Efficient 1-Hydroxy-2-Butanone Production from 1,2-Butanediol by Whole Cells of Engineered E. coli
Abstract
:1. Introduction
2. Results and Discussion
2.1. Screening of Enzymes for HB Production from 1,2-BD
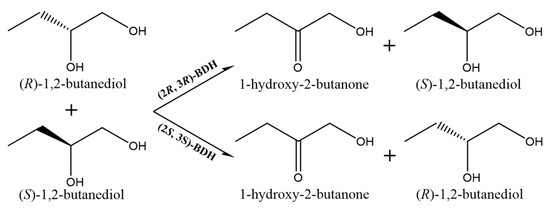
2.2. Analysis of Substrate Stereospecificity
2.3. Enzymatic Properties of (2R, 3R)-BDH and (2S, 3S)-BDH for 1,2-BD Mixture
2.4. Construction of Whole Cell Biocatalysts
2.5. HB Production from 1,2-BD Mixture by Whole-Cell Catalysis
2.6. Effects of VHB Expression on the Production of HB from 1,2-BD Mixture
3. Materials and Methods
3.1. Strains, Plasmids, Primers, and Chemicals
3.2. Recombinant Enzyme Expression and Purification
3.3. Enzyme Assays
3.4. Analysis of Substrate Stereospecificity
3.5. Construction of Whole Cell Biocatalysts
3.6. HB Production from 1,2-BD Mixture by Whole-Cell Catalysis
3.7. Effect of VHB Co-Expression on HB Production from 1,2-BD Mixture
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chen, F.F.; Cosgrove, S.C.; Birmingham, W.R.; Mangas-Sanchez, J.; Citoler, J.; Thompson, M.P.; Zheng, G.W.; Xu, J.H.; Turner, N.J. Enantioselective synthesis of chiral vicinal amino alcohols using amine dehydrogenases. ACS Catal. 2019, 9, 11813–11818. [Google Scholar] [CrossRef]
- Wang, H.Y.; Zhu, C.H.; Liu, Q.Y.; Tan, J.; Wang, C.G.; Liang, Z.; Ma, L.L. Selective Conversion of cellulose to hydroxyacetone and 1-hydroxy-2-butanone with Sn-Ni bimetallic catalysts. ChemSusChem 2019, 12, 2154–2160. [Google Scholar] [CrossRef]
- Sato, S.; Takahashi, R.; Sodesawa, T.; Fukuda, H.; Sekine, T.; Tsukuda, E. Synthesis of a-hydroxyketones from 1,2-diols over Cu-based catalyst. Catal. Commun. 2005, 6, 607–610. [Google Scholar] [CrossRef]
- Lee, L.G.; Whitesides, G.M. Preparation of optically active 1,2-diols and α-hydroxy ketones using glycerol dehydrogenase as catalyst: Limits to enzyme-catalyzed synthesis due to noncompetitive and mixed inhibition by product. J. Org. Chem. 1986, 51, 25–36. [Google Scholar] [CrossRef]
- Hu, N.J.; Li, S.Y.; Liu, Y.C. Recent advances in biocatalysis and metabolic engineering. Catalysts 2021, 11, 1052. [Google Scholar] [CrossRef]
- Hohn-Bentz, H.; Radler, F. Bacterial 2,3-butanediol dehydrogenases. Arch. Microbiol. 1983, 116, 197–203. [Google Scholar] [CrossRef]
- Xu, G.C.; Bian, Y.Q.; Han, R.Z.; Dong, J.J.; Ni, Y. Cloning, expression, and chararacterization of budC gene encoding meso-2,3-butanediol dehydrogenase from Bacillus licheniformis. Appl. Biochem. Biotechnol. 2016, 178, 604–717. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.W.; Zhao, X.H.; He, Y.Z.; Yang, T.X.; Gao, H.F.; Li, G.X.; Chen, F.X.; Sun, M.J.; Lee, J.K.; Zhang, L.Y. Efficient (3R)-acetoin production from meso-2,3-butanediol using a new whole-cell biocatalyst with co-expression of meso-2,3-butanediol dehydrogenase, NADH oxidase and Vitreoscilla hemoglobin. J. Microbiol. Biotechnol. 2017, 27, 92–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.Z.; Chen, F.X.; Sun, M.J.; Gao, H.F.; Guo, Z.W.; Lin, H.; Chen, J.B.; Jin, W.S.; Yang, Y.L.; Zhang, L.Y.; et al. Efficient (3S)-acetoin and (2S, 3S)-2,3-butanediol production from meso-2,3-butanediol using whole-cell biocatalysis. Molecules 2018, 23, 691. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.Y.; Singh, R.; Sivakumar, D.; Guo, Z.W.; Li, J.H.; Chen, F.B.; He, Y.Z.; Guan, X.; Kang, Y.C.; Lee, J.K. An artificial synthetic pathway for acetoin, 2,3-butanediol and 2-butanol production from ethanol using cell free multi-enzymes catalysis. Green Chem. 2018, 20, 230–242. [Google Scholar] [CrossRef]
- Cui, Z.Z.; Zhao, Y.J.; Mao, Y.F.; Shi, T.; Lu, L.X.; Ma, H.W.; Wang, Z.W.; Chen, T. In vitro biosynthesis of optically pure D-(-)-acetoin from meso-2,3-butanediol using 2,3-butanediol dehydrogenase and NADH oxidase. J. Chem. Technol. Biotechnol. 2019, 94, 2547–2554. [Google Scholar] [CrossRef]
- Subramanian, V.; Lunin, V.V.; Farmer, S.J.; Alahuhta, M.; Moore, K.T.; Ho, A.; Chaudhari, V.B.; Zhang, M.; Himmel, M.E.; Decker, S.R. Phylogenetics-based identification and characterization of a superior 2,3-butanediol dehydrogenase for Zymomonas mobilis expression. Biotechnol. Biofuels 2020, 13, 186. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Sun, J.B.; Bommareddy, R.R.; Song, L.F.; Zeng, A.P. Novel (2R, 3R)-2,3-butanediol dehydrogenase from potential industrial strain Paenibacillus polymyxa ATCC 12321. Appl. Environ. Microb. 2011, 77, 4230–4233. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.Y.; Xu, Q.M.; Zhan, S.R.; Li, Y.Y.; Lin, H.; Sun, S.J.; Sha, L.; Hu, K.H.; Guan, X.; Shen, Y.L. A new NAD(H)-dependent meso-2,3-butanediol dehydrogenase from an industrially potential strain Serratia marcescens H30. Appl. Microbiol. Biotechnol. 2014, 98, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wei, D.; Shi, J.P.; Wang, M.; Hao, J. Mechanism of 2,3-butanediol stereoisomer formation in Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 2014, 98, 4603–4613. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Guo, Z.W.; Chen, J.B.; Xu, Q.M.; Lin, H.; Hu, K.H.; Guan, X.; Shen, Y.L. Mechanism of 2,3-butanediol stereoisomers formation in a newly isolated Serratia sp. T241. Sci. Rep. 2016, 6, 19257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasaye-Anaya, L.; Vargas-Tah, A.; Martinez-Camara, C.; Castro-Montoya, A.J.; Campos-Garcia, J. Production of 2,3-butanediol by fermentation of enzymatic hydrolysed bagasse from agave mezcal-waste using the native Klebsiella oxytoca UM2-17 strain. J. Chem. Technol. Biotechnol. 2019, 94, 3915–3923. [Google Scholar] [CrossRef]
- Isobe, K.; Wakao, N. Thermostable NAD+-dependent (R)-specific secondary alcohol dehydrogenase from cholesterol-utilizing Burkholderia sp. AIU 652. J. Biosci. Bioeng. 2003, 96, 387–393. [Google Scholar] [CrossRef]
- Wang, Y.; Tao, F.; Xu, P. Glycerol dehydrogenase plays a dual role in glycerol metabolism and 2,3-butanediol formation in Klebsiella pneumoniae. J. Biol. Chem. 2014, 289, 6080–6090. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.Y.; Xu, Q.M.; Peng, X.Q.; Xu, B.H.; Wu, Y.H.; Yang, Y.L.; Sun, S.J.; Hu, K.H.; Shen, Y.L. Cloning, expression and characterization of glycerol dehydrogenase involved in 2,3-butanediol formation in Serratia marcescens H30. J. Ind. Microbiol. Biotechnol. 2014, 41, 1319–1327. [Google Scholar] [CrossRef]
- Marshall, J.H.; May, J.W.; Sloan, J. Purification and properties of glycerol: NAD+ 2-oxidoreductase (glycerol dehydrogenase) from Schizosaccharomyces pombe. Microbiology 1985, 131, 1581–1588. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.J.; Lee, C.C.; Liao, J.C. Enantioselective synthesis of pure (R,R)-2,3-butanediol in Escherichia coli with stereospecific secondary alcohol dehydrogenase. Org. Biomol. Chem. 2009, 7, 3914–3917. [Google Scholar] [CrossRef]
- Kopke, M.; Gerth, M.L.; Maddock, D.J.; Mueller, A.P.; Liew, F.; Simpson, S.D.; Patrick, W.M. Reconstruction of an acetogenic 2,3-butanediol pathway involving a novel NADPH-dependent primary-secondary alcohol dehydrogenase. Appl. Environ. Microb. 2014, 80, 3394–3403. [Google Scholar] [CrossRef] [Green Version]
- Maddock, D.J.; Patrick, W.M.; Gerth, M.L. Substitutions at the cofactor phosphate-binding site of a clostridial alcohol dehydrogenase lead to unexpected changes in substrate specificity. Protein. Eng. Des. Sel. 2015, 28, 251–258. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Z.J.; Lv, C.J.; Gao, C.; Qin, J.Y.; Ma, C.Q.; Liu, Z.; Liu, P.H.; Li, L.X.; Xu, P. A novel whole-cell biocatalyst with NAD+ regeneration for production of chiral chemicals. PLoS ONE 2010, 5, e8860. [Google Scholar] [CrossRef] [Green Version]
- Geueke, B.; Riebel, B.; Hummel, W. NADH oxidase from Lactobacillus brevis: A new catalyst for the regeneration of NAD. Enzyme Microb. Tech. 2003, 32, 205–211. [Google Scholar] [CrossRef]
- Takusagawa, Y.; Otagiri, M.; Ui, S.; Ohtsuki, T.; Mimura, A.; Ohkuma, M.; Kudo, T. Purification and characterization of L-2,3-butanediol dehydrogenase of Brevibacterium saccharolyticum C-1012 expression in Escherichia coli. Biosci. Biotechnol. Biochem. 2001, 65, 1876–1878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ui, S.; Otagiri, M.; Mimura, A.; Dohmae, N.; Takio, K.; Ohkuma, M.; Kudo, T. Cloning, expression and nucleotide sequence of the L-2,3-butanediol dehydrogenase gene from Brevibacterium saccharolyticum C-1012. J. Ferment. Bioeng. 1998, 86, 290–295. [Google Scholar] [CrossRef]






| Enzyme | Specific Activity (U/mg) |
|---|---|
| (2R, 3R)-BDH | 5.02 ± 0.31 |
| (2S, 3S)-BDH | 0.57 ± 0.07 |
| GDH | 2.28 ± 0.13 |
| meso-BDH | 0.41 ± 0.05 |
| CaADH | 0.23 ± 0.01 |
| CaADH:S199A | 0.90 ± 0.13 |
| CbADH | 1.43 ± 0.24 |
| Strains | (S)-1,2-BD (mM) | (R)-1,2-BD (mM) | Substrate Stereospecifity | HB (mM) |
|---|---|---|---|---|
| 1,2-BD mixture standard | 45.00 ± 1.17 | 55.00 ± 0.64 | ND | 0 |
| E. coli (pET28a) as control | 44.37 ± 2.34 | 54.13 ± 3.41 | ND | 1.32 ± 0.71 |
| E. coli (pET-gdh) | 45.71 ± 0.97 | 31.25 ± 1.35 | (R)-1,2-BD | 23.75 ± 0.72 |
| E. coli (pET-ssbdh) | 4.46 ± 1.44 | 51.37 ± 1.74 | (S)-1,2-BD | 40.54 ± 0.61 |
| E. coli (pET-mbdh) | 23.64 ±0.79 | 55.42 ± 2.11 | (S)-1,2-BD | 21.36 ± 0.92 |
| E. coli (pET-rrbdh) | 43.41 ± 1.31 | 2.61 ± 0.93 | (R)-1,2-BD | 51.09 ± 0.57 |
| E. coli (pET-Cbadh) | 32.34 ± 0.91 | 56.72 ± 0.85 | (S)-1,2-BD | 12.66 ± 0.64 |
| E. coli (pET-Caadh) | 36.23 ± 0.94 | 54.01 ± 0.69 | (S)-1,2-BD | 8.77 ± 0.78 |
| E. coli (pET-Caadh:S199A) | 34.42 ± 1.44 | 52.34 ± 1.34 | (S)-1,2-BD | 10.58 ± 0.83 |
| E. coli Strains | HB (mM) |
|---|---|
| E. coli (pET28a) | 2.08 ± 0.64 |
| E. coli (pET-ssbdh) | 76.78 ± 5.27 |
| E. coli (pET-ssbdh-nox) | 124.59 ± 7.53 |
| E. coli (pET-rrbdh) | 158.91 ± 7.41 |
| E. coli (pET-rrbdh-nox) | 202.98 ± 5.67 |
| Strains or Plasmids | Genotype or Properties | Source |
|---|---|---|
| Strains | - | - |
| Serratia sp. T241 | Wild type | Laboratory stock |
| E. coli DH5α | F−,φ80d/lacZΔM15, Δ(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17(rk-mk+), phoA, supE44, λ−, thi-1, gyrA96, relA1 | Tiangen Biotech |
| E. coli BL21(DE3) | F-, ompT, hsdSB(rB-mB-), gal(λ c I 857, ind1, Sam7, nin5, lacUV-T7 gene1), dcm(DE3) | Tiangen Biotech |
| Plasmids | - | - |
| pET28a | Kmr; expression vector | Laboratory stock |
| pET-nox | Kmr; nox in pET28a | Laboratory stock |
| pBR322-vgb | AMPr; pBR322 vector containing the vgb gene | Laboratory stock |
| pET-ssbdh | Kmr; ssbdh in pET28a | This study |
| pET-mbdh | Kmr; mbdh in pET28a | This study |
| pET-rrbdh | Kmr; rrbdh in pET28a | This study |
| pET-gdh | Kmr; gdh in pET28a | This study |
| pET-Cbadh | Kmr; Cbadh in pET28a | This study |
| pET-Caadh | Kmr; Caadh in pET28a | This study |
| pET-Caadh:S199A | Kmr; Caadh:S199A in pET28a | This study |
| pET-rrbdh-nox | Kmr; rrbdh and nox in pET28a | This study |
| pET-rrbdh-nox-vgb | Kmr; rrbdh, nox, and vgb in pET28a | This study |
| pET-ssbdh-nox | Kmr; ssbdh and nox in pET28a | This study |
| pET-ssbdh-nox-vgb | Kmr; ssbdh, nox, and vgb in pET28a | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, H.; Xu, J.; Sun, W.; Hu, W.; Gao, H.; Hu, K.; Qiu, J.; Huang, B.; Zhang, L. Efficient 1-Hydroxy-2-Butanone Production from 1,2-Butanediol by Whole Cells of Engineered E. coli. Catalysts 2021, 11, 1184. https://doi.org/10.3390/catal11101184
Lin H, Xu J, Sun W, Hu W, Gao H, Hu K, Qiu J, Huang B, Zhang L. Efficient 1-Hydroxy-2-Butanone Production from 1,2-Butanediol by Whole Cells of Engineered E. coli. Catalysts. 2021; 11(10):1184. https://doi.org/10.3390/catal11101184
Chicago/Turabian StyleLin, Hui, Jiayin Xu, Wenlian Sun, Wujia Hu, Huifang Gao, Kaihui Hu, Junzhi Qiu, Binbin Huang, and Liaoyuan Zhang. 2021. "Efficient 1-Hydroxy-2-Butanone Production from 1,2-Butanediol by Whole Cells of Engineered E. coli" Catalysts 11, no. 10: 1184. https://doi.org/10.3390/catal11101184
APA StyleLin, H., Xu, J., Sun, W., Hu, W., Gao, H., Hu, K., Qiu, J., Huang, B., & Zhang, L. (2021). Efficient 1-Hydroxy-2-Butanone Production from 1,2-Butanediol by Whole Cells of Engineered E. coli. Catalysts, 11(10), 1184. https://doi.org/10.3390/catal11101184