Characterization of a Solvent-Tolerant Amidohydrolase Involved in Natural Product Heterocycle Formation
Abstract
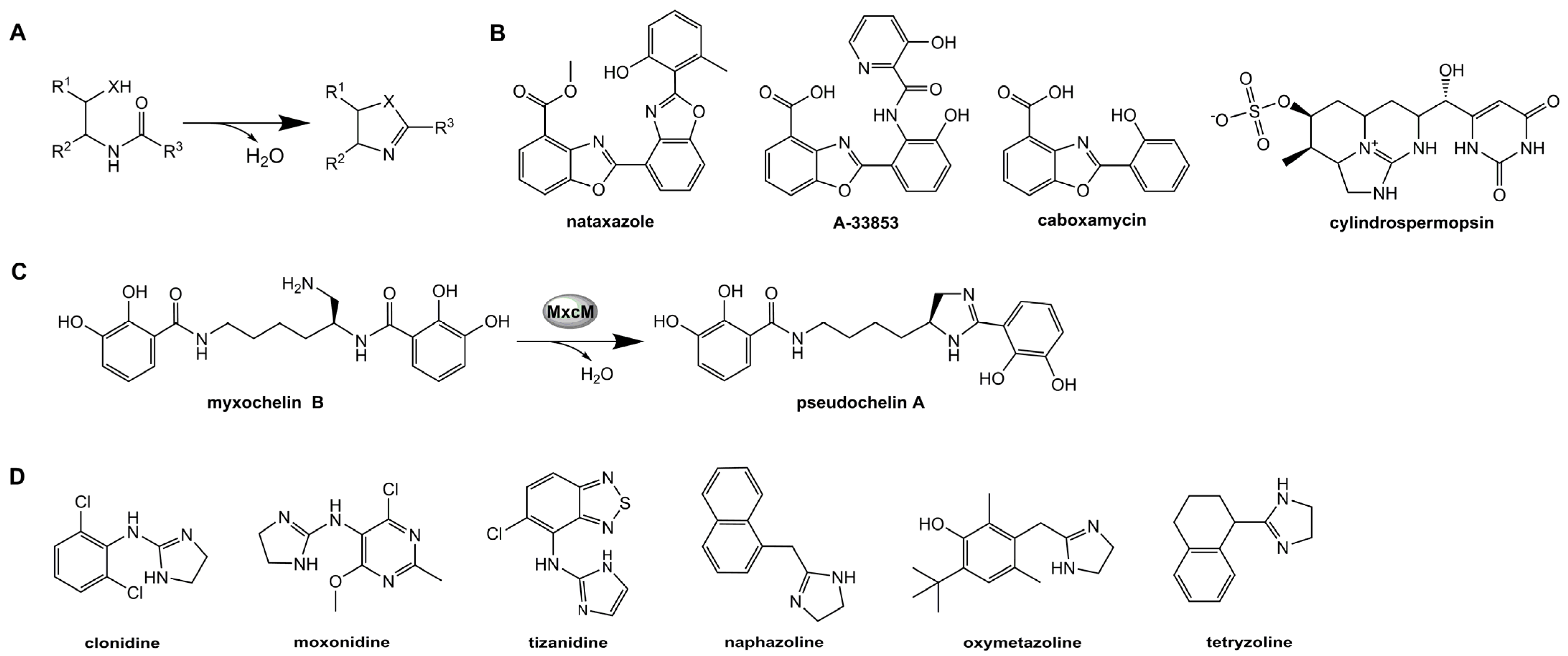
:1. Introduction
2. Results
2.1. Characterization of MxcM
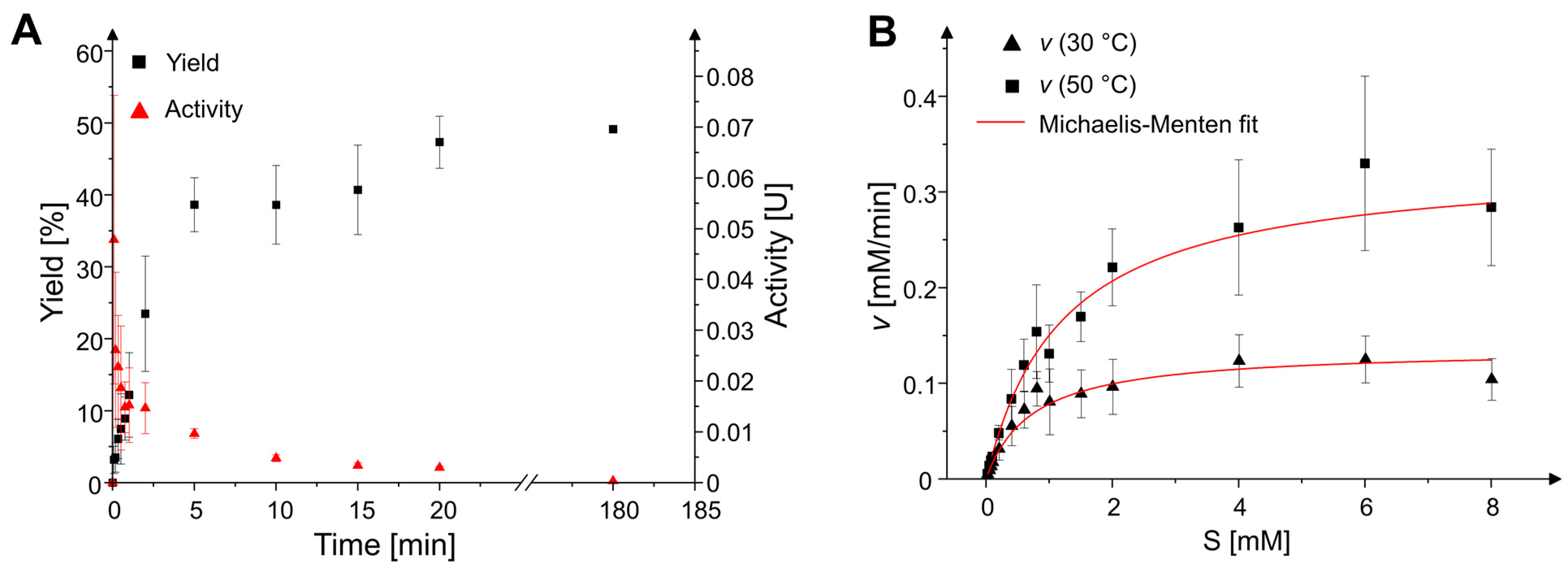
2.2. Reaction Time Course and Kinetic Parameters
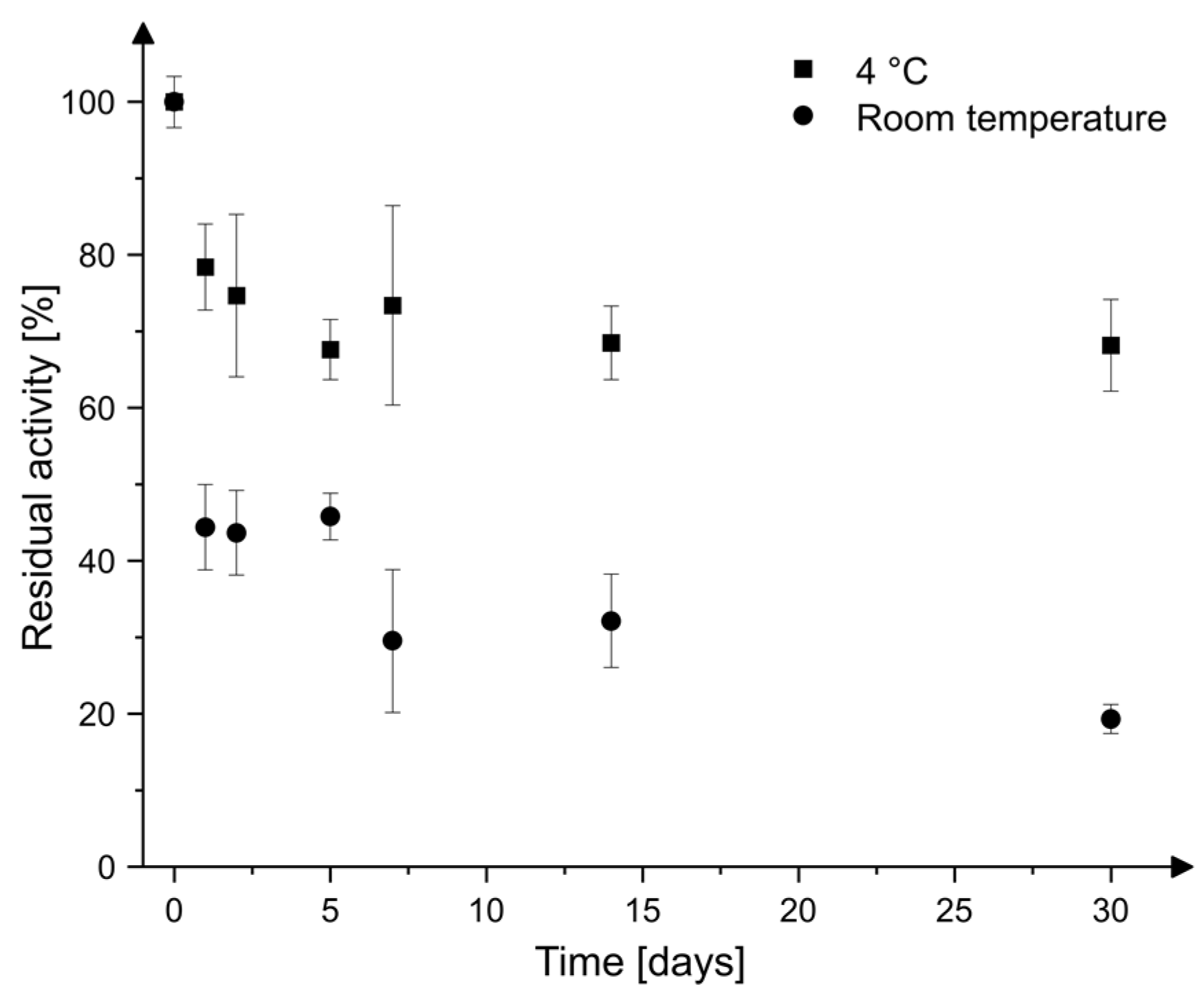
2.3. Storage Stability
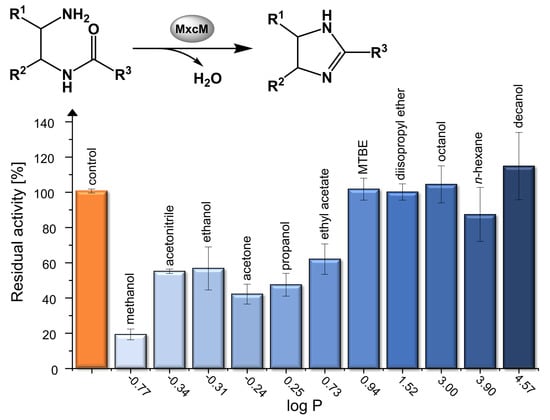
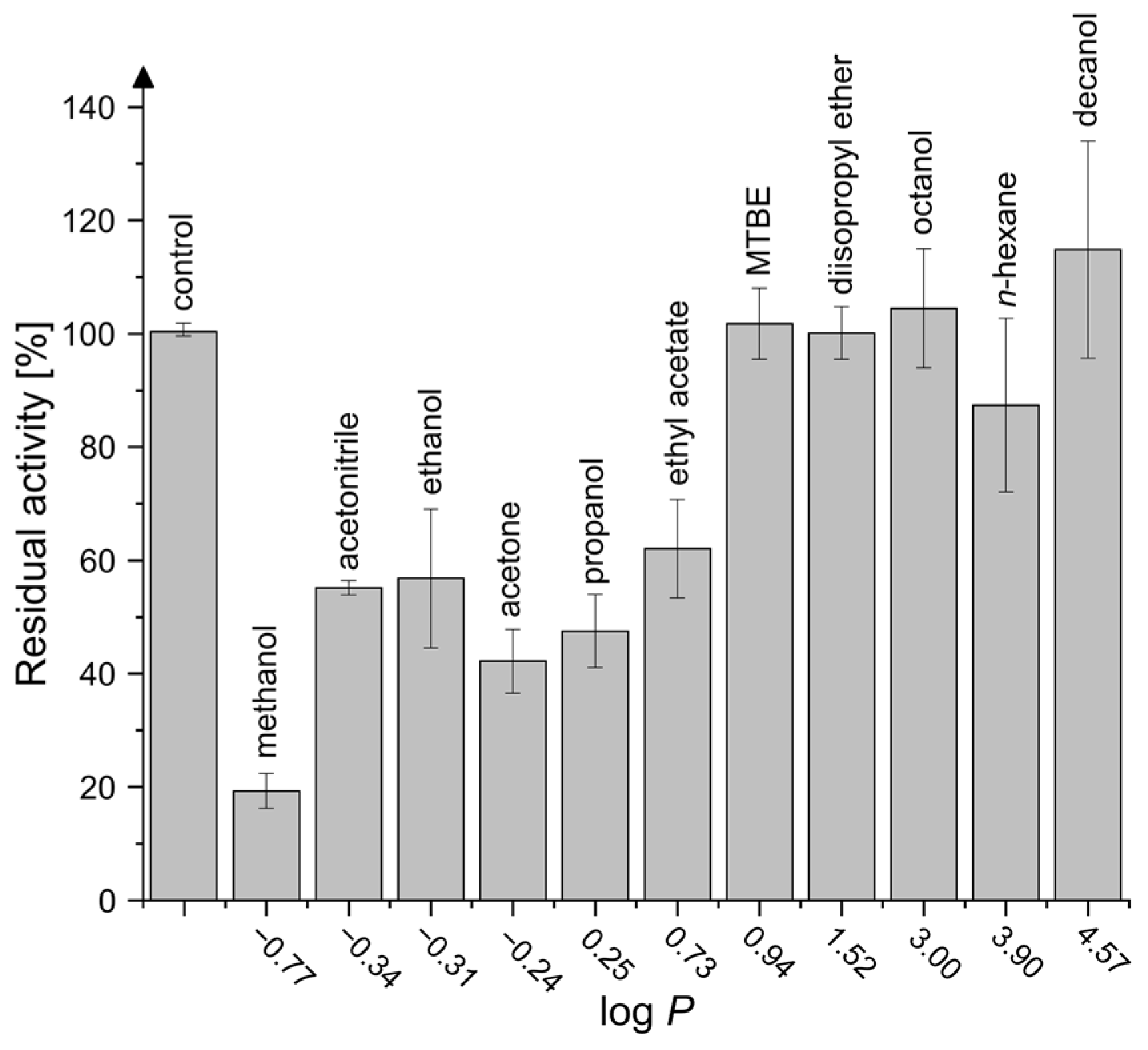
2.4. Activity in Organic Solvents and Salt-Containing Solutions
2.5. Substrate Specificity
3. Discussion
4. Materials and Methods
4.1. Enzyme Production and Purification
4.2. Activity Assay
4.3. Determination of Metal Dependency, pH and Temperature Optima, and Thermal Stability
4.4. Enzyme Kinetics
4.5. Storage Stability
4.6. Activity in Organic Solvents and Salt-Containing Solutions
4.7. Substrate Specificity
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Akiva, E.; Brown, S.; Almonacid, D.E.; Barber, A.E.; Custer, A.F.; Hicks, M.A.; Huang, C.C.; Lauck, F.; Mashiyama, S.T.; Meng, E.C.; et al. The Structure-Function Linkage Database. Nucleic Acids Res. 2014, 42, D521–D530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holm, L.; Sander, C. An evolutionary treasure: Unification of a broad set of amidohydrolases related to urease. Proteins 1997, 28, 72–82. [Google Scholar] [CrossRef]
- Seibert, C.M.; Raushel, F.M. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 2005, 44, 6383–6391. [Google Scholar] [CrossRef]
- Aimin, L.; Tingfeng, L.; Rong, F. Amidohydrolase Superfamily. In Encyclopedia of Life Sciences; John Wiley & Sons, Ltd.: Chichester, UK, 2001; ISBN 047001590X. [Google Scholar]
- Williams, L.; Nguyen, T.; Li, Y.; Porter, T.N.; Raushel, F.M. Uronate isomerase: A nonhydrolytic member of the amidohydrolase superfamily with an ambivalent requirement for a divalent metal ion. Biochemistry 2006, 45, 7453–7462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, M.; Zhao, J.; Deng, Z.; Yu, Y. Characterization of the Biosynthetic Gene Cluster for Benzoxazole Antibiotics A33853 Reveals Unusual Assembly Logic. Chem. Biol. 2015, 22, 1313–1324. [Google Scholar] [CrossRef] [Green Version]
- Losada, A.A.; Cano-Prieto, C.; García-Salcedo, R.; Braña, A.F.; Méndez, C.; Salas, J.A.; Olano, C. Caboxamycin biosynthesis pathway and identification of novel benzoxazoles produced by cross-talk in Streptomyces sp. NTK 937. Microb. Biotechnol. 2017, 10, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Rao, C.; Deng, Z.; Yu, Y.; Naismith, J.H. The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility. Angew. Chem. Int. Ed Engl. 2020, 59, 6054–6061. [Google Scholar] [CrossRef] [PubMed]
- Mihali, T.K.; Kellmann, R.; Muenchhoff, J.; Barrow, K.D.; Neilan, B.A. Characterization of the gene cluster responsible for cylindrospermopsin biosynthesis. Appl. Environ. Microbiol. 2008, 74, 716–722. [Google Scholar] [CrossRef] [Green Version]
- Korp, J.; Winand, L.; Sester, A.; Nett, M. Engineering Pseudochelin Production in Myxococcus xanthus. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [Green Version]
- Sester, A.; Winand, L.; Pace, S.; Hiller, W.; Werz, O.; Nett, M. Myxochelin- and Pseudochelin-Derived Lipoxygenase Inhibitors from a Genetically Engineered Myxococcus xanthus Strain. J. Nat. Prod. 2019, 82, 2544–2549. [Google Scholar] [CrossRef]
- Winand, L.; Sester, A.; Nett, M. Bioengineering of Anti-Inflammatory Natural Products. ChemMedChem 2021, 16, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Schieferdecker, S.; König, S.; Koeberle, A.; Dahse, H.-M.; Werz, O.; Nett, M. Myxochelins target human 5-lipoxygenase. J. Nat. Prod. 2015, 78, 335–338. [Google Scholar] [CrossRef]
- Sączewski, F.; Kornicka, A.; Balewski, Ł. Imidazoline scaffold in medicinal chemistry: A patent review (2012–2015). Expert Opin. Ther. Pat. 2016, 26, 1031–1048. [Google Scholar] [CrossRef]
- Guan, X.; Hu, Y. Imidazoline derivatives: A patent review (2006--present). Expert Opin. Ther. Pat. 2012, 22, 1353–1365. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Drug Approvals and Databases. Available online: https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm (accessed on 31 May 2021).
- Mehedi, M.S.A.; Tepe, J.J. Recent Advances in the Synthesis of Imidazolines (2009–2020). Adv. Synth. Catal. 2020, 362, 4189–4225. [Google Scholar] [CrossRef]
- Lechner, H.; Pressnitz, D.; Kroutil, W. Biocatalysts for the formation of three- to six-membered carbo- and heterocycles. Biotechnol. Adv. 2015, 33, 457–480. [Google Scholar] [CrossRef] [Green Version]
- Hemmerling, F.; Hahn, F. Biosynthesis of oxygen and nitrogen-containing heterocycles in polyketides. Beilstein J. Org. Chem. 2016, 12, 1512–1550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, A.; Jeske, L.; Ulbrich, S.; Hofmann, J.; Koblitz, J.; Schomburg, I.; Neumann-Schaal, M.; Jahn, D.; Schomburg, D. BRENDA, the ELIXIR core data resource in 2021: New developments and updates. Nucleic Acids Res. 2021, 49, D498–D508. [Google Scholar] [CrossRef]
- Wang, S.; Meng, X.; Zhou, H.; Liu, Y.; Secundo, F.; Liu, Y. Enzyme Stability and Activity in Non-Aqueous Reaction Systems: A Mini Review. Catalysts 2016, 6, 32. [Google Scholar] [CrossRef] [Green Version]
- Klibanov, A.M. Improving enzymes by using them in organic solvents. Nature 2001, 409, 241–246. [Google Scholar] [CrossRef]
- Doukyu, N.; Ogino, H. Organic solvent-tolerant enzymes. Biochem. Eng. J. 2010, 48, 270–282. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Hernick, M.; Fierke, C.A. Zinc hydrolases: The mechanisms of zinc-dependent deacetylases. Arch. Biochem. Biophys. 2005, 433, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Ramazzina, I.; Cendron, L.; Folli, C.; Berni, R.; Monteverdi, D.; Zanotti, G.; Percudani, R. Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases. J. Biol. Chem. 2008, 283, 23295–23304. [Google Scholar] [CrossRef] [Green Version]
- Bar-Even, A.; Noor, E.; Savir, Y.; Liebermeister, W.; Davidi, D.; Tawfik, D.S.; Milo, R. The moderately efficient enzyme: Evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 2011, 50, 4402–4410. [Google Scholar] [CrossRef]
- Laane, C.; Boeren, S.; Vos, K.; Veeger, C. Rules for optimization of biocatalysis in organic solvents. Biotechnol. Bioeng. 1987, 30, 81–87. [Google Scholar] [CrossRef]
- Zaks, A.; Klibanov, A.M. Enzymatic catalysis in nonaqueous solvents. J. Biol. Chem. 1988, 263, 3194–3201. [Google Scholar] [CrossRef]
- Sonnenschein, E.C.; Stierhof, M.; Goralczyk, S.; Vabre, F.M.; Pellissier, L.; Hanssen, K.Ø.; de La Cruz, M.; Díaz, C.; de Witte, P.; Copmans, D.; et al. Pseudochelin A, a siderophore of Pseudoalteromonas piscicida S2040. Tetrahedron 2017, 73, 2633–2637. [Google Scholar] [CrossRef] [Green Version]
- Richards, G.P.; Needleman, D.S.; Watson, M.A. Complete Genome Sequence of Pseudoalteromonas piscicida Strain DE2-B, a Bacterium with Broad Inhibitory Activity toward Human and Fish Pathogens. Genome Announc. 2017, 5, e00752-17. [Google Scholar] [CrossRef] [Green Version]
- Hua, L.; Zhou, R.; Thirumalai, D.; Berne, B.J. Urea denaturation by stronger dispersion interactions with proteins than water implies a 2-stage unfolding. Proc. Natl. Acad. Sci. USA 2008, 105, 16928–16933. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Temperature | vmax [mM/min] | Km [mM] | kcat [s−1] | kcat/Km [s−1 M−1] |
|---|---|---|---|---|
| 30 °C | 0.13 ± 0.01 | 0.74 ± 0.09 | 11.19 | 15,048.01 |
| 50 °C | 0.33 ± 0.02 | 1.21 ± 0.09 | 27.64 | 22,932.03 |
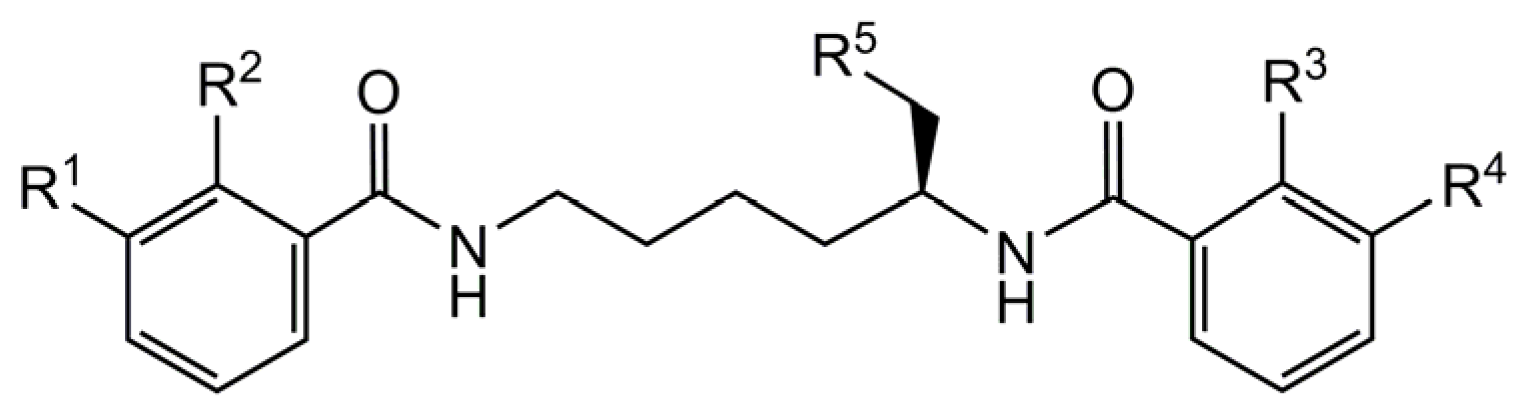
| Substrate | R1 | R2 | R3 | R4 | R5 | Residual Activity [%] |
|---|---|---|---|---|---|---|
| myxochelin B | OH | OH | OH | OH | NH2 | 100 ± 1.9 |
| myxochelin A | OH | OH | OH | OH | OH | n.d. |
| myxochelin B1 | H | H | OH | OH | NH2 | 73.3 ± 3.3 |
| myxochelin B2 | OH | OH | H | H | NH2 | 10.2 ± 1.9 |
| N-benzoylethylenediamine | - | - | - | - | - | n.d. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Winand, L.; Vollmann, D.J.; Hentschel, J.; Nett, M. Characterization of a Solvent-Tolerant Amidohydrolase Involved in Natural Product Heterocycle Formation. Catalysts 2021, 11, 892. https://doi.org/10.3390/catal11080892
Winand L, Vollmann DJ, Hentschel J, Nett M. Characterization of a Solvent-Tolerant Amidohydrolase Involved in Natural Product Heterocycle Formation. Catalysts. 2021; 11(8):892. https://doi.org/10.3390/catal11080892
Chicago/Turabian StyleWinand, Lea, Dustin Joshua Vollmann, Jacqueline Hentschel, and Markus Nett. 2021. "Characterization of a Solvent-Tolerant Amidohydrolase Involved in Natural Product Heterocycle Formation" Catalysts 11, no. 8: 892. https://doi.org/10.3390/catal11080892
APA StyleWinand, L., Vollmann, D. J., Hentschel, J., & Nett, M. (2021). Characterization of a Solvent-Tolerant Amidohydrolase Involved in Natural Product Heterocycle Formation. Catalysts, 11(8), 892. https://doi.org/10.3390/catal11080892