Application Potential of Cyanide Hydratase from Exidia glandulosa: Free Cyanide Removal from Simulated Industrial Effluents
Abstract
1. Introduction
2. Results
2.1. Enzyme Preparation and Properties
2.1.1. Enzyme Overproduction
2.1.2. Enzyme Purification and Activity
2.1.3. Effect of pH and Temperature
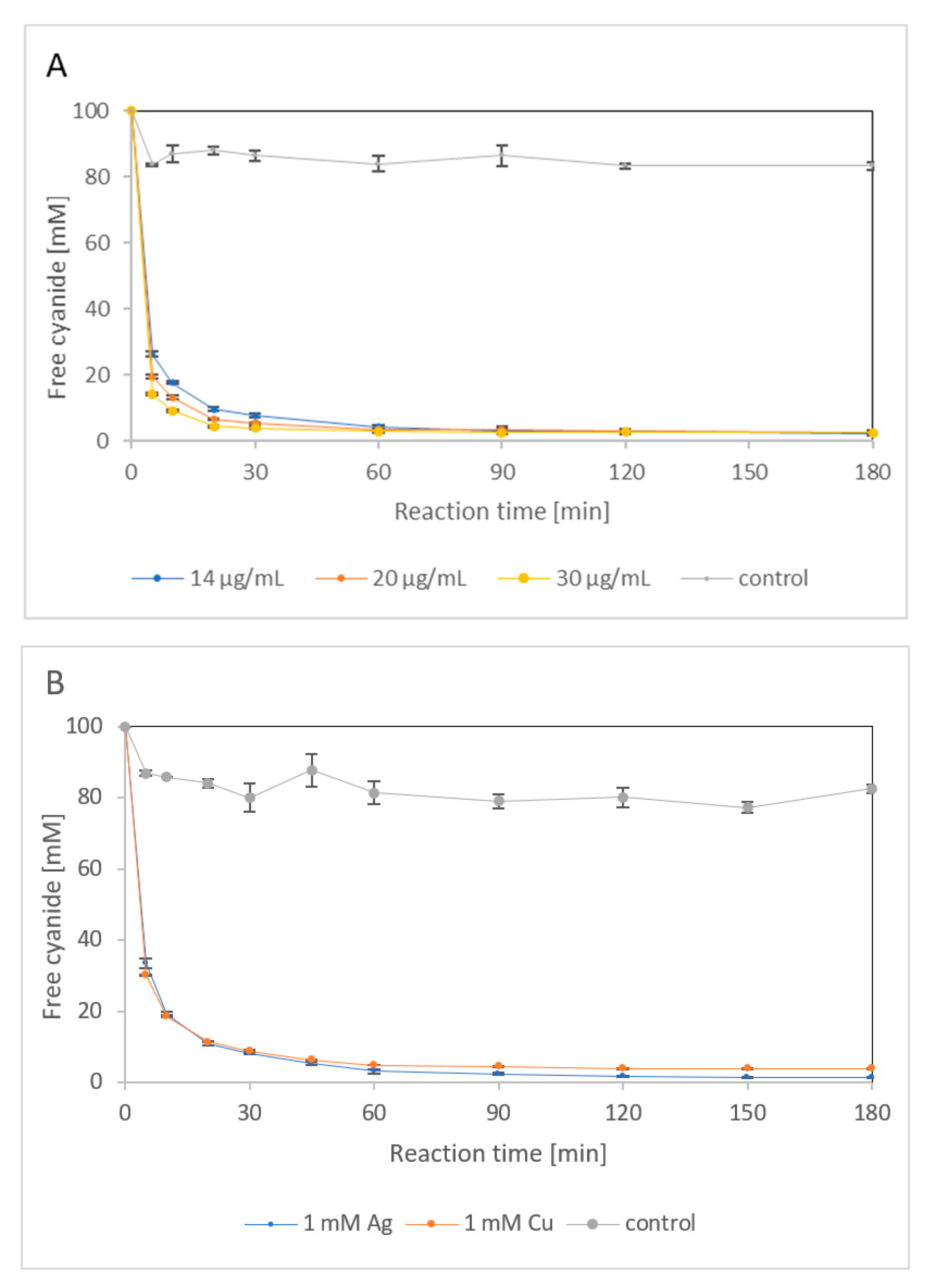
2.1.4. Effect of Silver and Copper
2.1.5. Temperature stability and shelf life
2.2. Enzyme Performance on Model Mixtures
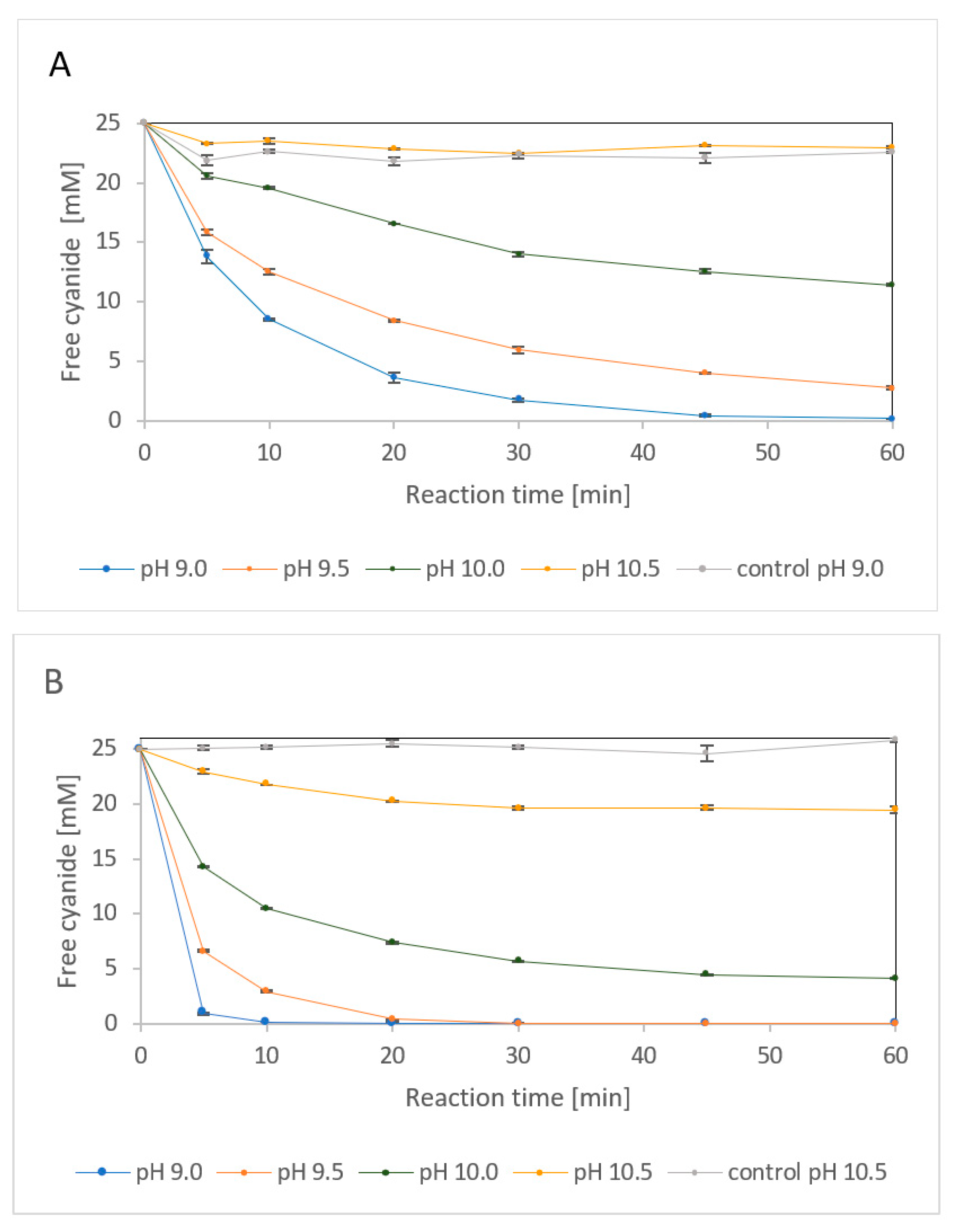
2.2.1. Effect of Alkaline Media
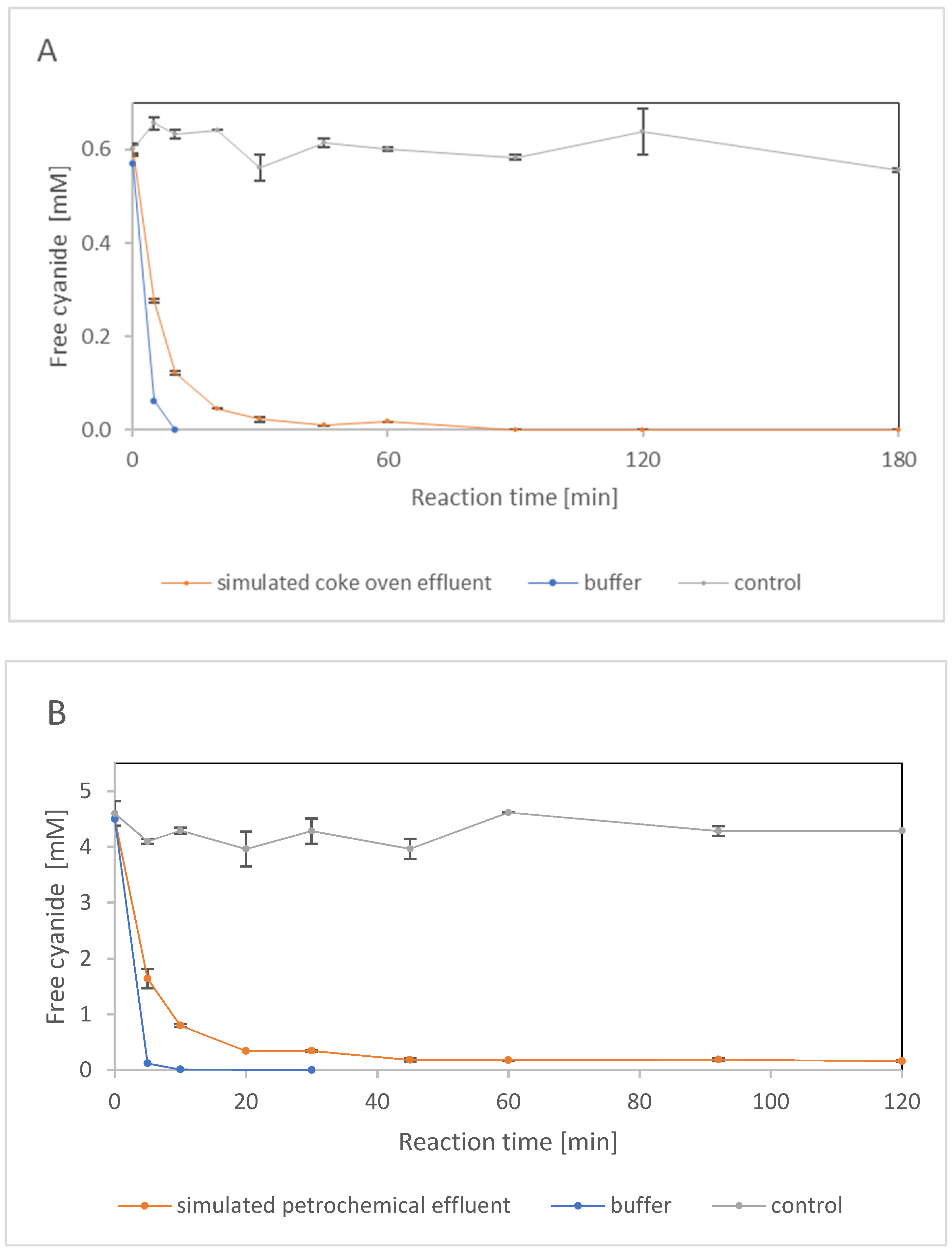
2.2.2. Enzyme Performance in the Presence of Phenol and Inorganic Salts
2.2.3. Enzyme Performance in the Presence of Heavy Metal Salts
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Strain
4.3. Enzyme Induction and Purification
4.4. Enzyme Assays
4.5. General Protocol for the Biocatalyzed Degradation of fCN
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kuyucak, N.; Akcil, A. Cyanide and removal options from effluents in gold mining and metallurgical processes. Miner. Eng. 2013, 50–51, 13–29. [Google Scholar] [CrossRef]
- Carmona-Orozco, M.L.; Panay, A.J. Immobilization of E. coli expressing Bacillus pumilus CynD in three organic polymer matrices. Appl. Microbiol. Biotechnol. 2019, 103, 5401–5410. [Google Scholar] [CrossRef]
- Basile, L.J.; Willson, R.C.; Sewell, B.T.; Benedik, M.J. Genome mining of cyanide-degrading nitrilases from filamentous fungi. Appl. Microbiol. Biotechnol. 2008, 80, 427–435. [Google Scholar] [CrossRef]
- Martínková, L.; Chmátal, M. The integration of cyanide hydratase and tyrosinase catalysts enables effective degradation of cyanide and phenol in coking wastewaters. Water Res. 2016, 102, 90–95. [Google Scholar] [CrossRef]
- Jarrah, N.; Mu’azu, N.D. Simultaneous electro-oxidation of phenol, CN−, S2− and NH4+ in synthetic wastewater using boron doped diamond anode. J. Environ. Chem. Eng. 2016, 4, 2656–2664. [Google Scholar] [CrossRef]
- Ibáñez, M.I.; Cabello, P.; Luque-Almagro, V.M.; Sáez, L.P.; Olaya, A.; Sánchez de Medina, V.; Luque de Castro, M.D.; Moreno-Vivián, C.; Roldán, M.D. Quantitative proteomic analysis of Pseudomonas pseudoalcaligenes CECT5344 in response to industrial cyanide-containing wastewaters using Liquid Chromatography-Mass Spectrometry/Mass Spectrometry (LC-MS/MS). PLoS ONE 2017, 12, e0172908. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.L.; Liu, G.S.; Chen, Y.H.; Miao, D.T.; Wei, Q.P.; Li, H.C.; Ma, L.; Zhou, K.C.; Liu, L.B.; Yu, Z.M. Persulfate enhanced electrochemical oxidation of highly toxic cyanide-containing organic wastewater using boron-doped diamond anode. Chemosphere 2020, 252, 126499. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou, C.A.; Samaras, P.; Sakellaropoulos, G.P. Comparative study of phenol and cyanide containing wastewater in CSTR and SBR activated sludge reactors. Bioresour. Technol. 2009, 100, 31–37. [Google Scholar] [CrossRef]
- Yu, X.B.; Xu, R.H.; Wei, C.H.; Wu, H.Z. Removal of cyanide compounds from coking wastewater by ferrous sulfate: Improvement of biodegradability. J. Hazard. Mater. 2016, 302, 468–474. [Google Scholar] [CrossRef]
- Makhado, E.; Pandey, S.; Nomngongo, P.N.; Ramontja, J. Preparation and characterization of xanthan gum-cl-poly(acrylic acid)/o-MWCNTs hydrogel nanocomposite as highly effective reusable adsorbent for removal of methylene blue from aqueous solutions. J. Colloid Interface Sci. 2018, 513, 700–714. [Google Scholar] [CrossRef]
- Pandey, S.; Do, J.Y.; Kim, J.; Kang, M. Fast and highly efficient catalytic degradation of dyes using κ-carrageenan stabilized silver nanoparticles nanocatalyst. Carbohydr. Polym. 2020, 230, 115597. [Google Scholar] [CrossRef]
- Electroplating and metal finishing point source categories; Effluent limitations guidelines, pretreatment, standards and new source performance standards. Fed. Regist. 1983, 48, 32487. Available online: https://www.epa.gov/sites/default/files/2015-10/documents/electroplating-and-metal-finishing_proposed-rule_08-31-1982_47-fr-38462.pdf (accessed on 12 November 2021).
- Code of Federal Regulations. 40, Subpart A-Cokemaking Subcategory. Available online: https://www.ecfr.gov/current/title-40/chapter-I/subchapter-N/part-420/subpart-A (accessed on 12 November 2021).
- Cosmos, A.; Erdenekhuyag, B.O.; Yao, G.; Li, H.J.; Zhao, J.G.; Wang, L.J.; Lyu, X.J. Principles and methods of bio detoxification of cyanide contaminants. J Mater. Cycles Waste Manag. 2020, 22, 939–954. [Google Scholar] [CrossRef]
- Rahdar, S.; Rahdar, A.; Sattari, M.; Hafshejani, L.D.; Tolkou, A.K.; Kyzas, G.Z. Barium/cobalt@polyethylene glycol nanocomposites for dye removal from aqueous solutions. Polymers 2021, 13, 1161. [Google Scholar] [CrossRef]
- Rahdar, S.; Pal, K.; Mohammadi, L.; Rahdar, A.; Goharniya, Y.; Samani, S.; Kyzas, G.Z. Response surface methodology for the removal of nitrate ions by adsorption onto copper oxide nanoparticles. J. Mol. Struct. 2021, 1231, 129686. [Google Scholar] [CrossRef]
- Baeissa, E.S.; Mohamed, R.M. Enhancement of photocatalytic properties of Ga2O3-SiO2 nanoparticles by Pt deposition. Chin. J. Catal. 2013, 34, 1167–1172. [Google Scholar] [CrossRef]
- Bagabas, A.; Alshammari, A.; Aboud, M.F.A.; Kosslick, H. Room-temperature synthesis of zinc oxide nanoparticles in different media and their application in cyanide photodegradation. Nanoscale Res. Lett. 2013, 8, 516. [Google Scholar] [CrossRef]
- Fan, L.; Yao, H.; Deng, S.; Jia, F.; Cai, W.; Hu, Z.; Guo, J.; Li, H. Performance and microbial community dynamics relationship within a step-feed anoxic/oxic/anoxic/oxic process (SF-A/O/A/O) for coking wastewater treatment. Sci. Total Environ. 2021, 792, 148263. [Google Scholar] [CrossRef]
- Martínková, L.; Veselá, A.B.; Rinágelová, A.; Chmátal, M. Cyanide hydratases and cyanide dihydratases: Emerging tools in the biodegradation and biodetection of cyanide. Appl. Microbiol. Biotechnol. 2015, 99, 8875–8882. [Google Scholar] [CrossRef]
- Fry, W.E.; Millar, R.L. Cyanide degradation by an enzyme from Stemphylium loti. Arch. Biochem. Biophys. 1972, 151, 468–474. [Google Scholar] [CrossRef]
- Wang, P.; VanEtten, H.D. Cloning and properties of a cyanide hydratase gene from the phytopathogenic fungus Gloeocercospora sorghi. Biochem. Biophys. Res. Commun. 1992, 187, 1048–1054. [Google Scholar] [CrossRef]
- Cluness, M.J.; Turner, P.D.; Clements, E.; Brown, D.T.; O’Reilly, C. Purification and properties of cyanide hydratase from Fusarium lateritium and analysis of the corresponding chy1 gene. J. Gen. Microbiol. 1993, 139, 1807–1815. [Google Scholar] [CrossRef][Green Version]
- Barclay, M.; Tett, V.A.; Knowles, C.J. Metabolism and enzymology of cyanide/metallocyanide biodegradation by Fusarium solani under neutral and acidic conditions. Enzym. Microb. Technol. 1998, 23, 321–330. [Google Scholar] [CrossRef]
- Yanase, H.; Sakamoto, A.; Okamoto, K.; Kita, K.; Sato, Y. Degradation of the metal-cyano complex tetracyanonickelate (II) by Fusarium oxysporum N-10. Appl. Microbiol. Biotechnol. 2000, 53, 328–334. [Google Scholar] [CrossRef]
- Kushwaha, M.; Kumar, V.; Mahajan, R.; Bhalla, T.C.; Chatterjee, S.; Akhter, Y. Molecular insights into the activity and mechanism of cyanide hydratase enzyme associated with cyanide biodegradation by Serratia marcescens. Arch. Microbiol. 2018, 200, 971–977. [Google Scholar] [CrossRef]
- Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov (accessed on 12 November 2021).
- Park, J.M.; Sewell, B.T.; Benedik, M.J. Cyanide bioremediation: The potential of engineered nitrilases. Appl. Microbiol. Biotechnol. 2017, 101, 3029–3042. [Google Scholar] [CrossRef]
- Nolan, L.M.; Harnedy, P.A.; Turner, P.; Hearne, A.B.; O’Reilly, C. The cyanide hydratase enzyme of Fusarium lateritium also has nitrilase activity. FEMS Microbiol. Lett. 2003, 221, 161–165. [Google Scholar] [CrossRef]
- Kaplan, O.; Veselá, A.B.; Petříčková, A.; Pasquarelli, F.; Pičmanová, M.; Rinágelová, A.; Bhalla, T.C.; Pátek, M.; Martínková, L. A comparative study of nitrilases identified by genome mining. Mol. Biotechnol. 2013, 54, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Veselá, A.B.; Rucká, L.; Kaplan, O.; Pelantová, H.; Nešvera, J.; Pátek, M.; Martínková, L. Bringing nitrilase sequences from databases to life: The search for novel substrate specificities with a focus on dinitriles. Appl. Microbiol. Biotechnol. 2016, 100, 2193–2202. [Google Scholar] [CrossRef]
- Rucká, L.; Chmátal, M.; Kulik, N.; Petrásková, L.; Pelantová, H.; Novotný, P.; Příhodová, R.; Pátek, M.; Martínková, L. Genetic and functional diversity of nitrilases in Agaricomycotina. Int. J. Mol. Sci. 2019, 20, 5990. [Google Scholar] [CrossRef]
- Rinágelová, A.; Kaplan, O.; Veselá, A.B.; Chmátal, M.; Křenková, A.; Plíhal, O.; Pasquarelli, F.; Cantarella, M.; Martínková, L. Cyanide hydratase from Aspergillus niger K10: Overproduction in Escherichia coli, purification, characterization and use in continuous cyanide degradation. Proc. Biochem. 2014, 49, 445–450. [Google Scholar] [CrossRef]
- Malmir, N.; Fard, N.A.; Mgwatyu, Y.; Mekuto, L. Cyanide hydratase modification using computational design and docking analysis for improved binding affinity in cyanide detoxification. Molecules 2021, 26, 1799. [Google Scholar] [CrossRef]
- Rucká, L.; Kulik, N.; Novotný, P.; Sedova, A.; Petrásková, L.; Příhodová, R.; Křístková, B.; Halada, P.; Pátek, M.; Martínková, L. Plant nitrilase homologues in fungi: Phylogenetic and functional analysis with focus on nitrilases in Trametes versicolor and Agaricus bisporus. Molecules 2020, 25, 3861. [Google Scholar] [CrossRef]
- Fisher, F.B.; Brown, J.S. Colorimetric determination of cyanide in stack gas and waste water. Anal. Chem. 1952, 24, 1440–1444. [Google Scholar] [CrossRef]
- Pérez-Cid, B.; Calvar, S.; Belén Moldes, A.; Cruz, J.M. Effective removal of cyanide and heavy metals from an industrial electroplating stream using calcium alginate hydrogels. Molecules 2020, 25, 5183. [Google Scholar] [CrossRef]
- Xue, Y.P.; Wang, Y.P.; Xu, Z.; Liu, Z.Q.; Shu, X.R.; Jia, D.X.; Zheng, Y.G.; Shen, Y.C. Chemoenzymatic synthesis of gabapentin by combining nitrilase-mediated hydrolysis with hydrogenation over Raney-nickel. Catal. Commun. 2015, 66, 121–125. [Google Scholar] [CrossRef]
- Wang, L.; Watermeyer, J.M.; Mulelu, A.E.; Sewell, B.T.; Benedik, M.J. Engineering pH-tolerant mutants of a cyanide dihydratase. Appl. Microbiol. Biotechnol. 2012, 94, 131–140. [Google Scholar] [CrossRef]
- Crum, M.A.; Park, J.M.; Mulelu, A.E.; Sewell, B.T.; Benedik, M.J. Probing C-terminal interactions of the Pseudomonas stutzeri cyanide-degrading CynD protein. Appl. Microbiol. Biotechnol. 2015, 99, 3093–3102. [Google Scholar] [CrossRef]
- Jandhyala, D.M.; Willson, R.C.; Sewell, B.T.; Benedik, M.J. Comparison of cyanide-degrading nitrilases. Appl. Microbiol. Biotechnol. 2005, 68, 327–335. [Google Scholar] [CrossRef] [PubMed]
- BRENDA. The Comprehensive Enzyme Information System. Available online: https://brenda-enzymes.org (accessed on 2 November 2021).
- Dennett, G.V.; Blamey, J.M. A new thermophilic nitrilase from an antarctic hyperthermophilic microorganism. Front. Bioeng. Biotechnol. 2016, 4, 5. [Google Scholar] [CrossRef]
- Mondal, A.; Sarkar, S.; Nair, U.G. Comparative characterization of cyanide-containing steel industrial wastewater. Water Sci. Technol. 2021, 83, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Ketterer, L.; Keusgen, M. Amperometric sensor for cyanide utilizing cyanidase and formate dehydrogenase. Anal. Chim. Acta 2010, 673, 54–59. [Google Scholar] [CrossRef] [PubMed]
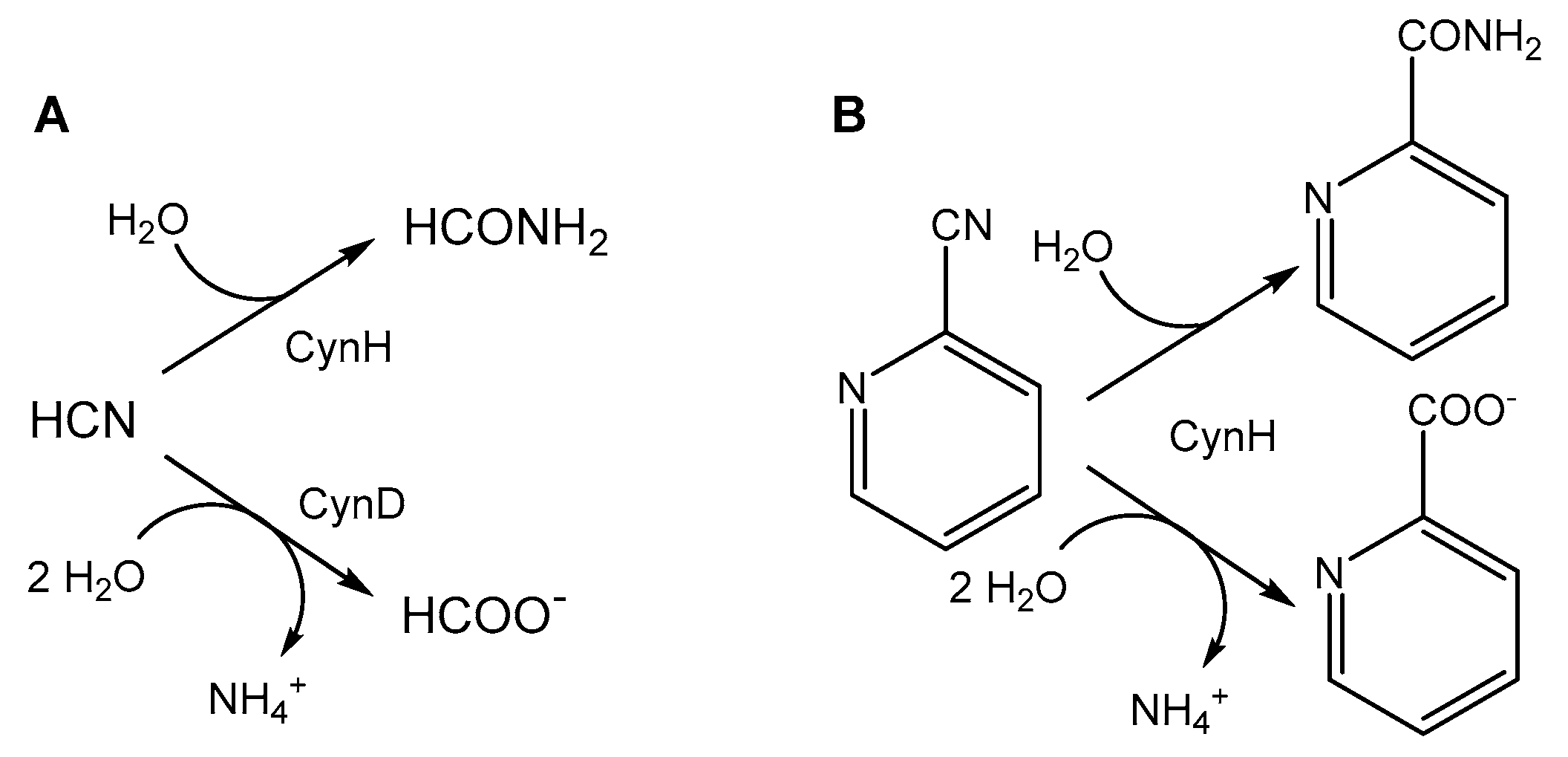
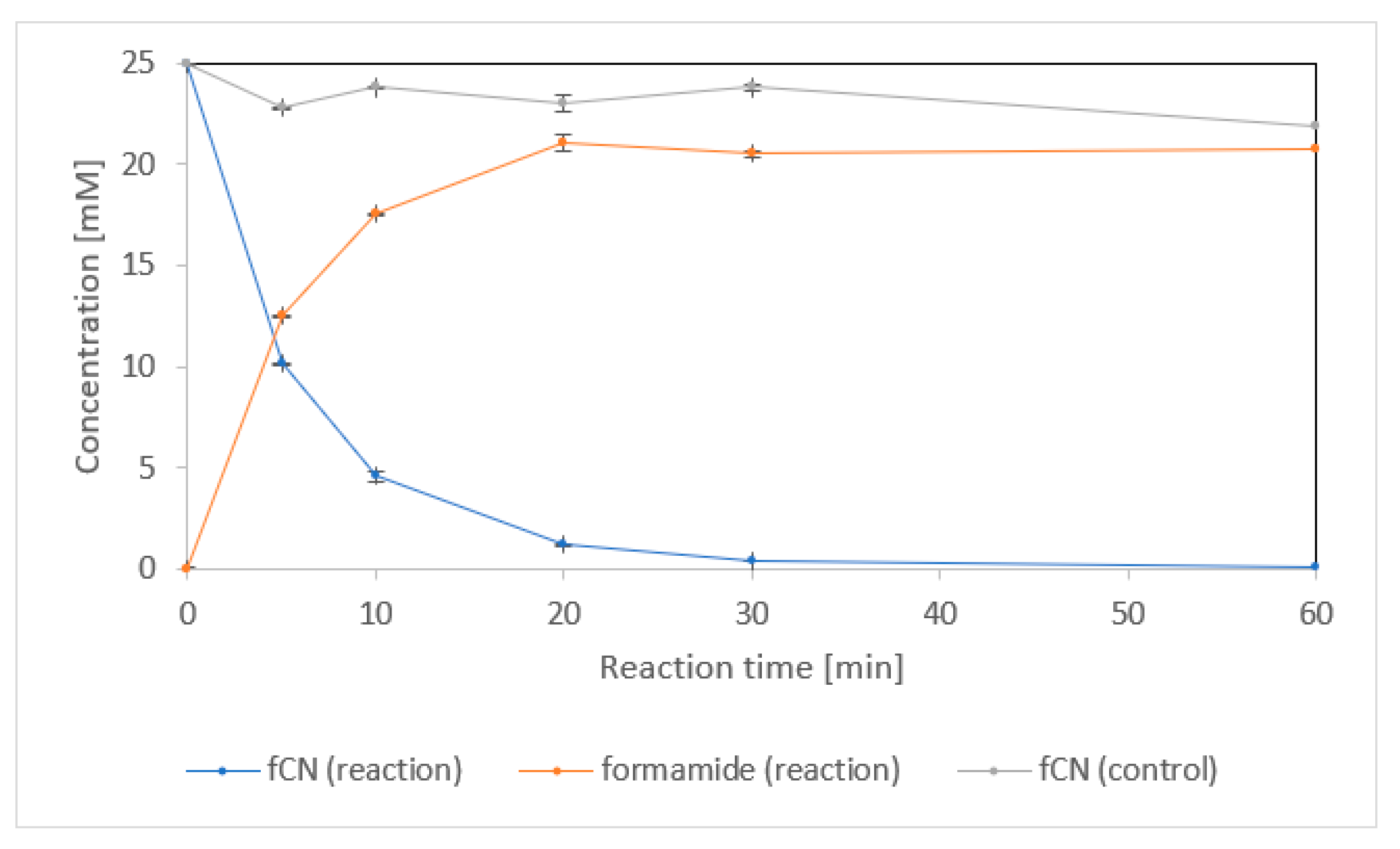
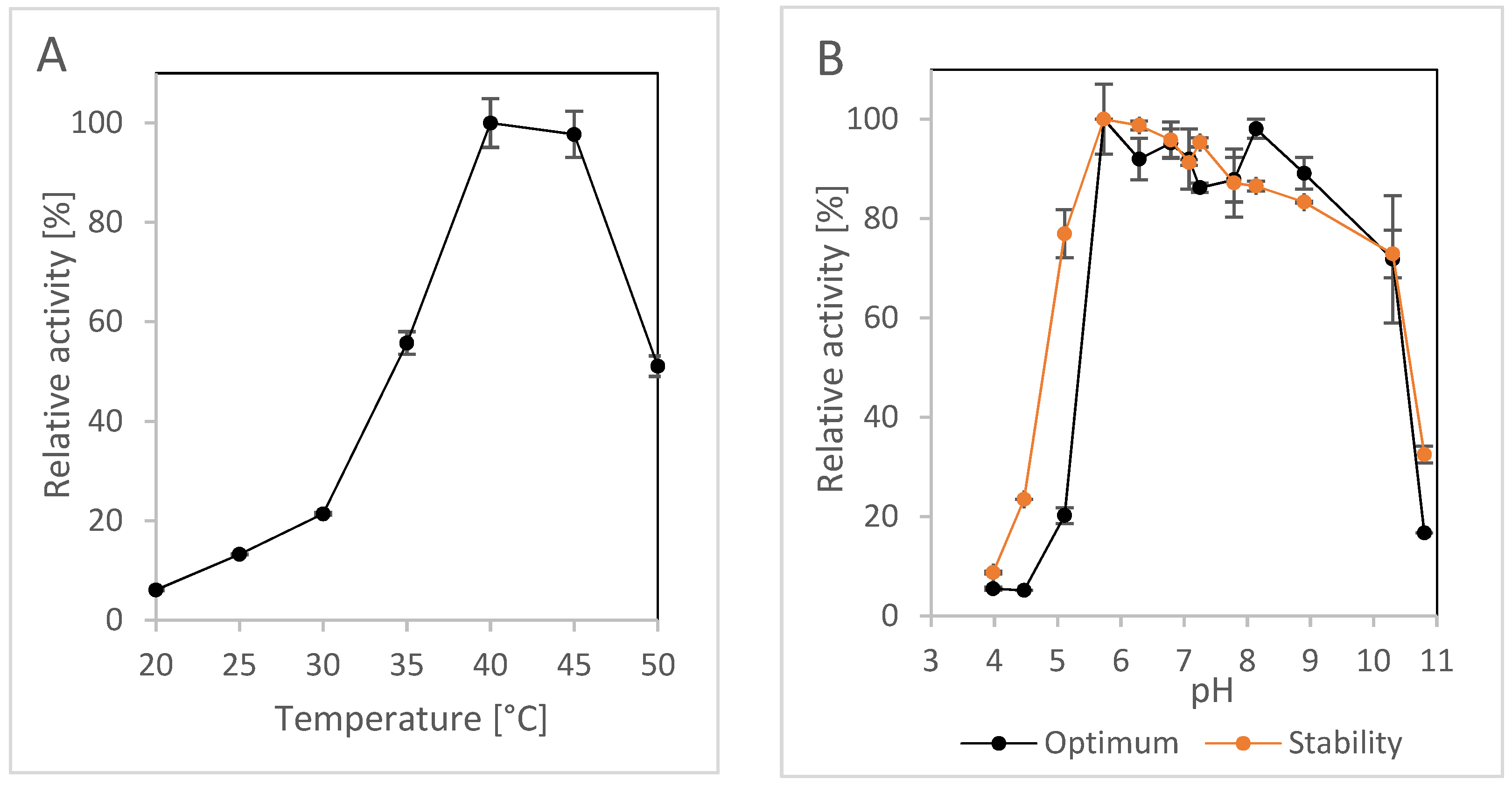
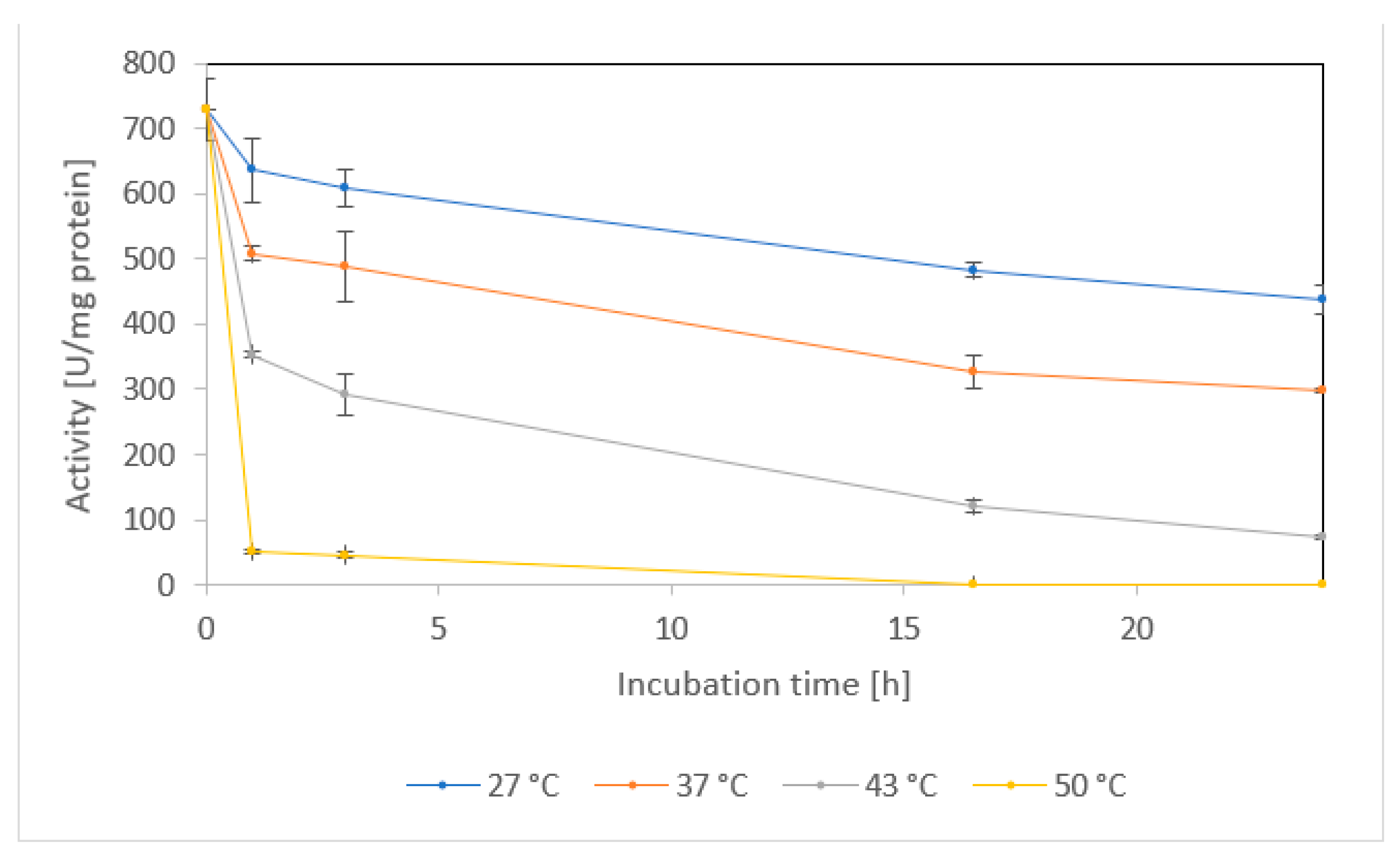
| Activity Assay | Specific Activity [U/mg] 1 | Vmax [U/mg] | KM [mM] | kcat [1/s] | kcat/KM [1/s/mM] |
|---|---|---|---|---|---|
| fCN consumption | 697 ± 95 | n.d. | n.d. | n.d. | n.d. |
| Formamide production | 784 ± 32 | 1335 ± 81 | 22.2 ± 2.4 | 927 ± 80 | 42 ± 10 |
| Metal Concentration (mM) | Relative Activity [%] | |
|---|---|---|
| Ag+ | Cu2+ | |
| - | 100 | 100 |
| 0.1 | 66 ± 5 | 93 ± 7 |
| 1 | 61 ± 5 | 68 ± 8 |
| 5 | 31 ± 10 1 | 14 ± 3 |
| 10 | n.d. 2 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sedova, A.; Rucká, L.; Bojarová, P.; Glozlová, M.; Novotný, P.; Křístková, B.; Pátek, M.; Martínková, L. Application Potential of Cyanide Hydratase from Exidia glandulosa: Free Cyanide Removal from Simulated Industrial Effluents. Catalysts 2021, 11, 1410. https://doi.org/10.3390/catal11111410
Sedova A, Rucká L, Bojarová P, Glozlová M, Novotný P, Křístková B, Pátek M, Martínková L. Application Potential of Cyanide Hydratase from Exidia glandulosa: Free Cyanide Removal from Simulated Industrial Effluents. Catalysts. 2021; 11(11):1410. https://doi.org/10.3390/catal11111410
Chicago/Turabian StyleSedova, Anastasia, Lenka Rucká, Pavla Bojarová, Michaela Glozlová, Petr Novotný, Barbora Křístková, Miroslav Pátek, and Ludmila Martínková. 2021. "Application Potential of Cyanide Hydratase from Exidia glandulosa: Free Cyanide Removal from Simulated Industrial Effluents" Catalysts 11, no. 11: 1410. https://doi.org/10.3390/catal11111410
APA StyleSedova, A., Rucká, L., Bojarová, P., Glozlová, M., Novotný, P., Křístková, B., Pátek, M., & Martínková, L. (2021). Application Potential of Cyanide Hydratase from Exidia glandulosa: Free Cyanide Removal from Simulated Industrial Effluents. Catalysts, 11(11), 1410. https://doi.org/10.3390/catal11111410









