: 24 mg/L chymosin | 1 | |||
| Control: 26 mg/L chymosin | - |
| Process | Modification | Performance | Improvement Factor | Reference |
|---|---|---|---|---|
| Carriers | Prochymosin sequence fused to A. niger GlaA signal peptide in A. nidulans | 146 µg/g dry weight chymosin compared to 93 µg/g dry weight in the control strain | 1.56 | [113] |
| Prochymosin sequence fused to codons for the GlaA signal peptide, propeptide, and 11 amino acids of mature glucoamylase in A. nidulans | 119 µg/g dry weight chymosin compared to 93 µg/g dry weight in the control strain | 1.27 | [113] | |
| Prochymosin sequence fused to GlaA signal peptide and propeptide in A. nidulans | 23 µg/g dry weight compared to 93 µg/g dry weight in the control strain | 0.24 | [113] | |
| Prochymosin sequence fused after the last codon of A. awamori GlaA in A. awamori | 140 µg/mL secreted chymosin compared to 8 µg/mL in the control strain (prochymosin + GlaA signal peptide) | 17.5 | [111,112] | |
| hlL6 fused to 1-514 nt of A. niger GlaA in A. niger | 15 mg/L hIL6 compared to less than 1 µg/L in the control strain (hlL6 fused to the GlaA signal peptide) | >15 | [114] | |
| E. coli uidA (13-glucuronidase) and the T. lanuginosa lipase fused to A. niger var. awamori endoxylanase II secretion signals | 798 Arbitrary units of glucuronidase activity/mg of total protein (the control did not carry uidA) | - | [120] | |
| 47.5 Arbitrary units of lipase activity compared to 47 Arbitrary units when the native lipase signal is used | 1 | |||
| ScFv-LYS encoding fragments fused to A. niger propeptide + GlaA (514nt) | 90 mg/L ScFv-LYS compared to 2–22 mg/L in the control strains (18 aa GlaA signal sequence + ScFv-LYS) | 4–45 | [115] | |
| Human antibodies (κ- and γ-chain of IgG1) fused to GlaA in A. niger | 0.9 g/L of trastuzumab IgG1 and 0.2 g/L of Hu1D10 | - | [116] | |
| Neoculin gene fused to α-amylase in A. oryzae | 1.3 mg/L of NCL | - | [40] | |
| Bovine chymosin gene fused to α-amylase in A. oryzae | 42 mg/L chymosin compared to 20 mg/L in strains with non-fused gene | 2.1 | [119] | |
| Hemicellulose degrading enzymes sequences fused to GlaA secretion peptide in A. nidulans strains | 50–100 mg/L xylanase B, xylanase C, xylosidase D, arabinofuranosidase B, ferulic acid esterase and arabinase | - | [117] |
| Process | Modification | Performance | Improvement Factor | Reference |
|---|---|---|---|---|
| Proteases | Deletion of pepA in A. awamori strains | Decreased extracellular proteolytic activity compared to the wild type (immunoassay using antibodies specific for PepA, but absolute values for PepA concentration were not determined) | - | [125] |
| Deletion of pepA in A. awamori | 430 mg/L of chymosin compared to 180 mg/L in the parental strain | 2.4 | [128] | |
| Deletion of pepA in A. niger (AB1.18) | 15–20% proteolytic activity compared to the parent strain AB4.1 | - | [126] | |
| Mutation on prtT (UV irradiation) in A. niger (AB1.13) | 1–2% proteolytic activity compared to the parent strain AB4.1 | - | [126] | |
| Deletion of prtR, pepA, cpI, tppA in A. oryzae | ΔprtR/pepA/cpI: 24.23 mg/L of Acremonium cellulolyticus cellobiohydrolase | 1.2 | [133] | |
| ΔprtR/pepA/tppA: 21.30 mg/L | 1.1 | |||
| ΔprtR/cpI/tppA: 22.08 mg/L | 1.1 | |||
| ΔprtR/pepA/cpI/tppA: 19.93 mg/L compared to 19.54 mg/L in the control strains | 1.02 | |||
| Deletion of alp and Npl in A. oryzae | 1041 U/g of Candida antarctica lipase B compared to 575 U/g in the parental strains | 1.8 | [132] | |
| Deletion of various proteases in A. niger | Δdpp4: 6% increase in Tramete laccase | 1.1 | [99] | |
| Δdpp5: 15.4% increase | 1.2 | |||
| ΔpepB: 8.6% increase | 1.1 | |||
| ΔpepD: 4.8% increase | 1.0 | |||
| ΔpepF: 5.3% increase | 1.1 | |||
| ΔpepAa: 0.5% increase | 1.1 | |||
| ΔpepAb: 13.4% increase | 1.1 | |||
| ΔpepAd: 2.7% increase | 1.0 | |||
| Δdpp4/dpp5: 26.6% increase | 1.3 | |||
| Disruption of tppA and pepE in A. oryzae strains | 25.4 mg/L of human lysozyme compared to 15 mg/L in the parental strains | 1.7 | [118] | |
| Disruption of tppA, pepE, nptB, dppIV and dppV in A. oryzae | 84.4 mg/L of chymosin compared to the 63.1 mg/L in the double protease gene disruptant (ΔtppA/pepE) | 1.3 | [130] | |
| Disruption of tppA, pepE, nptB, dppIV, and dppV, alpA, pepA, AopepAa, AopepAd and cpI in A. oryzae | 109.4 mg/L of chymosin and 35.8 mg/L of human lysozyme compared to the quintuple protease gene disruptant (ΔtppA/pepE/nptB/dppIV/dppV; 84.4 mg/L and 26.5 mg/L, respectively) | 1.3 and 1.35 | [131] | |
| Deletion of prtT in A. niger | 36.3–36.7 U/mL of mL G. cingulate cutinase compared to 21.2–20.4 U/mL in the parental strain | 1.7 | [127] | |
| Stability: Cutinase activity retained at 80% over the entire 14-day incubation period, while the parental lost more than 50% of their initial activities after six days of incubation and retained negligible activity after 14 days | - | |||
| Deletion of dppV and pepA in A. nidulans | P. sanguineus laccase activity 0.5 U/mL compared to 0.04 U/mL in the control strain | 12.5 | [51] | |
| Deletion of mnn9 and pepA in A. nidulans | P. sanguineus laccase activity 0.3 U/mL compared to 0.04 U/mL in the control strain | 7.5 | [51] |
| Process | Modification | Performance | Improvement Factor | Reference |
|---|---|---|---|---|
| Fungal morphology | Deletion of racA in A. niger | GlaA secreted into the culture medium is four times more compared to its parental strain, when ensuring continuous high-level expression of glaA. Quantification was done by dot blot analysis using a monoclonal antibody (Arbitrary units) | 4 | [102] |
| Overexpression of arfA in A. niger | Quantitative abundance of GlaA-dtomato reporter protein was 397.4 absolute fluorescence compared to 298.7 in control strain | 1.3 | [103] |
| Process | Modification | Performance | Improvement Factor | Reference |
|---|---|---|---|---|
| Fermentation conditions | Effect of growth medium and temperature on hen egg white lysozyme (HEWL) production in A. niger | 20–25 °C 8–10 mg/L HEWL while 30–37 °C 3–5 mg/L HEWL | Temperature: 2–2.6 | [141] |
| soluble starch: 8.0 mg/L HEWL | Carbon source: 1.7–2 | |||
| maltose: 4.5 mg/L HEWL | - | |||
| glucose: 4.0 mg/L HEWL | - | |||
| xylose: 0.2 mg/L HEWL | - | |||
| soy milk medium: 30–60 mg/L HEWL | Rich medium: 3.8–7.5 | |||
| Effect of organic nitrogen sources on recombinant glucoamylase production in A. niger | Unsupplemented: 44 mg glucoamylase/g biomass | - | [143] | |
| L-alanine: 32 mg glucoamylase/g biomass | 0.7 | |||
| L-methionine: 26 mg glucoamylase/g | 0.6 | |||
| casamino acids, yeast extract, peptone, and gelatin: 100 mg glucoamylase/g | 2.2 | |||
| Effect of agitation intensity on recombinant amyloglucosidase (AMG) production in A. oryzae | Titer at the end of the batch phase 525 rpm: 110 U/L AMG | - | [146] | |
| 675 rpm: 230 U/L AMG | 1.6 | |||
| 825 rpm: 370 U/L AMG | 3.3 | |||
| Effects of bioprocess parameters—agitation intensity, initial glucose concentration, initial yeast extract concentration, and dissolved oxygen tension (DO)—on heterologous protein production in A. oryzae | Highest GFP yields were achieved under these conditions: agitation 400 rpm, glucose 25 g/L, yeast extract 0 g/dm3, DO 15% | - | [142] | |
| Effect of agitation intensity on recombinant glucose oxidase production in A. niger | 200 rpm: 300 µkat/L of glucose oxidase | - | [144] | |
| 500 rpm: 800 µkat/L of glucose oxidase | 2.6 | |||
| 800 rpm: 600 µkat/L of glucose oxidase | 1.3 | |||
| Effect of temperature on Pleurotus eryngii versatile peroxidase production in A. nidulans and A. niger | -A. nidulans 31 °C: 24 U/L peroxidase activity | - | [145] | |
| 28 °C: 80 U/L peroxidase activity | 3.3 | |||
| 19 °C: 466 U/L peroxidase activity | 19.4 | |||
| -A. niger 28 °C: 107 U/L peroxidase activity | - | |||
| 19 °C: 412 U/L peroxidase activity | 3.8 | |||
| Fungal morphology | Effect of raising the viscosity of the medium by addition of polyvinylpyrrolidone-PVP (transition from aggregated mycelia (pellets) to dispersed mycelia) on hen egg white lysozyme (HEWL) in A. niger | Medium with no PVP: 110 mg/L fresh and 8 mg/g dry weight of HEWL | 1.7 | [147] |
| Medium with PVP: 190 mg/L fresh and 14 mg/g dry weight of HEWL | ||||
| Effect of addition of microparticles (linked to the formation of freely dispersed mycelium) on titers of native glucoamylase (GlaA) and recombinant fructofuranosidase (FF) produced in A. niger | No microparticles: 17 U/mL GlaA and 42 U/mL FF | 3.5 GlaA 2–3.8 FF | [148] | |
| Talc microparticles: 61 U/mL GlaA and 92 U/mL FF FF production can reach up to 160 U/mL (10 g/L talc microparticles of size 6 mm) | ||||
| Effect of addition of titanate microparticles (TiSiO4, 8 mm) on titers of native glucoamylase (GlaA) and recombinant fructofuranosidase (FF) produced in A. niger | No microparticles: 19 U/mL GlaA and 40 U/mL FF | 9.5 GlaA 3.7 FF | [149] | |
| Microparticles: 190 U/mL glucoamylase and 150 U/mL fructofuranosidase | ||||
| Effect of growth type on hen egg white lysozyme (HEWL) production and protease activity in A. niger | Free suspension: 5.8 mg/g HEWL 95.3 U/g Protease activity | 1.5 | [140] | |
| Mycelial pellets: 5.0 mg/g HEWL 58.6 U/g Protease activity | 1.2 | |||
| Celite-560-immobilized cultures: 4.1 mg/g HEWL 56.3 U/g Protease activity | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ntana, F.; Mortensen, U.H.; Sarazin, C.; Figge, R. Aspergillus: A Powerful Protein Production Platform. Catalysts 2020, 10, 1064. https://doi.org/10.3390/catal10091064
Ntana F, Mortensen UH, Sarazin C, Figge R. Aspergillus: A Powerful Protein Production Platform. Catalysts. 2020; 10(9):1064. https://doi.org/10.3390/catal10091064
Chicago/Turabian StyleNtana, Fani, Uffe Hasbro Mortensen, Catherine Sarazin, and Rainer Figge. 2020. "Aspergillus: A Powerful Protein Production Platform" Catalysts 10, no. 9: 1064. https://doi.org/10.3390/catal10091064
APA StyleNtana, F., Mortensen, U. H., Sarazin, C., & Figge, R. (2020). Aspergillus: A Powerful Protein Production Platform. Catalysts, 10(9), 1064. https://doi.org/10.3390/catal10091064






