Optimizing the Production of Recombinant Hydroperoxide Lyase in Escherichia coli Using Statistical Design
Abstract
1. Introduction
2. Results
2.1. Optimizing the Culture Conditions Using RSM
2.1.1. Model Fitting and Statistical Analysis
2.1.2. 3D Response Surface Plots and Experimental Validation
2.2. Large-Scale Production
2.3. Protein Expression Analysis by SDS–PAGE and Western Blot
3. Discussion
4. Material and Methods
4.1. Bacterial Strains and Expression Vectors
4.2. Molecular Cloning and Construction of Expression Clones
4.3. Statistical Analyses for Optimization of Culture Conditions Using Response Surface Methodology (RSM)
4.4. Culture in Microplates
4.5. Culture in 1 L Erlenmeyer Flask
4.6. Enzymatic Assays
4.7. Protein Expression Analysis by SDS–PAGE and Western Blot
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gardner, H.W.; Weisleder, D.; Plattner, R.D. Hydroperoxide lyase and other hydroperoxide-metabolizing activity in tissues of soybean. Glycine Max. Plant Physiol. 1991, 97, 1059–1072. [Google Scholar] [CrossRef][Green Version]
- Delcarte, J.; Fauconnier, M.-L.; Hoyaux, P.; Jacques, P.; Thonart, P.; Marlier, M. Revue bibliographique: L’hydroperoxyde lyase. Biotechnol. Agron. Soc. Environ. 2000, 4, 157–167. [Google Scholar]
- Noordermeer, M.A.; Veldink, G.A.; Vliegenthart, J.F. Fatty acid hydroperoxide lyase: A plant cytochrome p450 enzyme involved in wound healing and pest resistance. Chembiochem. 2001, 2, 494–504. [Google Scholar] [CrossRef]
- Feussner, I.; Wasternack, C. The lipoxygenase pathway. Annu. Rev. Plant Biol. 2002, 53, 275–297. [Google Scholar] [CrossRef] [PubMed]
- Howe, G.A.; Schilmiller, A.L. Oxylipin metabolism in response to stress. Curr. Opin. Plant Biol. 2002, 5, 230–236. [Google Scholar] [CrossRef]
- Matsui, K.; Minami, A.; Hornung, E.; Shibata, H.; Kishimoto, K.; Ahnert, V.; Kindl, H.; Kajiwara, T.; Feussner, I. Biosynthesis of fatty acid derived aldehydes is induced upon mechanical wounding and its products show fungicidal activities in cucumber. Phytochemistry 2006, 67, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R.K.; De Domenico, S.; Santino, A. Plant cytochrome CYP74 family: Biochemical features, endocellular localisation, activation mechanism in plant defence and improvements for industrial applications. ChemBioChem 2009, 10, 1122–1133. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.C.; Schwab, W. Cloning and characterization of a 9-lipoxygenase gene induced by pathogen attack from Nicotiana benthamiana for biotechnological application. BMC Biotechnol. 2011, 11, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Savchenko, T.; Dehesh, K. Drought stress modulates oxylipin signature by eliciting 12-OPDA as a potent regulator of stomatal aperture. Plant Signal. Behav. 2014, 9, e28304. [Google Scholar] [CrossRef] [PubMed]
- Savchenko, T.V.; Zastrijnaja, O.M.; Klimov, V.V. Oxylipins and plant abiotic stress resistance. Biochem. Mosc. 2014, 79, 362–375. [Google Scholar] [CrossRef]
- Hassan, M.N.; Zainal, Z.; Ismail, I. Green leaf volatiles: Biosynthesis, biological functions and their applications in biotechnology. Plant Biotechnol. J. 2015, 13, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Rustgi, S.; Springer, A.; Kang, C.; von Wettstein, D.; Reinbothe, C.; Reinbothe, S.; Pollmann, S. ALLENE OXIDE SYNTHASE and HYDROPEROXIDE LYASE, Two Non-Canonical Cytochrome P450s in Arabidopsis thaliana and Their Different Roles in Plant Defense. Int. J. Mol. Sci. 2019, 20, 3064. [Google Scholar] [CrossRef] [PubMed]
- Liavonchanka, A.; Feussner, I. Lipoxygenases: Occurrence, functions and catalysis. J. Plant Physiol. 2006, 163, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Schiller, D.; Contreras, C.; Vogt, J.; Dunemann, F.; Defilippi, B.G.; Beaudry, R.; Schwab, W. A dual positional specific lipoxygenase functions in the generation of flavor compounds during climacteric ripening of apple. Hortic. Res. 2015, 2, 15003. [Google Scholar] [CrossRef]
- Grechkin, A.N.; Hamberg, M. The “heterolytic hydroperoxide lyase” is an isomerase producing a short-lived fatty acid hemiacetal. Biochim. Biophys. Acta 2004, 1636, 47–58. [Google Scholar] [CrossRef]
- Mita, G.; Quarta, A.; Fasano, P.; De Paolis, A.; Di Sansebastiano, G.P.; Perrotta, C.; Iannacone, R.; Belfield, E.; Hughes, R.; Tsesmetzis, N.; et al. Molecular cloning and characterization of an almond 9-hydroperoxide lyase, a new CYP74 targeted to lipid bodies. J. Exp. Bot. 2005, 56, 2321–2333. [Google Scholar] [CrossRef]
- Grechkin, A.N.; Bruhlmann, F.; Mukhtarova, L.S.; Gogolev, Y.V.; Hamberg, M. Hydroperoxide lyases (CYP74C and CYP74B) catalyze the homolytic isomerization of fatty acid hydroperoxides into hemiacetals. Biochim. Biophys. Acta 2006, 1761, 1419–1428. [Google Scholar] [CrossRef]
- Lee, D.S.; Nioche, P.; Hamberg, M.; Raman, C.S. Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature 2008, 455, 363–368. [Google Scholar] [CrossRef]
- Vincenti, S.; Mariani, M.; Alberti, J.-C.; Jacopini, S.; Brunini-Bronzini de Caraffa, V.; Berti, L.; Maury, J. Biocatalytic Synthesis of Natural Green Leaf Volatiles Using the Lipoxygenase Metabolic Pathway. Catalysts 2019, 9, 873. [Google Scholar] [CrossRef]
- Suurmeijer, C.N.; Perez-Gilabert, M.; van Unen, D.J.; van der Hijden, H.T.; Veldink, G.A.; Vliegenthart, J.F. Purification, stabilization and characterization of tomato fatty acid hydroperoxide lyase. Phytochemistry 2000, 53, 177–185. [Google Scholar] [CrossRef][Green Version]
- Tijet, N.; Wäspi, U.; Gaskin, D.J.; Hunziker, P.; Muller, B.L.; Vulfson, E.N.; Slusarenko, A.; Brash, A.R.; Whitehead, I.M. Purification, molecular cloning, and expression of the gene encoding fatty acid 13-hydroperoxide lyase from guava fruit (Psidium guajava). Lipids 2000, 35, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Fukushige, H.; Hildebrand, D.F. Watermelon (Citrullus lanatus) hydroperoxide lyase greatly increases C6 aldehyde formation in transgenic leaves. J. Agric. Food Chem. 2005, 53, 2046–2051. [Google Scholar] [CrossRef]
- Rabetafika, H.N.; Gigot, C.; Fauconnier, M.-L.; Ongena, M.; Destain, J.; Du Jardin, P.; Wathelet, J.-P.; Thonart, P. Sugar beet leaves as new source of hydroperoxide lyase in a bioprocess producing green-note aldehydes. Biotechnol. Lett. 2008, 30, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Padilla, M.N.; Hernandez, M.L.; Perez, A.G.; Sanz, C.; Martinez-Rivas, J.M. Isolation, expression, and characterization of a 13-hydroperoxide lyase gene from olive fruit related to the biosynthesis of the main virgin olive oil aroma compounds. J. Agric. Food Chem. 2010, 58, 5649–5657. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Xue, Q.; Jiang, B.; Hua, Y. Molecular cloning, expression, and enzymatic characterization of Solanum tuberosum hydroperoxide lyase. Eur. Food Res. Technol. 2012, 234, 723–731. [Google Scholar] [CrossRef]
- Matsui, K.; Ujita, C.; Fujimoto, S.; Wilkinson, J.; Hiatt, B.; Knauf, V.; Kajiwara, T.; Feussner, I. Fatty acid 9- and 13-hydroperoxide lyases from cucumber. FEBS Lett. 2000, 481, 183–188. [Google Scholar] [CrossRef]
- Tijet, N.; Schneider, C.; Muller, B.L.; Brash, A.R. Biogenesis of volatile aldehydes from fatty acid hydroperoxides: Molecular cloning of a hydroperoxide lyase (CYP74C) with specificity for both the 9- and 13-hydroperoxides of linoleic and linolenic acids. Arch. Biochem. Biophys. 2001, 386, 281–289. [Google Scholar] [CrossRef]
- Kuroda, H.; Oshima, T.; Kaneda, H.; Takashio, M. Identification and functional analyses of two cDNAs that encode fatty acid 9-/13-hydroperoxide lyase (CYP74C) in rice. Biosci. Biotechnol. Biochem. 2005, 69, 1545–1554. [Google Scholar] [CrossRef]
- Hatanaka, A.; Kajiwara, T.; Sekiya, J. Biosynthetic pathway for C6-aldehydes formation from linolenic acid in green leaves. Chem. Phys. Lipids 1987, 44, 341–361. [Google Scholar] [CrossRef]
- Hatanaka, A. The biogeneration of green odour by green leaves. Phytochemistry 1993, 34, 1201–1218. [Google Scholar] [CrossRef]
- Lanciotti, R.; Gianotti, A.; Patrignani, F.; Belletti, N.; Guerzoni, M.E.; Gardini, F. Use of natural aroma compounds to improve shelf-life and safety of minimally processed fruits. Trends Food Sci. Technol. 2004, 15, 201–208. [Google Scholar] [CrossRef]
- Gigot, C.; Ongena, M.; Fauconnier, M.-L.; Wathelet, J.-P.; Du Jardin, P.; Thonart, P. The lipoxygenase metabolic pathway in plants: Potential for industrial production of natural green leaf volatiles. Biotechnol. Agron. Soc. Environ. 2010, 14, 451–460. [Google Scholar]
- Muller, B.; Gautier, A.; Dean, C.; Kuhn, J.C. Process for the Enzymatic Preparation of Aliphatic Alcohols and Aldehydes from Linoleic Acid, Linoleic Acid, or a Natural Precursor. U.S. Patent 5,464,761 A, 7 November 1995. [Google Scholar]
- Márczy, J.S.; Németh, Á.S.; Samu, Z.; Háger-Veress, Á.; Szajáni, B. Production of hexanal from hydrolyzed sunflower oil by lipoxygenase and hydroperoxide lyase enzymes. Biotechnol. Lett. 2002, 24, 1673–1675. [Google Scholar] [CrossRef]
- Németh, Á.S.; Márczy, J.S.; Samu, Z.; Háger-Veress, Á.; Szajáni, B. Biocatalytic production of 2(E)-hexenal from hydrolysed linseed oil. Enzym. Microb. Technol. 2004, 34, 667–672. [Google Scholar] [CrossRef]
- Whitehead, I.M.; Slusarenko, A.J.; Waspi, U.; Gaskin, D.J.H.; Brash, A.R.; Tijet, N. Guava (Psidium Guajava) 13-Hydroperoxide Lyase and Uses Thereof. U.S. Patent 6,780,621 B2, 24 August 2004. [Google Scholar]
- Noordermeer, M.A.; Van Der Goot, W.; Van Kooij, A.J.; Veldsink, J.W.; Veldink, G.A.; Vliegenthart, J.F. Development of a biocatalytic process for the production of c6-aldehydes from vegetable oils by soybean lipoxygenase and recombinant hydroperoxide lyase. J. Agric. Food Chem. 2002, 50, 4270–4274. [Google Scholar] [CrossRef] [PubMed]
- Delcarte, J.; Fauconnier, M.; Jacques, P.; Matsui, K.; Thonart, P.; Marlier, M. Optimisation of expression and immobilized metal ion affinity chromatographic purification of recombinant (His)6-tagged cytochrome P450 hydroperoxide lyase in Escherichia coli. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2003, 786, 229–236. [Google Scholar] [CrossRef]
- Bourel, G.; Nicaud, J.-M.; Nthangeni, B.; Santiago-Gomez, P.; Belin, J.-M.; Husson, F. Fatty acid hydroperoxide lyase of green bell pepper: Cloning in Yarrowia lipolytica and biogenesis of volatile aldehydes. Enzym. Microb. Technol. 2004, 35, 293–299. [Google Scholar] [CrossRef]
- Santiago-Gomez, M.P.; Thanh, H.T.; De Coninck, J.; Cachon, R.; Kermasha, S.; Belin, J.-M.; Gervais, P.; Husson, F. Modeling hexanal production in oxido-reducing conditions by the yeast Yarrowia lipolytica. Process Biochem. 2009, 44, 1013–1018. [Google Scholar] [CrossRef]
- Gigot, C.; Ongena, M.; Fauconnier, M.-L.; Muhovski, Y.; Wathelet, J.-P.; Du Jardin, P.; Thonart, P. Optimization and scaling up of a biotechnological synthesis of natural green leaf volatiles using Beta vulgaris hydroperoxide lyase. Process Biochem. 2012, 47, 2547–2551. [Google Scholar] [CrossRef]
- Jacopini, S.; Mariani, M.; Brunini-Bronzini de Caraffa, V.; Gambotti, C.; Vincenti, S.; Desjobert, J.-M.; Muselli, A.; Costa, J.; Berti, L.; Maury, J. Olive recombinant hydroperoxide lyase, an efficient biocatalyst for synthesis of green leaf volatiles. Appl. Biochem. Biotechnol. 2016, 179, 671–683. [Google Scholar] [CrossRef]
- Jacopini, S.; Vincenti, S.; Mariani, M.; Brunini-Bronzini de Caraffa, V.; Gambotti, C.; Desjobert, J.M.; Muselli, A.; Costa, J.; Tomi, F.; Berti, L.; et al. Activation and Stabilization of Olive Recombinant 13-Hydroperoxide Lyase Using Selected Additives. Appl. Biochem. Biotechnol. 2017, 182, 1000–1013. [Google Scholar] [CrossRef] [PubMed]
- Baneyx, F. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 1999, 10, 411–421. [Google Scholar] [CrossRef]
- Baneyx, F. Protein Expression Technologies: Current Status and Future Trends; Garland Science: New York, NY, USA, 2004; p. 546. [Google Scholar]
- Thomas, J.G.; Baneyx, F. Protein Misfolding and Inclusion Body Formation in Recombinant Escherichia coli Cells Overexpressing Heat-shock Proteins. J. Biol. Chem. 1996, 271, 11141–11147. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.G.; Ayling, A.; Baneyx, F. Molecular chaperones, folding catalysts, and the recovery of active recombinant proteins from E. coli. Appl. Biochem. Biotechnol. 1997, 66, 197–238. [Google Scholar] [CrossRef]
- Mertens, J.A.; Skory, C.D. Isolation and characterization of two genes that encode active glucoamylase without a starch binding domain from Rhizopus oryzae. Curr. Microbiol. 2007, 54, 462–466. [Google Scholar] [CrossRef]
- Villaverde, A.; Mar Carrió, M. Protein aggregation in recombinant bacteria: Biological role of inclusion bodies. Biotechnol. Lett. 2003, 25, 1385–1395. [Google Scholar] [CrossRef]
- Schrödel, A.; de Marco, A. Characterization of the aggregates formed during recombinant protein expression in bacteria. BMC Biochem. 2005, 6, 10. [Google Scholar] [CrossRef]
- Machida, S.; Yu, Y.; Singh, S.P.; Kim, J.-D.; Hayashi, K.; Kawata, Y. Overproduction of β-glucosidase in active form by an Escherichia coli system coexpressing the chaperonin GroEL/ES. FEMS Microbiol. Lett. 1998, 159, 41–46. [Google Scholar] [CrossRef]
- De Bernardez Clark, E.; Schwarz, E.; Rudolph, R. Inhibition of aggregation side reactions during in vitro protein folding. Methods Enzymol. 1999, 309, 217–236. [Google Scholar]
- Palmer, I.; Wingfield, P.T. Preparation and Extraction of Insoluble (Inclusion-Body) Proteins from Escherichia coli. Curr. Protoc. Protein Sci. 2012, 70, 6.3.1–6.3.20. [Google Scholar] [CrossRef]
- Singh, S.M.; Panda, A.K. Solubilization and refolding of bacterial inclusion body proteins. J. Biosci. Bioeng. 2005, 99, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Ventura, S.; Villaverde, A. Protein quality in bacterial inclusion bodies. Trends Biotechnol. 2006, 24, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Schein, C.H. Production of Soluble Recombinant Proteins in Bacteria. Bio Technol. 1989, 7, 1141–1149. [Google Scholar] [CrossRef]
- Feller, G.; Le Bussy, O.; Gerday, C. Expression of Psychrophilic Genes in Mesophilic Hosts: Assessment of the Folding State of a Recombinant α-Amylase. Appl. Environ. Microbiol. 1998, 64, 1163–1165. [Google Scholar] [CrossRef] [PubMed]
- Mujacic, M.; Cooper, K.W.; Baneyx, F. Cold-inducible cloning vectors for low-temperature protein expression in Escherichia coli: Application to the production of a toxic and proteolytically sensitive fusion protein. Gene 1999, 238, 325–332. [Google Scholar] [CrossRef]
- Song, J.M.; An, Y.J.; Kang, M.H.; Lee, Y.-H.; Cha, S.-S. Cultivation at 6–10 °C is an effective strategy to overcome the insolubility of recombinant proteins in Escherichia coli. Protein Expr. Purif. 2012, 82, 297–301. [Google Scholar] [CrossRef]
- Vasina, J.A.; Baneyx, F. Expression of Aggregation-Prone Recombinant Proteins at Low Temperatures: A Comparative Study of the Escherichia coli cspA and tac Promoter Systems. Protein Expr. Purif. 1997, 9, 211–218. [Google Scholar] [CrossRef]
- Vuillemin, M.; Malbert, Y.; Laguerre, S.; Remaud-Siméon, M.; Moulis, C. Optimizing the production of an α-(1→2) branching sucrase in Escherichia coli using statistical design. Appl. Microbiol. Biotechnol. 2014, 98, 5173–5184. [Google Scholar] [CrossRef]
- Betton, J.-M.; Chaffotte, A. Repliement et production de protéines recombinantes. Méd. Sci. 2005, 21, 613–617. [Google Scholar] [CrossRef][Green Version]
- Ferrer, M.; Chernikova, T.N.; Yakimov, M.M.; Golyshin, P.N.; Timmis, K.N. Chaperonins govern growth of Escherichia coli at low temperatures. Nat. Biotechnol. 2003, 21, 1266–1267. [Google Scholar] [CrossRef]
- Kimata, K.; Yamaguchi, M.; Saito, Y.; Hata, H.; Miyake, K.; Yamane, T.; Nakagawa, Y.; Yano, A.; Ito, K.; Kawarasaki, Y. High cell-density expression system: A novel method for extracellular production of difficult-to-express proteins. J. Biosci. Bioeng. 2012, 113, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Vick, B.A. A spectrophotometric assay for hydroperoxide lyase. Lipids 1991, 26, 315–320. [Google Scholar] [CrossRef]
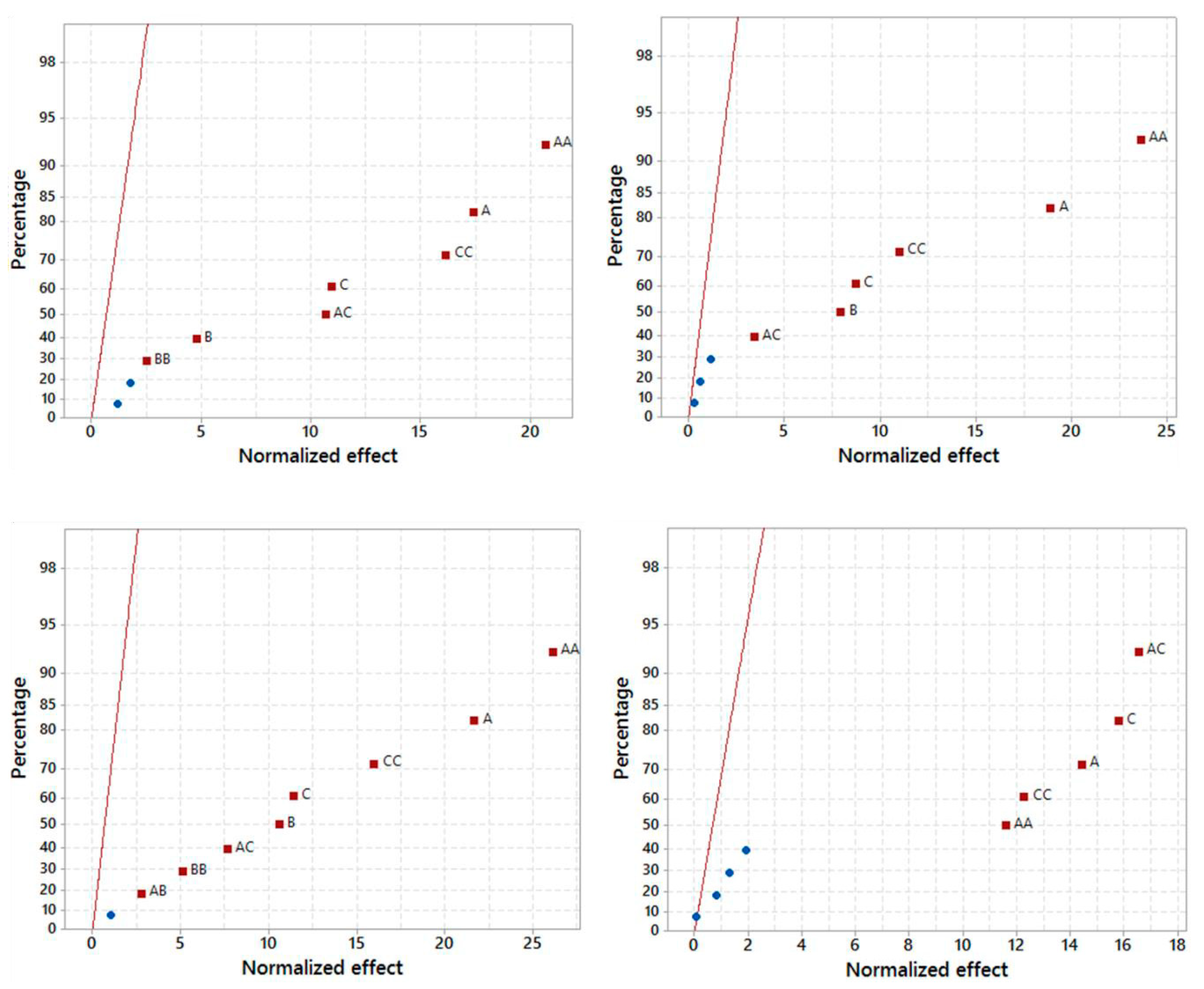
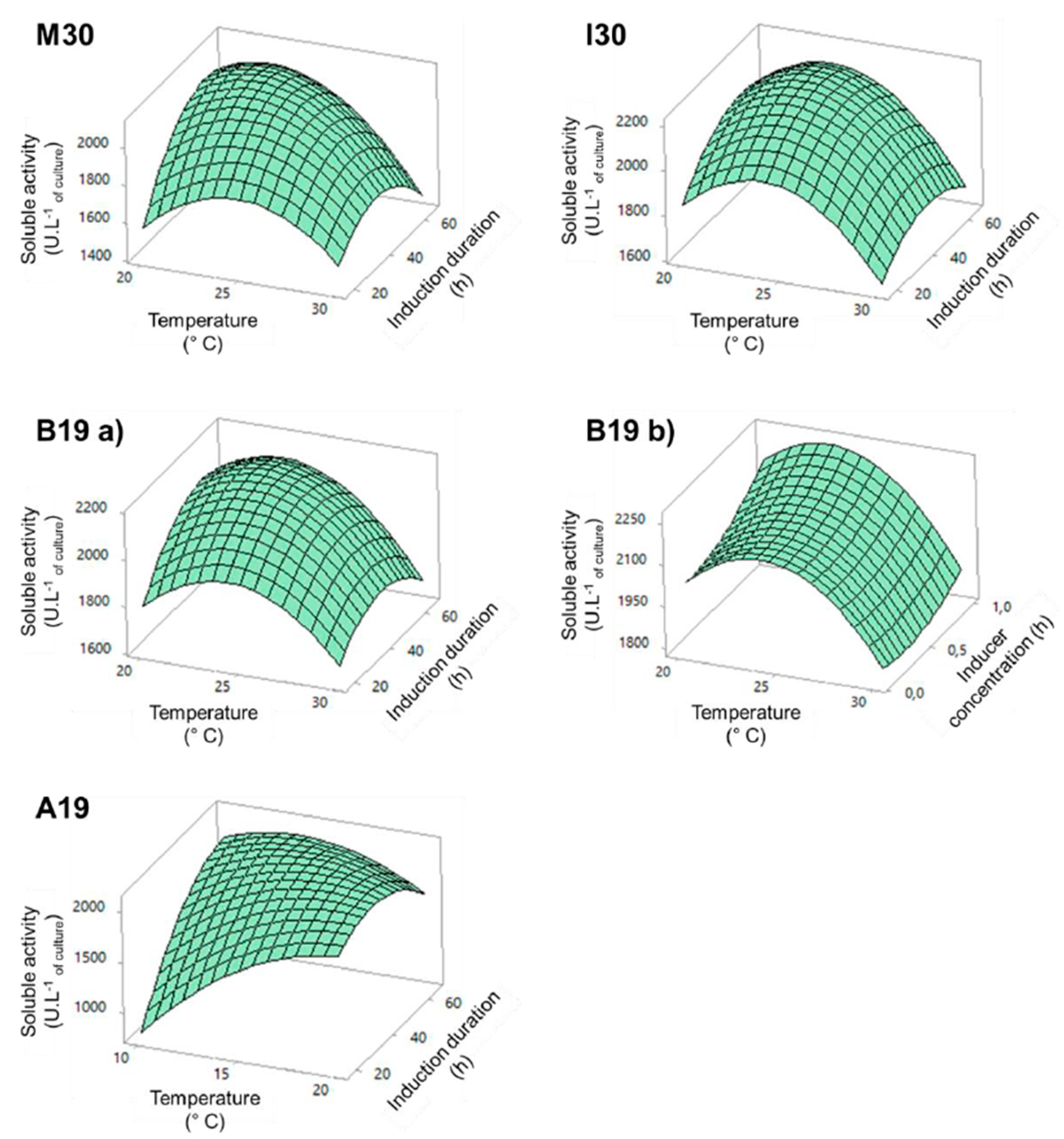

| Expression System | Equation a | R2 | Adj R2 |
|---|---|---|---|
| M30 | Soluble activity (U L−1of culture) = −5346 + 516.1 × T + 97 × I + 56.89 × D − 9.929 × T2 + 146.7 × I2 − 0.3363 × D2 − 8.99 × T × I − 1.0205 × T × D + 1.27 × I × D | 0.9724 | 0.9653 |
| I30 | Soluble activity (U L−1of culture) = −4477 + 500.8 × T + 185 × I + 28.38 × D − 10.336 × T2 −32.2 × I2 − 0.2090 × D2 + 1.08 × T × I − 0.3006 × T × D − 1.090 × I × D | 0.9706 | 0.9630 |
| B19 | Soluble activity (U L−1of culture) = −3809 + 430.8 × T + 176 × I + 34.64 × D − 8.573 × T2 + 207 × I2 − 0.2275 × D2 − 9.70 × T × I − 0.4994 × T × D − 0.724 × I × D | 0.9801 | 0.9750 |
| A19 | Soluble activity (U L−1of culture) = −3571 + 442.1 × T − 152 × I + 86.35 × D − 9.566 × T2 + 132 × I2 − 0.4398 × D2 + 0.27 × T × I − 2.734 × T × D + 1.49 × I × D | 0.9665 | 0.9578 |
| Source a | Sum of Squares | Degree of Freedom | F Value | p-Value b | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M30 | I30 | B19 | A19 | M30 | I30 | B19 | A19 | M30 | I30 | B19 | A19 | M30 | I30 | B19 | A19 | |
| Model | 1,963,026 | 1,532,237 | 1,284,812 | 4,758,653 | 9 | 9 | 9 | 9 | 136.81 | 128.21 | 191.51 | 112.09 | 0.000 | 0.000 | 0.000 | 0.000 |
| T | 482,971 | 473,401 | 350,658 | 979,862 | 1 | 1 | 1 | 1 | 302.93 | 356.50 | 470.40 | 207.73 | 0.000 | 0.000 | 0.000 | 0.000 |
| I | 36,465 | 83,497 | 83,355 | 17,275 | 1 | 1 | 1 | 1 | 22.87 | 62.88 | 111.82 | 3.66 | 0.000 | 0.000 | 0.000 | 0.064 |
| D | 190,478 | 101,023 | 96,520 | 1,175,413 | 1 | 1 | 1 | 1 | 119.47 | 76.08 | 129.48 | 249.19 | 0.000 | 0.000 | 0.000 | 0.000 |
| T2 | 682,550 | 739,644 | 508,860 | 633,549 | 1 | 1 | 1 | 1 | 428.12 | 557.00 | 682.63 | 134.31 | 0.000 | 0.000 | 0.000 | 0.000 |
| I2 | 9777 | - | 19,462 | - | 1 | - | 1 | - | 6.13 | - | 26.11 | - | 0.018 | - | 0.000 | - |
| D2 | 415,704 | 160,588 | 190,188 | 710,970 | 1 | 1 | 1 | 1 | 260.74 | 120.93 | 255.13 | 150.73 | 0.000 | 0.000 | 0.000 | 0.000 |
| T × I | - | - | 5720 | - | - | - | 1 | - | - | - | 7.67 | - | - | - | 0.009 | - |
| T × D | 181,548 | 15,610 | 43,104 | 1,291,336 | 1 | 1 | 1 | 1 | 113.87 | 11.76 | 57.82 | 273.77 | 0.000 | 0.002 | 0.000 | 0.000 |
| I × D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Lack of fit | 49,275 | 33,761 | 18,529 | 156,098 | 3 | 3 | 3 | 3 | 80.54 | 28.32 | 26.14 | 185.12 | 0.000 | 0.000 | 0.000 | 0.000 |
| Pure error | 6526 | 12,715 | 7561 | 8994 | 32 | 32 | 32 | 32 | - | - | - | - | - | - | - | - |
| Total | 2,018,827 | 1,578,714 | 1,310,903 | 4,923,745 | 44 | 44 | 44 | 44 | - | - | - | - | - | - | - | - |
| Expression System | Optimal Expression Conditions | Predicted HPL Activity a (U L−1of culture) | Measured HPL Activity b (U L−1of culture) | ||
|---|---|---|---|---|---|
| T (°C) | I (mM) | D (h) | |||
| M30 | 23 | 1 | 51.5 | 2197 | 2254 ± 19 |
| I30 | 23.5 | 1 | 48.5 | 2250 | 2177 ± 20 |
| B19 | 23 | 1 | 49 | 2277 | 2285 ± 12 |
| A19 | 16 | 1 | 50.5 | 2143 | 2209 ± 16 |
| Factor | E. coli Strain/Vector Construction | |||||
|---|---|---|---|---|---|---|
| M30 I30 B19 | A19 | |||||
| Codded Factor Levels | Codded Factor Levels | |||||
| −1 (Low) | 0 (Middle) | +1 (High) | −1 (Low) | 0 (Middle) | +1 (High) | |
| Temperature (T), °C | 20 | 25 | 30 | 10 | 15 | 20 |
| Inducer conc. (I), mM | 0.1 | 0.55 | 1 | 0.1 | 0.55 | 1 |
| Duration (D), h | 18 | 42 | 66 | 18 | 42 | 66 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vincenti, S.; Mariani, M.; Croce, J.; Faillace, E.; de Caraffa, V.B.-B.; Berti, L.; Maury, J. Optimizing the Production of Recombinant Hydroperoxide Lyase in Escherichia coli Using Statistical Design. Catalysts 2021, 11, 176. https://doi.org/10.3390/catal11020176
Vincenti S, Mariani M, Croce J, Faillace E, de Caraffa VB-B, Berti L, Maury J. Optimizing the Production of Recombinant Hydroperoxide Lyase in Escherichia coli Using Statistical Design. Catalysts. 2021; 11(2):176. https://doi.org/10.3390/catal11020176
Chicago/Turabian StyleVincenti, Sophie, Magali Mariani, Jessica Croce, Eva Faillace, Virginie Brunini-Bronzini de Caraffa, Liliane Berti, and Jacques Maury. 2021. "Optimizing the Production of Recombinant Hydroperoxide Lyase in Escherichia coli Using Statistical Design" Catalysts 11, no. 2: 176. https://doi.org/10.3390/catal11020176
APA StyleVincenti, S., Mariani, M., Croce, J., Faillace, E., de Caraffa, V. B.-B., Berti, L., & Maury, J. (2021). Optimizing the Production of Recombinant Hydroperoxide Lyase in Escherichia coli Using Statistical Design. Catalysts, 11(2), 176. https://doi.org/10.3390/catal11020176






