Immobilization-Stabilization of β-Glucosidase for Implementation of Intensified Hydrolysis of Cellobiose in Continuous Flow Reactors
Abstract
:1. Introduction
2. Results and Discussion
2.1. Design of Immobilized Preparations of β-glucosidase
2.2. Kinetic and Stability Characterization of Immobilized β-glucosidase
2.3. Implementation of the Hydrolysis in Discontinuous Stirred Tank Reactor: Identification of Substrate and Product Inhibition
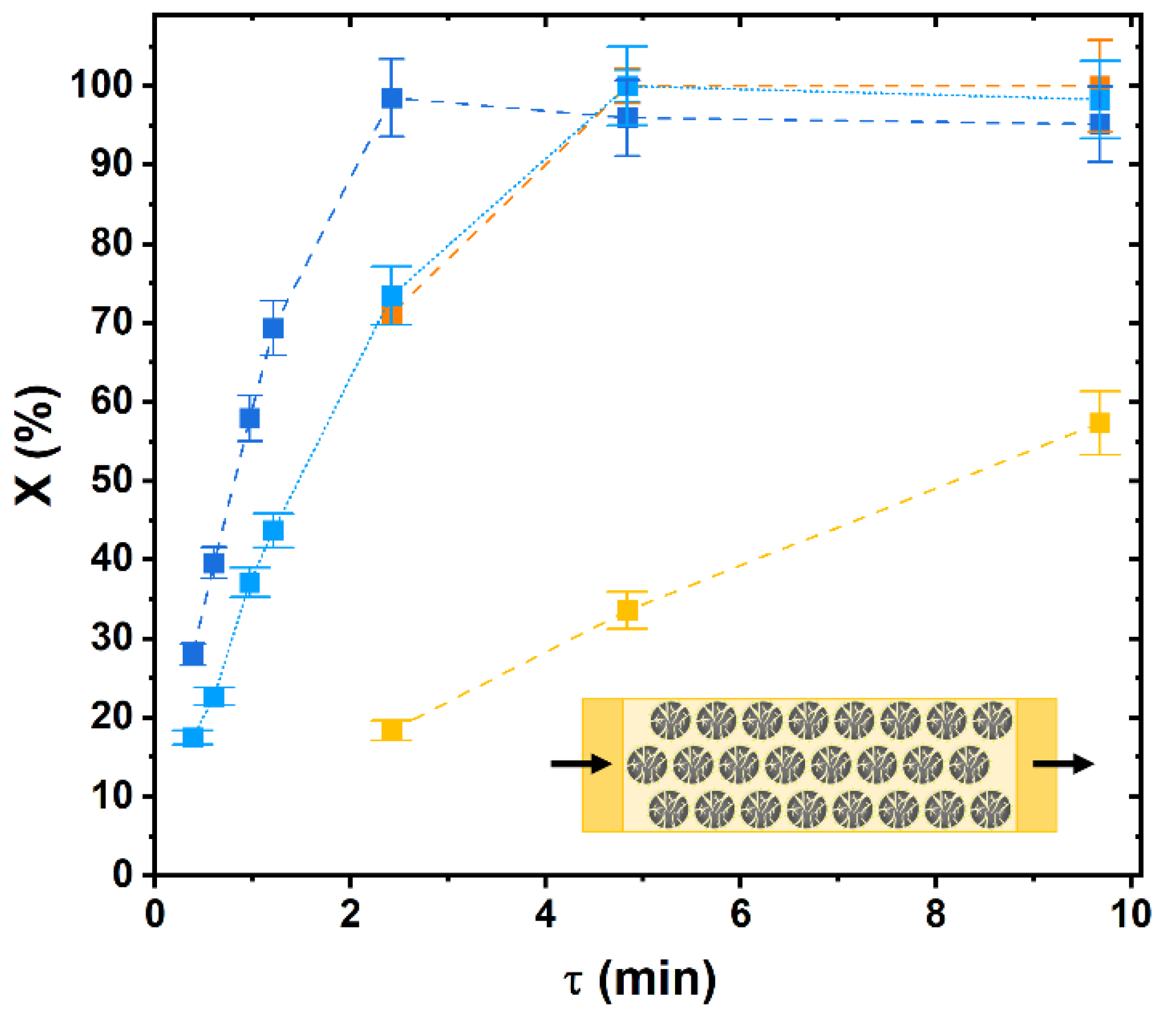
2.4. From Batch to Continuous: Intensification of Continuous Hydrolysis of Cellobiose Catalyzed by Immobilized β-glucosidase
2.5. Implementation of the Hydrolysis of Cellobiose in a Fixed-Bed Continuous Reactor
3. Materials and Methods
3.1. Materials
3.2. Characterization of Free Enzyme Formulation
3.3. Enzyme Activity Assay
3.4. Enzyme Immobilization
3.4.1. Material Preparation
3.4.2. Reversible Immobilization by Anionic Exchange Ionic Adsorption
3.4.3. Covalent Immobilization
3.5. Characterization of Immobilized Enzymes
3.5.1. Immobilization
3.5.2. Stability
3.5.3. Kinetic Analysis
3.6. Implementation of the Hydrolysis of Cellobiose Enzyme-Immobilized Reactors
3.6.1. Discontinuous Stirred Tank Reactor
3.6.2. Fixed Bed Reactor
3.7. Kinetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Venkata Mohan, S.; Dahiya, S.; Amulya, K.; Katakojwala, R.; Vanitha, T.K. Can Circular Bioeconomy Be Fueled by Waste Biorefine ies—A Closer Look. Bioresour. Technol. Rep. 2019, 7, 100277. [Google Scholar] [CrossRef]
- Mak, T.M.W.; Xiong, X.; Tsang, D.C.W.; Yu, I.K.M.; Poon, C.S. Sustainable Food Waste Management towards Circular Bioeconomy: Policy Review, Limitations and Opportunities. Bioresour. Technol. 2020, 297, 122497. [Google Scholar] [CrossRef]
- Ubando, A.T.; Felix, C.B.; Chen, W.-H. Biorefineries in Circular Bioeconomy: A Comprehensive Review. Bioresour. Technol. 2020, 299, 122585. [Google Scholar] [CrossRef]
- Cantzler, J.; Creutzig, F.; Ayargarnchanakul, E.; Javaid, A.; Wong, L.; Haas, W. Saving Resources and the Climate? A Systematic Review of the Circular Economy and Its Mitigation Potential. Environ. Res. Lett. 2020, 15, 123001. [Google Scholar] [CrossRef]
- Garcia-Ochoa, F.; Vergara, P.; Wojtusik, M.; Gutiérrez, S.; Santos, V.E.; Ladero, M.; Villar, J.C. Multi-Feedstock Lignocellulosic Biorefineries Based on Biological Processes: An Overview. Ind. Crops Prod. 2021, 172, 114062. [Google Scholar] [CrossRef]
- Vu, H.P.; Nguyen, L.N.; Vu, M.T.; Johir, M.A.H.; McLaughlan, R.; Nghiem, L.D. A Comprehensive Review on the Framework to Valorise Lignocellulosic Biomass as Biorefinery Feedstocks. Sci. Total Environ. 2020, 743, 140630. [Google Scholar] [CrossRef]
- Méndez-Líter, J.A.; Gil-Muñoz, J.; Nieto-Domínguez, M.; Barriuso, J.; de Eugenio, L.I.; Martínez, M.J. A Novel, Highly Efficient β-Glucosidase with a Cellulose-Binding Domain: Characterization and Properties of Native and Recombinant Proteins. Biotechnol. Biofuels 2017, 10, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Xiao, Y.; Feng, Y.; Li, B.; Li, W.; Cui, Q. The Spatial Proximity Effect of Beta-Glucosidase and Cellulosomes on Cellulose Degradation. Enzym. Microb. Technol. 2018, 115, 52–61. [Google Scholar] [CrossRef]
- Wang, Y.; Milewska, M.; Foster, H.; Chapman, R.; Stenzel, M.H. The Core–Shell Structure, Not Sugar, Drives the Thermal Stabilization of Single-Enzyme Nanoparticles. Biomacromolecules 2021, 22, 4569–4581. [Google Scholar] [CrossRef] [PubMed]
- Wojtusik, M.; Vergara, P.; Villar, J.C.; Garcia-Ochoa, F.; Ladero, M. Thermal and Operational Deactivation of Aspergillus Fumigatus β-Glucosidase in Ethanol/Water Pretreated Wheat Straw Enzymatic Hydrolysis. J. Biotechnol. 2019, 292, 32–38. [Google Scholar] [CrossRef]
- Andrić, P.; Meyer, A.S.; Jensen, P.A.; Dam-Johansen, K. Reactor Design for Minimizing Product Inhibition during Enzymatic Lignocellulose Hydrolysis: I. Significance and Mechanism of Cellobiose and Glucose Inhibition on Cellulolytic Enzymes. Biotechnol. Adv. 2010, 28, 308–324. [Google Scholar] [CrossRef]
- Huang, C.; Feng, Y.; Patel, G.; Xu, X.; Qian, J.; Liu, Q.; Kai, G. Production, Immobilization and Characterization of Beta-Glucosidase for Application in Cellulose Degradation from a Novel Aspergillus Versicolor. Int. J. Biol. Macromol. 2021, 177, 437–446. [Google Scholar] [CrossRef]
- Uchiyama, T.; Miyazaki, K.; Yaoi, K. Characterization of a Novel β-Glucosidase from a Compost Microbial Metagenome with Strong Transglycosylation Activity. J. Biol. Chem. 2013, 288, 18325–18334. [Google Scholar] [CrossRef] [Green Version]
- Sørensen, A.; Lübeck, M.; Lübeck, P.; Ahring, B. Fungal Beta-Glucosidases: A Bottleneck in Industrial Use of Lignocellulosic Materials. Biomolecules 2013, 3, 612–631. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, N.; Rathour, R.; Jha, S.; Pandey, K.; Srivastava, M.; Thakur, V.K.; Sengar, R.S.; Gupta, V.K.; Mazumder, P.B.; Khan, A.F.; et al. Microbial Beta Glucosidase Enzymes: Recent Advances in Biomass Conversation for Biofuels Application. Biomolecules 2019, 9, 220. [Google Scholar] [CrossRef] [Green Version]
- Singhania, R.R.; Patel, A.K.; Sukumaran, R.K.; Larroche, C.; Pandey, A. Role and Significance of Beta-Glucosidases in the Hydrolysis of Cellulose for Bioethanol Production. Bioresour. Technol. 2013, 127, 500–507. [Google Scholar] [CrossRef]
- Andrades, D.; Graebin, N.G.; Ayub, M.A.Z.; Fernandez-Lafuente, R.; Rodrigues, R.C. Preparation of Immobilized/Stabilized Biocatalysts of Β-glucosidases from Different Sources: Importance of the Support Active Groups and the Immobilization Protocol. Biotechnol. Prog. 2019, 35. [Google Scholar] [CrossRef] [PubMed]
- Andrades, D.; Graebin, N.G.; Ayub, M.A.Z.; Fernandez-Lafuente, R.; Rodrigues, R.C. Physico-Chemical Properties, Kinetic Parameters, and Glucose Inhibition of Several Beta-Glucosidases for Industrial Applications. Process Biochem. 2019, 78, 82–90. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Fernández-Lafuente, R. Modifying Enzyme Activity and Selectivity by Immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef] [PubMed]
- Guisan, J.M.; López-Gallego, F.; Bolivar, J.M.; Rocha-Martín, J.; Fernandez-Lorente, G. The Science of Enzyme Immobilization. In Immobilization of Enzymes and Cells; Guisan, J.M., Bolivar, J.M., López-Gallego, F., Rocha-Martín, J., Eds.; Springer: New York, NY, USA, 2020; Volume 2100, pp. 1–26. ISBN 978-1-07-160214-0. [Google Scholar]
- Bolivar, J.M.; Eisl, I.; Nidetzky, B. Advanced Characterization of Immobilized Enzymes as Heterogeneous Biocatalysts. Catal. Today 2016, 259, 66–80. [Google Scholar] [CrossRef]
- Valikhani, D.; Bolivar, J.M.; Pelletier, J.N. An Overview of Cytochrome P450 Immobilization Strategies for Drug Metabolism Studies, Biosensing, and Biocatalytic Applications: Challenges and Opportunities. ACS Catal. 2021, 11, 9418–9434. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of Different Enzyme Immobilization Strategies to Improve Enzyme Performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Jung, Y.R.; Shin, H.Y.; Song, Y.S.; Kim, S.B.; Kim, S.W. Enhancement of Immobilized Enzyme Activity by Pretreatment of β-Glucosidase with Cellobiose and Glucose. J. Ind. Eng. Chem. 2012, 18, 702–706. [Google Scholar] [CrossRef]
- Ahirwar, R.; Sharma, J.G.; Nahar, P.; Kumar, S. Immobilization Studies of Cellulase on Three Engineered Polymer Surfaces. Biocatal. Agric. Biotechnol. 2017, 11, 248–251. [Google Scholar] [CrossRef]
- Morais Junior, W.G.; Pacheco, T.F.; Gao, S.; Martins, P.A.; Guisán, J.M.; Caetano, N.S. Sugarcane Bagasse Saccharification by Enzymatic Hydrolysis Using Endocellulase and β-Glucosidase Immobilized on Different Supports. Catalysts 2021, 11, 340. [Google Scholar] [CrossRef]
- Ahmed, S.A.; El-Shayeb, N.M.A.; Hashem, A.M.; Saleh, S.A.; Abdel-Fattah, A.F. Biochemical Studies on Immobilized Fungal β-Glucosidase. Braz. J. Chem. Eng. 2013, 30, 747–758. [Google Scholar] [CrossRef] [Green Version]
- Figueira, J.A.; Dias, F.F.G.; Sato, H.H.; Fernandes, P. Screening of Supports for the Immobilization of β-Glucosidase. Enzym. Res. 2011, 2011, 642460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piñuel, L.; Mazzaferro, L.S.; Breccia, J.D. Operational Stabilization of Fungal α-Rhamnosyl-β-Glucosidase by Immobilization on Chitosan Composites. Process Biochem. 2011, 46, 2330–2335. [Google Scholar] [CrossRef]
- Califano, V.; Costantini, A.; Silvestri, B.; Venezia, V.; Cimino, S.; Sannino, F. The Effect of Pore Morphology on the Catalytic Performance of β-Glucosidase Immobilized into Mesoporous Silica. Pure Appl. Chem. 2019, 91, 1583–1592. [Google Scholar] [CrossRef]
- Wu, X.; Qu, B.; Liu, Y.; Ren, X.; Wang, S.; Quan, Y. Highly Enhanced Activity and Stability via Affinity Induced Immobilization β-Glucosidase from Aspergillus Niger onto Amino-Based Silica for the Biotransformation of Ginsenoside Rb1. J. Chromatogr. A 2021, 1653, 462388. [Google Scholar] [CrossRef]
- Alftrén, J.; Hobley, T.J. Covalent Immobilization of β-Glucosidase on Magnetic Particles for Lignocellulose Hydrolysis. Appl. Biochem. Biotechnol. 2013, 169, 2076–2087. [Google Scholar] [CrossRef] [PubMed]
- Wojtusik, M.; Yepes, C.M.; Villar, J.C.; Cordes, A.; Arroyo, M.; Garcia-Ochoa, F.; Ladero, M. Kinetic Modeling of Cellobiose by a β-Glucosidase from Aspergillus Fumigatus. Chem. Eng. Res. Des. 2018, 136, 502–512. [Google Scholar] [CrossRef]
- Dias Gomes, M.; Woodley, J.M. Considerations When Measuring Biocatalyst Performance. Molecules 2019, 24, 3573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuusk, S.; Väljamäe, P. When Substrate Inhibits and Inhibitor Activates: Implications of β-Glucosidases. Biotechnol. Biofuels 2017, 10, 7. [Google Scholar] [CrossRef] [Green Version]
- Lorente-Arevalo, A.; Garcia-Martin, A.; Ladero, M.; Bolivar, J.M. Chemical Reaction Engineering to Understand Applied Kinetics in Free Enzyme Homogeneous Reactors. In Enzyme Engineering; Magnani, F., Marabelli, C., Paradisi, F., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2022; Volume 2397, pp. 277–320. ISBN 978-1-07-161825-7. [Google Scholar]
- Venezia, V.; Califano, V.; Pota, G.; Costantini, A.; Landi, G.; Di Benedetto, A. CFD Simulations of Microreactors for the Hydrolysis of Cellobiose to Glucose by β-Glucosidase Enzyme. Micromachines 2020, 11, 790. [Google Scholar] [CrossRef] [PubMed]
- Venezia, V.; Costantini, A.; Landi, G.; Di Benedetto, A.; Sannino, F.; Califano, V. Immobilization of β-Glucosidase over Structured Cordierite Monoliths Washcoated with Wrinkled Silica Nanoparticles. Catalysts 2020, 10, 889. [Google Scholar] [CrossRef]
- Wei, C.; Zhou, Y.; Zhuang, W.; Li, G.; Jiang, M.; Zhang, H. Improving the Performance of Immobilized β-Glucosidase Using a Microreactor. J. Biosci. Bioeng. 2018, 125, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.C.S.; Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Importance of the Support Properties for Immobilization or Purification of Enzymes. ChemCatChem 2015, 7, 2413–2432. [Google Scholar] [CrossRef] [Green Version]
- López-Gallego, F.; Fernandez-Lorente, G.; Rocha-Martín, J.; Bolivar, J.M.; Mateo, C.; Guisan, J.M. Multi-Point Covalent Immobilization of Enzymes on Glyoxyl Agarose with Minimal Physico-Chemical Modification: Stabilization of Industrial Enzymes. In Immobilization of Enzymes and Cells; Guisan, J.M., Bolivar, J.M., López-Gallego, F., Rocha-Martín, J., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; Volume 2100, pp. 93–107. ISBN 978-1-07-160214-0. [Google Scholar]
- Mateo, C.; Palomo, J.M.; Fuentes, M.; Betancor, L.; Grazu, V.; López-Gallego, F.; Pessela, B.C.C.; Hidalgo, A.; Fernández-Lorente, G.; Fernández-Lafuente, R.; et al. Glyoxyl Agarose: A Fully Inert and Hydrophilic Support for Immobilization and High Stabilization of Proteins. Enzym. Microb. Technol. 2006, 39, 274–280. [Google Scholar] [CrossRef]
- Mateo, C.; Abian, O.; Fernandez-Lafuente, R.; Guisan, J.M. Reversible Enzyme Immobilization via a Very Strong and Nondistorting Ionic Adsorption on Support-Polyethylenimine Composites. Biotechnol. Bioeng. 2000, 68, 98–105. [Google Scholar] [CrossRef]
- Virgen-Ortíz, J.J.; dos Santos, J.C.S.; Berenguer-Murcia, Á.; Barbosa, O.; Rodrigues, R.C.; Fernandez-Lafuente, R. Polyethylenimine: A Very Useful Ionic Polymer in the Design of Immobilized Enzyme Biocatalysts. J. Mater. Chem. B 2017, 5, 7461–7490. [Google Scholar] [CrossRef] [Green Version]
- Vieira, M.F.; Vieira, A.M.S.; Zanin, G.M.; Tardioli, P.W.; Mateo, C.; Guisán, J.M. β-Glucosidase Immobilized and Stabilized on Agarose Matrix Functionalized with Distinct Reactive Groups. J. Mol. Catal. B Enzym. 2011, 69, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Vazquez-Ortega, P.G.; Alcaraz-Fructuoso, M.T.; Rojas-Contreras, J.A.; López-Miranda, J.; Fernandez-Lafuente, R. Stabilization of Dimeric β-Glucosidase from Aspergillus Niger via Glutaraldehyde Immobilization under Different Conditions. Enzym. Microb. Technol. 2018, 110, 38–45. [Google Scholar] [CrossRef]
- Da Silva, T.M.; Pessela, B.C.; da Silva, J.C.R.; Lima, M.S.; Jorge, J.A.; Guisan, J.M.; Maria de Lourdes, T.M. Immobilization and High Stability of an Extracellular β-Glucosidase from Aspergillus Japonicus by Ionic Interactions. J. Mol. Catal. B Enzym. 2014, 104, 95–100. [Google Scholar] [CrossRef]
- De Andrades, D.; Graebin, N.G.; Kadowaki, M.K.; Ayub, M.A.Z.; Fernandez-Lafuente, R.; Rodrigues, R.C. Immobilization and Stabilization of Different β-Glucosidases Using the Glutaraldehyde Chemistry: Optimal Protocol Depends on the Enzyme. Int. J. Biol. Macromol. 2019, 129, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Teixeira da Silva, V.C.; de Souza Coto, A.L.; de Carvalho Souza, R.; Bertoldi Sanchez Neves, M.; Gomes, E.; Bonilla-Rodriguez, G.O. Effect of pH, Temperature, and Chemicals on the Endoglucanases and β-Glucosidases from the Thermophilic Fungus Myceliophthora Heterothallica F.2.1.4. Obtained by Solid-State and Submerged Cultivation. Biochem. Res. Int. 2016, 2016, 9781216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Zhang, R.; Yang, X.; Zhang, Z.; Song, S.; Miao, Y.; Shen, Q. Characterization of a Thermostable β-Glucosidase from Aspergillus Fumigatus Z5, and Its Functional Expression in Pichia Pastoris X33. Microb. Cell Fact. 2012, 11, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolivar, J.M.; Tribulato, M.A.; Petrasek, Z.; Nidetzky, B. Let the Substrate Flow, Not the Enzyme: Practical Immobilization of d-Amino Acid Oxidase in a Glass Microreactor for Effective Biocatalytic Conversions. Biotechnol. Bioeng. 2016, 113, 2342–2349. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Valikhani, D.; Nidetzky, B. Demystifying the Flow: Biocatalytic Reaction Intensification in Microstructured Enzyme Reactors. Biotechnol. J. 2019, 14, 1800244. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of Enzyme Activity, Stability and Selectivity via Immobilization Techniques. Enzym. Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Mateo, C.; Bolivar, J.M.; Godoy, C.A.; Rocha-Martin, J.; Pessela, B.C.; Curiel, J.A.; Muñoz, R.; Guisan, J.M.; Fernández-Lorente, G. Improvement of Enzyme Properties with a Two-Step Immobilizaton Process on Novel Heterofunctional Supports. Biomacromolecules 2010, 11, 3112–3117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Lafuente, R.; Rosell, C.M.; Rodriguez, V.; Santana, C.; Soler, G.; Bastida, A.; Guisán, J.M. Preparation of Activated Supports Containing Low PK Amino Groups. A New Tool for Protein Immobilization via the Carboxyl Coupling Method. Enzym. Microb. Technol. 1993, 15, 546–550. [Google Scholar] [CrossRef]
- Guisán, J.M. Aldehyde-Agarose Gels as Activated Supports for Immobilization-Stabilization of Enzymes. Enzym. Microb. Technol. 1988, 10, 375–382. [Google Scholar] [CrossRef]
- Illanes, A.; Wilson, L.; Vera, C. Enzyme Kinetics in a Heterogeneous System. In Problem Solving in Enzyme Biocatalysis; John Wiley and Sons Ltd.: Chichester, UK, 2013; pp. 87–140. ISBN 978-1-118-34174-2. [Google Scholar]








| Immobilized Biocatalyst | Activity Offered (U_pNPG/gsupport) | Immobilization Yield (%) | Measured Activity (U_pNPG/gsupport) | Recovered Activity (%) | Effectiveness Factor |
|---|---|---|---|---|---|
| Ag-MANAE | 19.4 | 38.5 | 5.7 | 29.4 | 0.76 |
| 193 | 42.3 | 28.3 | 14.7 | 0.35 | |
| 1871 | 43.2 | 137 | 7.3 | 0.17 | |
| Ag-PEI | 19.4 | 100 | 19.4 | 103 | 1.00 |
| 1923 | 97.7 | 153 | 79.6 | 0.81 | |
| 1871 | 93.6 | 1111 | 59.4 | 0.63 | |
| Pur-PEI | 19.4 | 100 | 14.1 | 72.6 | 0.73 |
| 193 | 89.8 | 119 | 61.7 | 0.69 | |
| 1871 | 57.3 | 423 | 22.6 | 0.39 | |
| Ag-Gly 4BCL | 286 | 92.7 | 119 | 41.76 | 0.45 |
| 2127 | 42.8 | 159 | 7.57 | 0.18 | |
| Ag-Gly 6BCL | 286 | 85.6 | 101 | 35.2 | 0.41 |
| 2127 | 39.2 | 189 | 8.86 | 0.23 | |
| Pur-Gly | 286 | 91.8 | 87.9 | 30.8 | 0.33 |
| 2127 | 39.4 | 77.7 | 3.66 | 0.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez-Gonzalez, C.; Santos, V.E.; Ladero, M.; Bolivar, J.M. Immobilization-Stabilization of β-Glucosidase for Implementation of Intensified Hydrolysis of Cellobiose in Continuous Flow Reactors. Catalysts 2022, 12, 80. https://doi.org/10.3390/catal12010080
Alvarez-Gonzalez C, Santos VE, Ladero M, Bolivar JM. Immobilization-Stabilization of β-Glucosidase for Implementation of Intensified Hydrolysis of Cellobiose in Continuous Flow Reactors. Catalysts. 2022; 12(1):80. https://doi.org/10.3390/catal12010080
Chicago/Turabian StyleAlvarez-Gonzalez, Celia, Victoria E. Santos, Miguel Ladero, and Juan M. Bolivar. 2022. "Immobilization-Stabilization of β-Glucosidase for Implementation of Intensified Hydrolysis of Cellobiose in Continuous Flow Reactors" Catalysts 12, no. 1: 80. https://doi.org/10.3390/catal12010080






