Cutinases: Characteristics and Insights in Industrial Production
Abstract
:1. Introduction
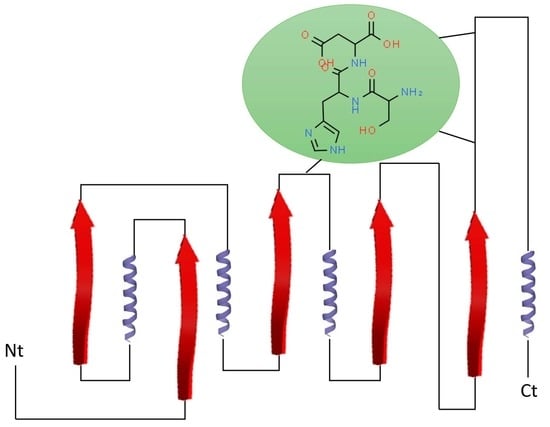
2. Classification and Structure
3. Evolutionary History
4. Biology
5. Use of Biocatalysts in Industrial Production
5.1. Immobilization
5.2. Modification of Cutinases
5.3. Industrial Applications of Cutinases
5.3.1. Textile Industry
Cotton Fibers
Synthetic Fibers
Polyester Fabrics
Polyamide Fabrics
Wool Fabrics
5.3.2. Detergents
5.3.3. Food Industry
Fruits and Vegetables
Aromas and Flavors
Plastics
6. Production of Cutinases
6.1. Purification of Cutinases
6.1.1. Purification by Ion Exchange Chromatography
6.1.2. Purification by Affinity Chromatography
6.1.3. Purification by Means of Two Aqueous Phase Systems
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| LGT | Lateral gene transfer |
| DOSS | Dioctyl sulfosuccinate sodium surfactant |
| DEAE | Diethylaminoethyl |
| MBTFP | 1,1,1-Trifluoro-3-((4-mercaptobutyl) thio) -2-propanone |
| TFK | 1,1,1-Trifluoro-2-propane |
| OTFP | 3-n-Octylthiio-1,1,1-trifluoro-2-propanone |
| PTFP | 3-n-Pentthio-1,1,1-trifluoro-2-propanone |
| SDS-PAGE | Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis |
| ATPS | Aqueous two phase systems |
| PET | Polyethylene386terephthalate |
| PA | Polyamide |
| PEG | Polyethylene glycol |
References
- Dutta, K.; Sen, S.; Veeranki, V.D. Production characterization and applications of microbial cutinases. Process. Biochem. 2009, 44, 127–134. [Google Scholar] [CrossRef]
- Dantzig, A.H.; Zuckerman, S.H.; Andonov-Roland, M.M. Isolation of a Fusarium solani mutant reduced in cutinase activity and virulence. J. Bacteriol. 1986, 2, 911–916. [Google Scholar] [CrossRef] [Green Version]
- Dickman, M.B.; Patil, S.S. Cutinase deficient mutants of Colletotrichum gloeosporioides are nonpathogenic to papaya fruit. Physiol. Mol. Plant Pathol. 1986, 28, 235–242. [Google Scholar] [CrossRef]
- Deising, H.; Nicholson, R.L.; Haug, M.; Howard, R.J.; Mendgen, K. Adhesion pad formation and the involvement of cutinase and esterases in the attachment of uredospores to the host cuticle. Plant Cell 1992, 4, 1101–1111. [Google Scholar] [CrossRef] [Green Version]
- Francis, S.A.; Dewey, F.M.; Gurr, S. The role of cutinase in germling development and infection by Erysiphe graminis f. sp. hordei. Physiol. Mol. Plant Pathol. 1996, 49, 201–211. [Google Scholar] [CrossRef]
- Skamnioti, P.; Gurr, S.J. Magnaporthe grisea cutinase mediates appressorium differentiation and host penetration and is required for full virulence. Plant Cell 2007, 19, 2674–2689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stahl, D.J.; Schafer, W. Cutinase is not required for fungal pathogenicity on pea. Plant Cell 1992, 4, 621–629. [Google Scholar] [PubMed] [Green Version]
- Yao, C.; Koller, W. Diversity of cutinases from plant pathogenic fungi: Different cutinases are expressed during saprophytic and pathogenic stages of Alternaria brassicicola. Mol. Plant Microbe Interact. 1995, 8, 122–130. [Google Scholar] [CrossRef]
- Chen, S.; Su, L.; Chen, J.; Wu, J. Cutinase: Characteristics preparation, and application. Biotechnol. Adv. 2013, 31, 1754–1767. [Google Scholar] [CrossRef]
- Egmond, M.R.; de Vlieg, J. Fusarium solani pisi cutinase. Biochimie 2000, 82, 1015–1021. [Google Scholar] [CrossRef]
- Chang, A.; Jeske, L.; Ulbrich, S.; Hofmann, J.; Koblitz, J.; Schomburg, I.; Neumann-Schaal, M.; Jahn, D.; Schomburg, D. BRENDA, the ELIXIR core data resource in 2021: New developments and updates. Nucleic Acids Res. 2020, 49, 498–508. [Google Scholar] [CrossRef]
- Roussel, A.; Amara, S.; Nyyssölä, A.; Mateos-Diaz, E.; Blangy, S.; Kontkanen, H.; Westerholm-Parvinen, A.; Carrière, F.; Cambillau, C. A cutinase from Trichoderma reesei with a lid-covered active site and kinetic properties of true lipases. J. Mol. Biol. 2014, 426, 3757–3772. [Google Scholar] [CrossRef]
- Longhi, S.; Czjzek, M.; Lamzin, V.; Nicolas, A.; Cambillau, C. Atomic resolution (1.0 A) crystal structure of Fusarium solani. Stereochemical analysis. J. Mol. Biol. 1997, 268, 779–799. [Google Scholar] [CrossRef] [PubMed]
- Bornscheuer, U.T. Microbial carboxyl esterases: Classification, properties and application in biocatalysis. Fems Microbiol. Rev. 2002, 26, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Nikolaivits, E.; Kanelli, M.; Dimarogona, M.; Topakas, E. A middle-aged enzyme still in its prime: Recent advances in the field of cutinases. Catalysts 2018, 8, 612. [Google Scholar] [CrossRef] [Green Version]
- Skamnioti, P.; Furlong, R.F.; Gurr, S.J. Evolutionary history of the ancient cutinase family in five filamentous Ascomycetes reveals differential gene duplications and losses and in Magnaporthe grisea shows evidence of sub-and neo-functionalization. New Phytol. 2008, 180, 711–721. [Google Scholar] [CrossRef]
- Belbahri, L.; Calmin, G.; Mauch, F.; Andersson, J.O. Evolution of the cutinase gene family: Evidence for lateral gene transfer of a candidate phytophthora virulence factor. Gene 2008, 408, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, K.; De Lorono, I.; Foster, S.; Li, D.; Johnstone, K.; Ashby, A. Evidence for a role of cutinase in pathogenicity of Pyrenopeziza brassicae on brassicas. Physiol. Mol. Plant Pathol. 2000, 57, 63–75. [Google Scholar] [CrossRef]
- Shaykh, M.; Soliday, C.; Kolattukudy, P.E. Proof for the production of cutinase byFusarium solani f. pisi during penetration into its host Pisum sativum. Plant Physiol. 1977, 60, 170–172. [Google Scholar] [CrossRef] [Green Version]
- Dickman, M.; Podila, G.; Kolattukudy, P. Insertion of cutinase gene into a wound pathogen enables it to infect intact host. Nature 1989, 342, 446–448. [Google Scholar] [CrossRef]
- Li, D.; Ashby, A.M.; Johnstone, K. Molecular evidence that the extracellular cutinase Pbc1 is required for pathogenicity of Pyrenopeziza brassicae on oilseed rape. Mol.-Plant-Microbe Interact. 2003, 16, 545–552. [Google Scholar] [CrossRef] [Green Version]
- Dickman, M.B.; Patil, S.S.; Kolattukudy, P. Purification, characterization and role in infection of an extracellular cutinolytic enzyme from Colletotrichum gloeosporioides penz on Carica papaya L. Physiol. Plant Pathol. 1982, 20, 333–347. [Google Scholar] [CrossRef]
- Stahl, D.J.; Theuerkauf, A.; Heitefuss, R.; Schafer, W. Cutinase of Nectria haematococca (Fusarium solani f. sp. pisi) is not required for fungal virulence or organ specificity on pea. Mol. Plant Microbe Interact. 1994, 7, 713–725. [Google Scholar] [CrossRef]
- Sweigard, J.A.; Chumley, F.G.; Valent, B. Disruption of a Magnaporthe grisea cutinase gene. Mol. Gen. Genet. 1992, 232, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Van Kan, J.; van’t Klooster, J.W.; Wagemakers, C.A.; Dees, D.C.; Van der Vlugt-Bergmans, C. Cutinase A of Botrytis cinerea is expressed, but not essential, during penetration of gerbera and tomato. Mol.-Plant-Microbe Interact. 1997, 10, 30–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köller, W.; Parker, D.; Becker, C. Role of cutinase in the penetration of apple leaves by Venturia inaequalis. Phytopathology 1991, 81, 1375–1379. [Google Scholar] [CrossRef]
- Liu, T.; Hou, J.; Wang, Y.; Jin, Y.; Borth, W.; Zhao, F.; Liu, Z.; Hu, J.; Zuo, Y. Genome-wide identification, classification and expression analysis in fungal-plant interactions of cutinase gene family and functional analysis of a putative ClCUT in Curvularia lunata. Mol. Genet. Genom. 2016, 7, 1–11. [Google Scholar] [CrossRef]
- Fan, C.; Köller, W. Diversity of cutinases from plant pathogenic fungi: Differential and sequential expression of cutinolytic esterases by Alternaria brassicicola. Fems Microbiol. Lett. 1998, 158, 33–38. [Google Scholar] [CrossRef]
- Arroyo, M.I.d.e. Fundamentos, métodos y aplicaciones. Ars Pharm. 1998, 39, 23–39. [Google Scholar]
- Su, A.; Shirke, A.; Baik, J.; Zou, Y.; Gross, R. Immobilized cutinases: Preparation, solvent tolerance and thermal stability. Enzym. Microb Technol. 2018, 116, 33–40. [Google Scholar] [CrossRef]
- Gonçalves, A.; Lopes, J.; Lemos, F.; Ribeiro, F.R.; Prazeres, D.; Cabral, J.; Aires-Barros, M.R. Effect of the immobilization support on the hydrolytic activity of a cutinase from Fusarium solani pisi. Enzym. Microb. Technol. 1997, 20, 93–101. [Google Scholar] [CrossRef]
- Serralha, F.; Lopes, J.; Lemos, F.; Prazeres, D.; Aires-Barros, M.; Cabral, J.; Aires-Barros, M.R. Zeolites as supports for an enzymatic alcoholysis reaction. J. Mol. Catal. B Enzym. 1998, 4, 303–311. [Google Scholar] [CrossRef]
- Serralha, F.; Lopes, J.; Aires-Barros, M.; Prazeres, D.; Cabral, J.; Lemos, F.; Ramôa Ribeiro, F. Stability of a recombinant cutinase immobilized on zeolites. Enzym. Microb. Technol. 2002, 31, 29–34. [Google Scholar] [CrossRef]
- Serralha, F.; Lopes, J.; Lemos, F.; Ribeiro, F.R.; Prazeres, D.; Aires-Barros, M.; Cabral, J.M.S. Application of factorial design to the study of an alcoholysis transformation promoted by cutinase immobilized on NaY zeolite and accurel PA6. J. Mol. Catal. B Enzym. 2004, 27, 19–27. [Google Scholar] [CrossRef]
- Bedade, D.K.; Muley, A.B.S.R.S. Magnetic cross-linked enzyme aggregates of acrylamidase from Cupriavidus oxalaticus ICTDB921 for biodegradation of acrylamide from industrial waste water. Bioresour. Technol. 2019, 272, 137–145. [Google Scholar] [CrossRef]
- Lucena, G.N.; dos Santos, C.; Pinto, G.; da Rocha, C.; Brandt, J.; de Paula, A.; Junior, M.; Marques, R. Magnetic cross-linked enzyme aggregates (MCLEAs) applied to biomass conversion. J. Solid State Chem. 2019, 10, 58–70. [Google Scholar] [CrossRef]
- Muley, A.B.; Thorat, A.S.; Singhal, R.S.; Babu, K.H. A tri-enzyme co-immobilized magnetic complex: Process details, kinetics, thermodynamics and applications. Int. J. Biol. Macromol. 2018, 118, 1781–1795. [Google Scholar] [CrossRef] [PubMed]
- Al-Tammar, A.; Omar, K.; Murad, O.; Munir, A.; Bakar, A.; Diba, A. Expression and and characterization of a cutinase (AnCUT2) from Aspergillus niger. Open Life Sci. 2016, 11, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Araujo, R.; Silva, C.; O’Neill, A.; Micaelo, N.; Guebitz, G.; Soares, C.M.; Casal, M.; Cavaco-Paulo, A. Tailoring cutinase activity towards polyethylene terephthalate and polyamide 6, 6 fibers. J. Biotechnol. 2007, 128, 849–857. [Google Scholar] [CrossRef] [Green Version]
- Brissos, V.; Eggert, T.; Cabral, J.M.; Jaeger, K.E. Improving activity and stability of cutinase towards the anionic detergent AOT by complete saturation mutagenesis. Protein Eng. Des. Sel. 2008, 21, 387–393. [Google Scholar] [CrossRef]
- Pio, T.; Macedo, G. Chapter 4 Cutinases: Properties and Industrial Applications. Adv. Appl. Microbiol. 2009, 66, 77–95. [Google Scholar] [CrossRef] [PubMed]
- Nyyssola, A. Which properties of cutinases are important for applications? Appl. Microbiol. Biotechnol. 2015, 99, 4931–4942. [Google Scholar] [CrossRef] [PubMed]
- Degani, O.; Gepstein, S.; Dosoretz, C.G. Potential use of cutinase in enzymatic scouring of cotton fiber cuticle. Appl. Biochem. Biotechnol. 2002, 102, 277–289. [Google Scholar] [CrossRef]
- Agrawal, P.; Agrawal, P.; Nierstrasz, V.; Bouwhuis, G.; Warmoeskerken, M. Biocatalysis and Biotransformation. Cutinase Pectinase Cotton Bioscouring Innov. Fast Bioscouring Process. 2008, 26, 412–421. [Google Scholar]
- Agrawal, P.; Nierstrasz, V.; Warmoeskerken, M. Role of mechanical action in low-temperature cotton scouring with F. solani pisicutinase and pectate lyase. Enzym. Microb. Technol. 2008, 42, 473–482. [Google Scholar] [CrossRef]
- Agrawal, P.; Nierstrasz, V.A.; Warmoeskerken, M. Ultrasound-boosted enzymatic cotton scouring process with cutinase and pectate lyase. Biocatal. Biotransform. 2010, 28, 320–328. [Google Scholar] [CrossRef]
- Vertommen, M.; Nierstrasz, V.; van der Veer, D.; Warmoeskerken, M.M.C.G. Enzymatic surface modification of poly (ethylene terephthalate). J. Biotechnol. 2005, 120, 376–386. [Google Scholar] [CrossRef]
- Kanelli, M.; Vasilakos, S.; Nikolaivits, E.; Ladas, S.; Christakopoulos, P.; Topakas, E. Surface modification of poly (ethylene terephthalate) (PET) fibers by a cutinase from Fusarium oxysporum. Process. Biochem. 2015, 50, 1885–1892. [Google Scholar] [CrossRef]
- Silva, C.M.; Carneiro, F.; O’Neill, A.; Fonseca, L.P.; Cabral, J.S.; Guebitz, G.; Cavaco-Paulo, A. Cutinase-A new tool for biomodification of synthetic fibers. J. Polym. Sci. Part Polym. Chem. 2005, 43, 2448–2450. [Google Scholar] [CrossRef] [Green Version]
- Ribitsch, D.; Herrero Acero, E.; Greimel, K.; Dellacher, A.; Zitzenbacher, S.; Marold, A.; Rodriguez, R.D.; Steinkellner, G.; Gruber, K.; Schwab, H.; et al. A new esterase from Thermobifida halotolerans hydrolyses polyethylene terephthalate (PET) and polylactic acid (PLA). Polymers 2012, 4, 617–629. [Google Scholar] [CrossRef]
- Oeser, T.; Wei, R.; Baumgarten, T.; Billig, S.; Föllner, C.; Zimmermann, W. High level expression of a hydrophobic poly (ethylene terephthalate)-hydrolyzing carboxylesterase fromThermobifida fusca KW3 in Escherichia coli BL21 (DE3). J. Biotechnol. 2010, 146, 100–104. [Google Scholar] [CrossRef]
- Ribitsch, D.; Herrero-Acero, E.; Prylucka, A.; Zitzenbacher, S.; Marold, A.; Gamerith, A.C.; Tscheließnig, R.; Jungbauer, A.; Rennhofer, H.; Lichtenegger, H.; et al. Enhanced cutinase-catalyzed hydrolysis of polyethylene terephthalate by covalent fusion to hydrophobins. Appl. Environ. Microbiol. 2015, 81, 3586–3592. [Google Scholar] [CrossRef] [Green Version]
- Donelli, I.; Taddei, P.; Smet, P.F.; Poelman, D.; Nierstrasz, V.A.; Freddi, G. Enzymatic surface modification and functionalization of PET: A water contact angle, FTIR, and fluorescence spectroscopy study. Biotechnol. Bioeng. 2009, 103, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.; Araujo, R.; Casal, M.; Gübitz, G.M.; Cavaco-Paulo, A. Influence of mechanical agitation on cutinases and protease activity towards polyamide substrates. Enzym. Microb. Technol. 2007, 40, 1678–1685. [Google Scholar] [CrossRef] [Green Version]
- García, S.; Olivera, N. Oportunidades de innovación biotecnológica en la industria textil lanera. In Proceedings of the VII Congreso De Ingeniería Industrial y Afines, Chubut, Argentina, 2014. [Google Scholar]
- Wang, P.; Wang, Q.; Fan, X.; Cui, L.; Yuan, J.; Chen, S.; Wu, J. Effects of cutinase on the enzymatic shrink-resist finishing of wool fabrics. Enzym. Microb. Technol. 2009, 44, 302–308. [Google Scholar] [CrossRef]
- Aehle, W. Enzymes in Household Detergents. In Enzymes in Industry: Products and Applications; Aehle, W., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2006; pp. 154–177. [Google Scholar]
- Silva, C.; Azoia, N.G.; Martins, M.; Matamá, T.; Wu, J.; Cavaco-Paulo, A. Molecular recognition of esterase plays a major role on the removal of fatty soils during detergency. J. Biotechnol. 2012, 161, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Flipsen, J.; Appel, A.; Van der Hijden, H.; Verrips, C. Mechanism of removal of immobilized triacylglycerol by lipolytic enzymes in a sequential laundry wash process. Enzym. Microb. Technol. 1998, 23, 274–280. [Google Scholar] [CrossRef]
- Poulose, A.J.; Boston, M. Enzyme Assisted Degradation of Surface Membranes of Harvested Fruits and Vegetables. U.S. Patent 5298265, 5 August 1991. [Google Scholar]
- De Barros, D.P.; Fonseca, L.P.; Fernandes, P.; Cabral, J.M.; Mojovic, L. Biosynthesis of ethyl caproate and other short ethyl esters catalyzed by cutinase in organic solvent. J. Mol. Catal. Enzym. 2009, 60, 178–185. [Google Scholar] [CrossRef]
- De Barros, D.P.; Azevedo, A.M.; Cabral, J.; Fonseca, L.P. Optimization of flavor esters synthesis by Fusarium solani pisi cutinase. J. Food Biochem. 2012, 36, 275–284. [Google Scholar] [CrossRef]
- Su, L.; Hong, R.; Guo, X.; Wu, J.; Xia, Y. Short-chain aliphatic ester synthesis using Thermobifida fusca cutinase. Food Chem. 2016, 206, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Regado, M.A.; Cristóvao, B.M.; Moutinho, C.G.; Balcao, V.M.; Aires-Barros, R.; Ferreira, J.P.M.; Malcata, F.X. Flavour development via lipolysis of milkfats: Changes in free fatty acid pool. Int. J. Food Sci. Technol. 2007, 42, 961–968. [Google Scholar] [CrossRef]
- Horii, K.; Adachi, T.; Tanino, T.; Tanaka, T.; Kotaka, A.; Sahara, H.; Hashimoto, T.; Kuratani, N.; Shibasaki, S.; Ogino, C.; et al. Fatty acid production from butter using novel cutinase-displaying yeast. Enzym. Microb. Technol. 2010, 46, 194–199. [Google Scholar] [CrossRef]
- Xu, Z. Research Progress on bacterial cutinases for plastic pollution. IOP Conf. Ser. Earth Environ. Sci. 2020, 450, 012077. [Google Scholar] [CrossRef]
- Gomes, D.S.; Matamá, T.; Cavaco-Paulo, A.; Campos-Takaki, G.M.; Salgueiro, A.A. Production of heterologous cutinases by E. coli and improved enzyme formulation for application on plastic degradation. Electron. J. Biotechnol. 2013, 16, 3. [Google Scholar] [CrossRef]
- Fett, W.; Wijey, C.; Moreau, R.; Osman, S. Production of cutinase by Thermomonospora fusca ATCC 27730. J. Appl. Microbiol. 1999, 86, 561–568. [Google Scholar] [CrossRef] [Green Version]
- Fett, W.; Wijey, C.; Moreau, R.; Osman, S. Production of cutinolytic esterase by filamentous bacteria. Lett. Appl. Microbiol. 2000, 31, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.S.; Kolattukudy, P.E. Structural studies on cutinase, a glycoprotein containing novel amino acids and glucuronic acid amide at the N terminus. Eur. J. Biochem. 1980, 106, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Fett, W.F.; Gerard, H.C.; Moreau, R.A.; Osman, S.F.; Jones, L.E. Cutinase production by Streptomyces spp. Curr. Microbiol. 1992, 25, 165–171. [Google Scholar] [CrossRef]
- Sebastian, J.; Kolattukudy, P.E. Purification and and characterization of cutinase from a fluorescent Pseudomonas putida bacterial strain isolated from phyllosphere. Arch Biochem. Biophys. 1988, 263, 77–85. [Google Scholar] [CrossRef]
- Kim, Y.H.; Lee, J.; Moon, S.H. Uniqueness of microbial cutinases in hydrolysis of p-nitrophenyl esters. J. Microbiol. Biotechnol. 2003, 13, 57–63. [Google Scholar]
- Hijden, V.; Htjd, M.; Warr, J.F.; Klugkist, J.; Musters, W.; DHA, H. Enzyme-Containing Surfactants Compositions. Unilever Patent WO943578.94.2, 1994. [Google Scholar]
- Chen, Z.; Franco, C.F.; Baptista, R.P.; Cabral, J.M.; Coelho, A.V.; Rodrigues, C.J.; Melo, E.P. Purification and identification of cutinases from Colletotrichum kahawae and Colletotrichum gloeosporioides. Appl. Microbiol. Biotechnol. 2007, 73, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Michailides, T.J.; Hammock, B.D.; Lee, Y.; Bostock, R.M. Affinity purification and characterization of a cutinase from the fungal plant pathogen Monilinia fructicola (Wint.) Honey. Arch. Biochem. Biophys. 2000, 382, 31–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villalba, M.; Verdasco-Martín, C.M.; dos Santos, J.C.; Fernandez-Lafuente, R.; Otero, C. Operational stabilities of different chemical derivatives of Novozym 435 in an alcoholysis reaction. Enzym. Microb. Technol. 2016, 90, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Feder, D.; Gross, R. Exploring chain length selectivity in HIC-catalyzed polycondensation reactions. Biomacromolecules 2010, 3, 690–697. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lee, J.; Seung-Hyeon, M. Increasing Pharmacological Effect of Agricultural Chemicals. U.S. Patent 88-08945, 1988. [Google Scholar]
- Kolattukudy, P.; Poulose, A.J. Cutinase for Use in Detergent Composition Produced by Culturing Pseudomonas Putida. U.S. Patent 89-02922, 1989. [Google Scholar]
- Voet, D.P.; Voet, J.G.; Pratt, C.W. Purificación y análisis de proteínas. In Fundamentos de Bioquímica: La Vida a Nivel Molecular; Editorial Médica Panamericana: Madrid, Spain, 2007; pp. 97–107. [Google Scholar]
- Almeida, M.; Venancio, A.; Teixeira, J.; Aires-Barros, M. Cutinase purification on poly (ethylene glycol)-hydroxypropyl starch aqueous two-phase systems. J. Chromatogr. Biomed. Sci. Appl. 1998, 711, 151–159. [Google Scholar] [CrossRef] [Green Version]
- Rito-Palomares, M. Practical application of aqueous two-phase partition to process development for the recovery of biological products. J. Chromatogr. B 2004, 807, 3–11. [Google Scholar] [CrossRef]






| Zeolite | Enzymatic Activity (U mg) | Amount of Water (%) | Water Activity |
|---|---|---|---|
| CaA | 0 | 0.25 | 0.48 |
| 9.10 | 0.46 | 0.95 | |
| 9.90 | 0.75 | 0.95 | |
| 7.10 | 0.77 | ≈1 | |
| 1.52 | 1.50 | ≈1 | |
| LiA | 0 | 0.28 | 0.40 |
| 11.8 | 0.44 | 0.97 | |
| 5.90 | 0.71 | ≈1 | |
| 0.99 | 1.30 | ≈1 | |
| 1.00 | 1.50 | ≈1 | |
| NaA | 3.3 | 0.21 | 0.68 |
| 5.1 | 0.27 | 0.77 | |
| 8.7 | 0.32 | 0.90 | |
| 6.9 | 0.51 | 0.96 | |
| 4.3 | 0.64 | ≈1 | |
| 1.1 | 1.00 | ≈1 | |
| 1.4 | 1.03 | ≈1 | |
| KA | 6.0 | 0.07 | 0.68 |
| 6.5 | 0.12 | 0.70 | |
| 7.3 | 0.38 | 0.96 | |
| 7.0 | 0.52 | 0.97 | |
| 1.7 | 1.30 | ≈1 | |
| CsA | 0 | 0.13 | 0.35 |
| 9.8 | 0.32 | 0.96 | |
| 9.7 | 0.37 | 0.97 | |
| 8.6 | 0.38 | ≈1 | |
| 7.6 | 0.41 | ≈1 | |
| 7.0 | 0.54 | ≈1 | |
| 1.5 | 1.50 | ≈1 |
| Length of the Acid Chain (Carbon Atoms) | Initial Velocity (mol min mg) | Time Delay at the Start of the Reaction (Lag) (min) |
|---|---|---|
| 2 | 0.30 | 0 |
| 4 | 1.15 | 0 |
| 5 | 1.06 | 0 |
| 6 | 0.41 | 0 |
| 8 | 0.48 | 10 |
| 10 | 0.44 | 20 |
| 18 | 0.24 | 40 |
| Origin of Cutinase | Cow’s Milk Fat | Cow’s Milk Fat | Goat’s Milk Fat |
|---|---|---|---|
| Candida cylindracea | 14:26:60 | 14:27:59 | 17:28:55 |
| Pseudomonas fluorescens | 11:22:67 | 11:25:64 | 08:23:69 |
| Fusarium solani pisii | 34:36:30 | 34:37:29 | 33:37:30 |
| Humicola lanuginosa | 18:31:51 | 20:34:46 | 14:29:57 |
| Rhizopus delemar | 13:23:64 | 13:27:60 | 15:25:60 |
| Penicillium camembertii | 24:32:44 | 17:38:45 | 31:38:31 |
| Geotrichum candidum | 17:24:59 | 18:26:56 | 11:14:75 |
| Candida lipolytica | 15:24:61 | 14:25:61 | 17:28:55 |
| Mucor circinelloides | 17:27:56 | 16:28:56 | 17:25:58 |
| Rhizopus niveus | 15:26:59 | 13:26:61 | 12:24:64 |
| Penicillium roquefortii | 25:34:41 | 25:34:41 | 29:39:32 |
| Origin | Genus | Species | Ref. |
|---|---|---|---|
| Bacteria | Streptomyces | scabies | [70] |
| acidiscabies | [71] | ||
| badius | [71] | ||
| Pseudomonas | putida | [72] | |
| mendocina | [73] | ||
| aeruginosa | [71] | ||
| Thermomonospora | fusca | [68] | |
| Thermoactinomyces | vulgaris | [69] | |
| Botrytis | cinerea | [74] | |
| Fungi | Fusarium | solani pisi | [10] |
| oxysporium | [48] | ||
| roseum culmorum | [48] | ||
| roseum sambucinum | [48] | ||
| Colletotrichum | kahawae | [75] | |
| gloeosporioides | [75] | ||
| capsici | [75] | ||
| Monilinia | fructicola | [76] | |
| Venturia | inaequalis | [26] | |
| Alternaria | brassicicola | [8] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez, A.; Maicas, S. Cutinases: Characteristics and Insights in Industrial Production. Catalysts 2021, 11, 1194. https://doi.org/10.3390/catal11101194
Martínez A, Maicas S. Cutinases: Characteristics and Insights in Industrial Production. Catalysts. 2021; 11(10):1194. https://doi.org/10.3390/catal11101194
Chicago/Turabian StyleMartínez, Alejandro, and Sergi Maicas. 2021. "Cutinases: Characteristics and Insights in Industrial Production" Catalysts 11, no. 10: 1194. https://doi.org/10.3390/catal11101194
APA StyleMartínez, A., & Maicas, S. (2021). Cutinases: Characteristics and Insights in Industrial Production. Catalysts, 11(10), 1194. https://doi.org/10.3390/catal11101194