Biotransformation of Oleanolic Acid Using Rhodococcus rhodochrous IEGM 757
Abstract
1. Introduction
2. Results and Discussion
2.1. Biotransformation of OA and Effects of OA on Rhodococcal Cells
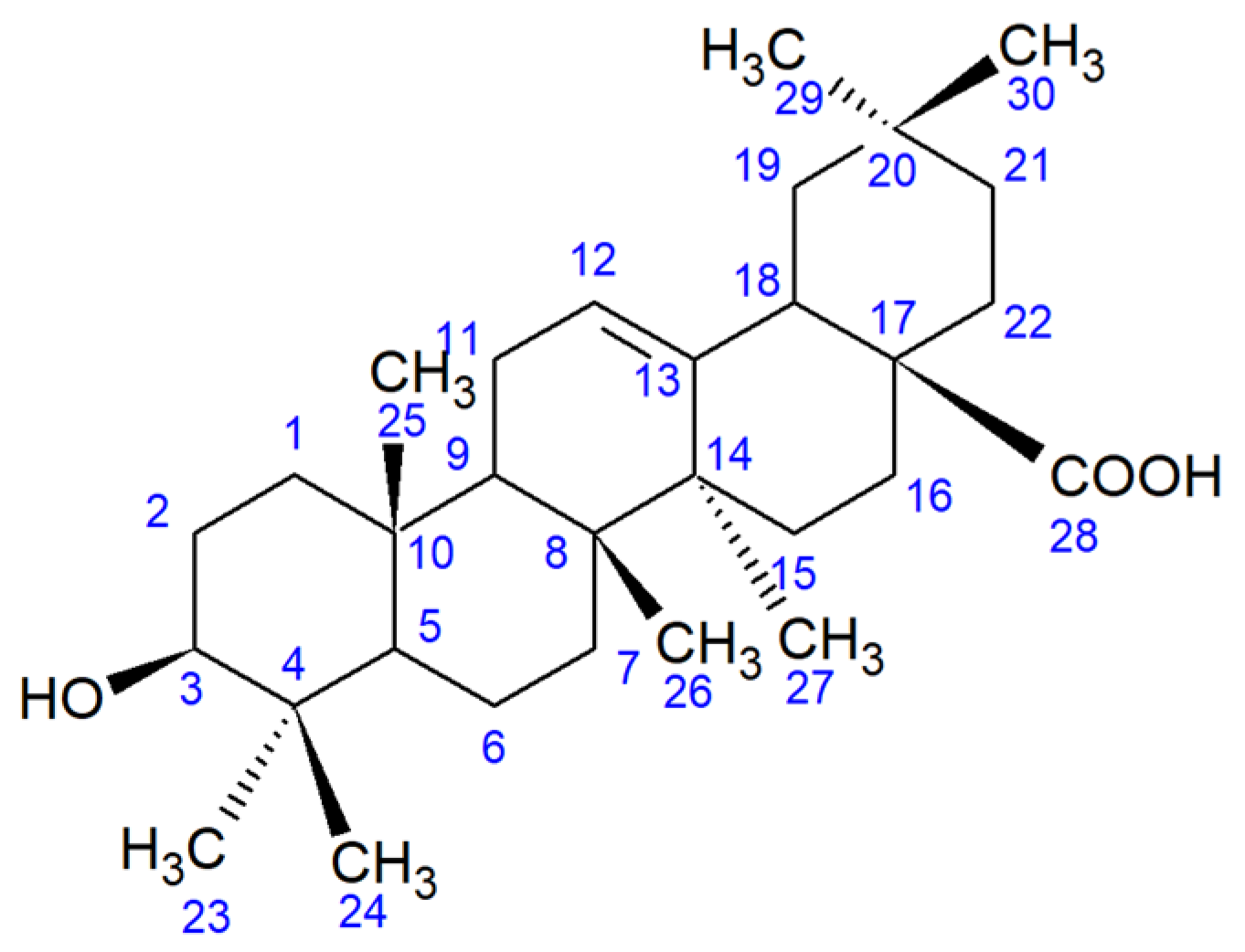
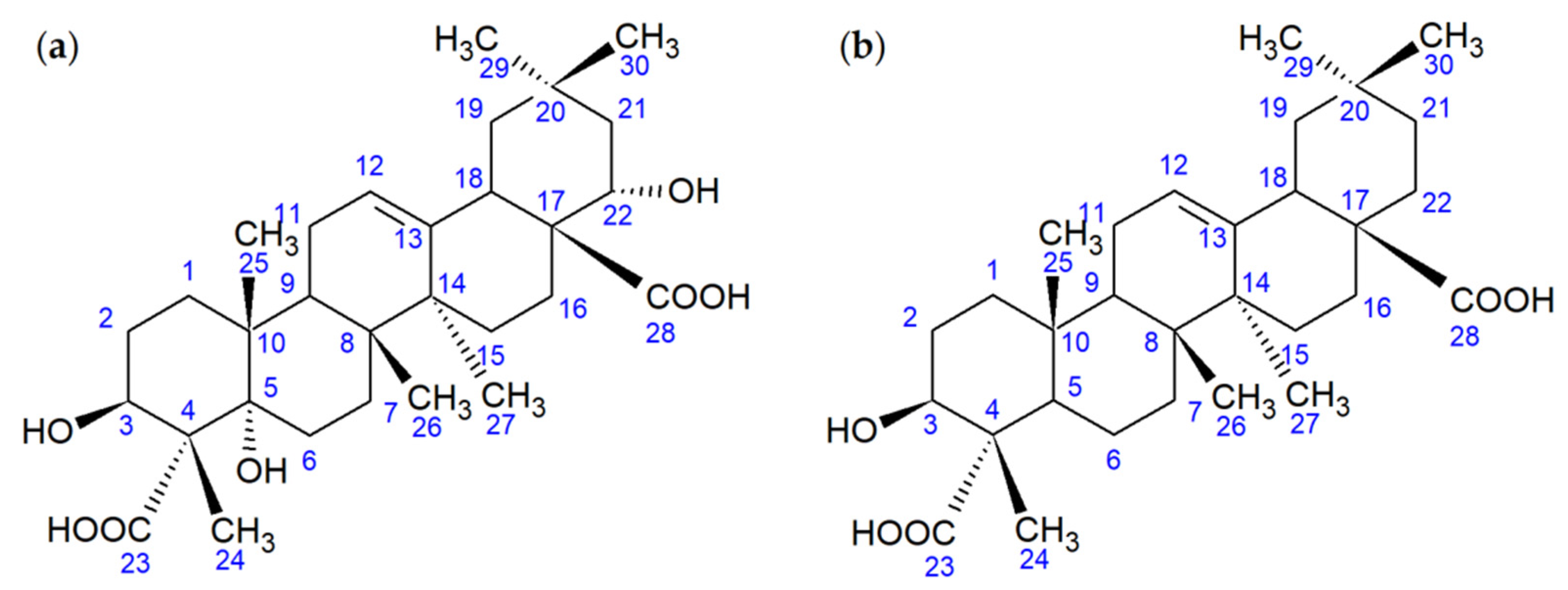
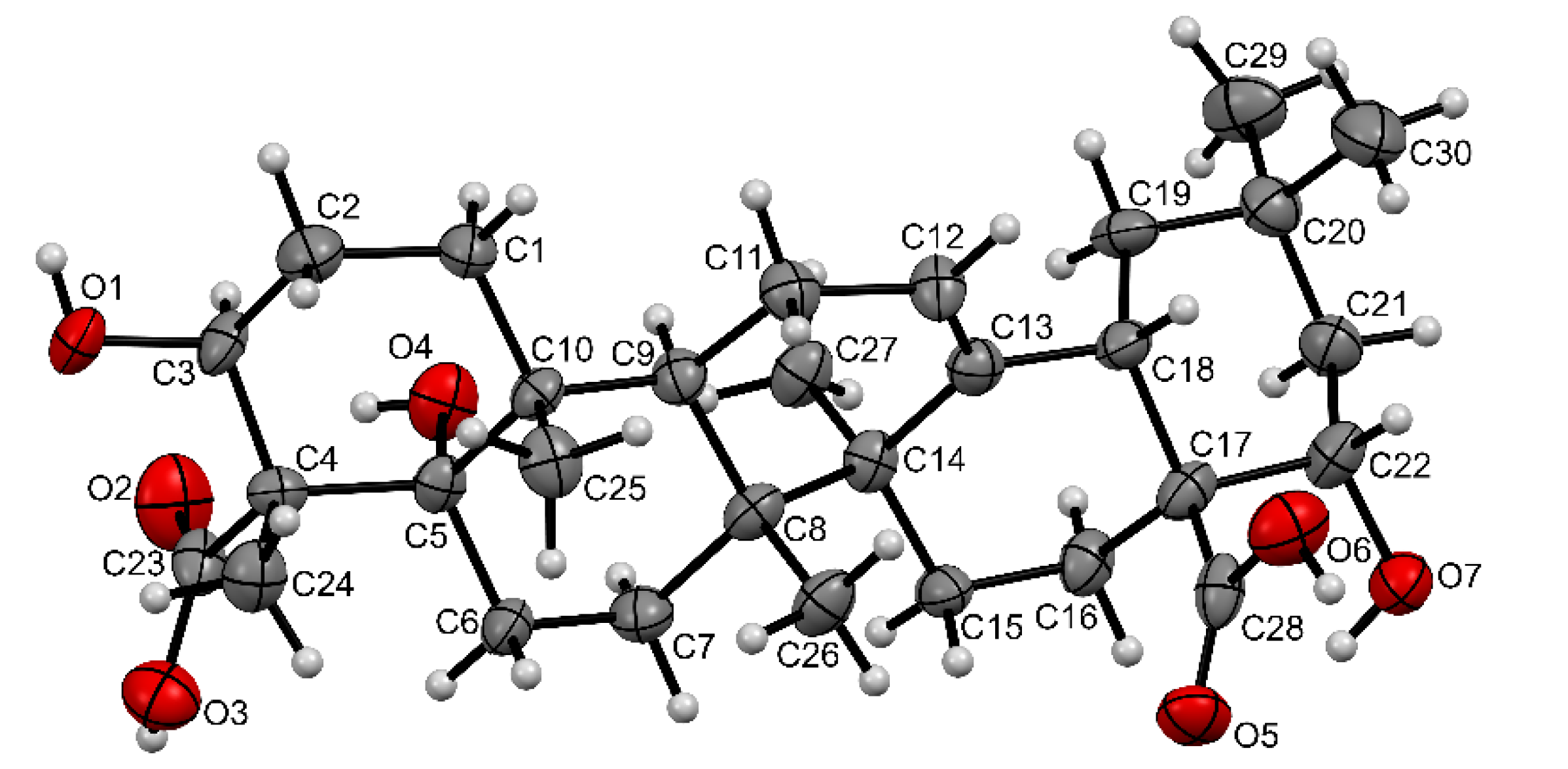
2.2. Identification and In Silico Analysis of OA Metabolite
2.3. Determination of Enzyme Systems Involved in OA Bioconversion
3. Materials and Methods
3.1. Microorganisms
3.2. Reagents
3.3. Cultivation Conditions
3.4. Controls
3.5. Preparation of Individual Cell Fractions
3.6. Microscopy
3.6.1. Phase-Contrast and Fluorescent Microscopy
3.6.2. Scanning and Transmission Electron Microscopy
3.7. Cell Viability Tests
3.8. Nile Red Staining of Bacterial Cells
3.9. Energy-Dispersive X-Ray Spectroscopy with Elemental Mapping
3.10. Respiration Activity
3.11. New-Generation Sequencing
3.12. Bioinformatics Analysis
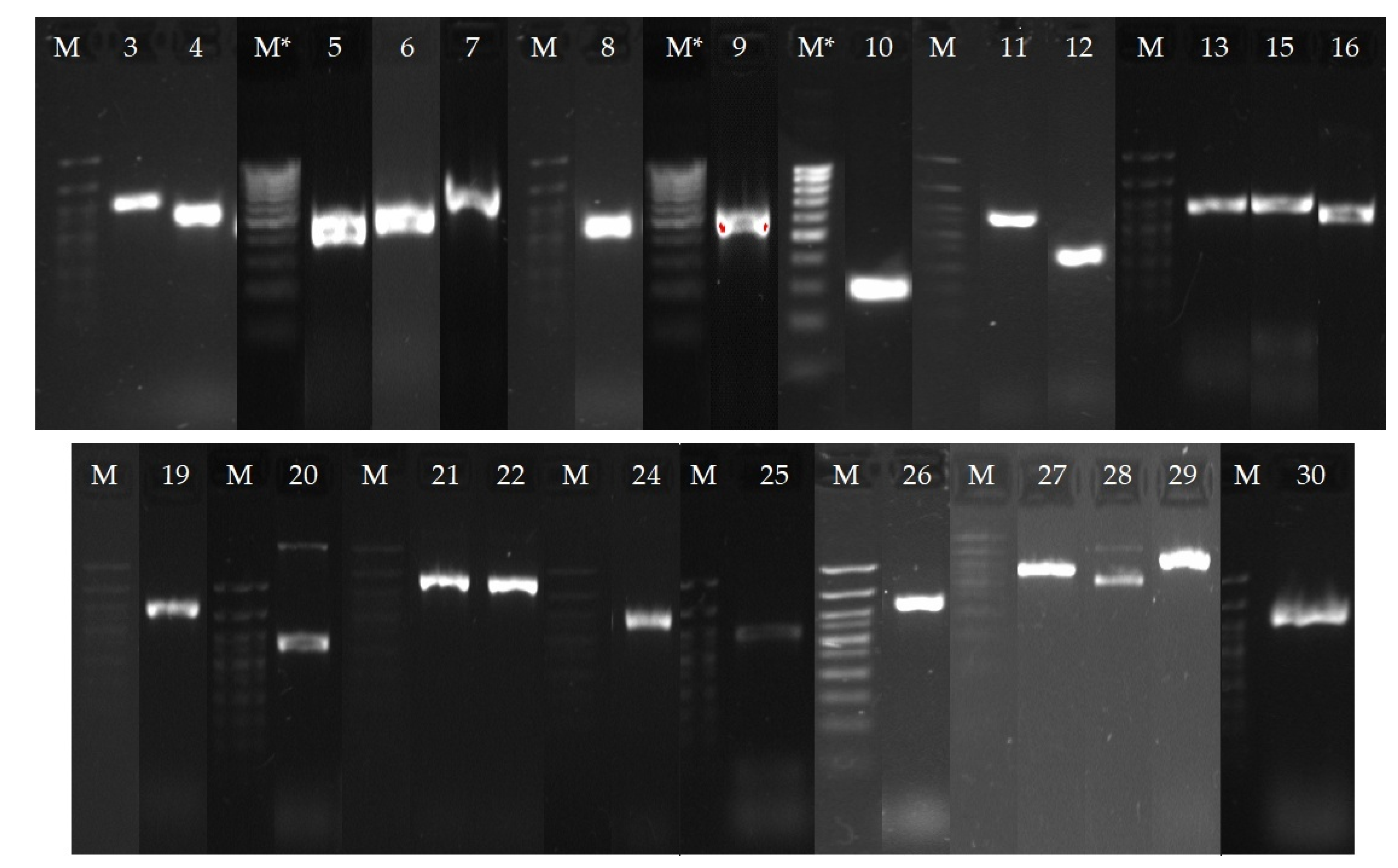
3.13. Real-Time PCR and Gel Electrophoresis
- Step 1
- 95.0 °C; 3 min.
- Step 2
- 95.0 °C; 30 s.
- Step 3
- Gradient 55.0–65.0 °C; 30 s.
- Step 4
- 72.0 °C; 1:30 min.
- Step 5
- Melt Curve from 65.0 to 95.0 °C, increment of 0.5 °C; 5 s.
- Step 6
- 72.0 °C; 10 min.
3.14. Quantitative and Qualitative Analysis of OA and Its Derivative
3.15. Isolation and Identification of OA Metabolite
3.16. In Silico Analysis of OA and Its Metabolite
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calixto, J.B. The role of natural products in modern drug discovery. An. Acad. Bras. Cienc. 2019, 91, e20190105. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Dubey, K.K. Hybrid approach for transformation for betulin (an anti-HIV molecule). In New and Future Developments in Microbial Biotechnology and Bioengineering; Gupta, V., Pandey, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 193–203. ISBN 9780444635044. [Google Scholar]
- Huang, L.R.; Luo, H.; Yang, X.S.; Chen, L.; Zhang, J.X.; Wang, D.P.; Hao, X.J. Enhancement of anti-bacterial and anti-tumor activities of pentacyclic triterpenes by introducing exocyclic α,β-unsaturated ketone moiety in ring A. Med. Chem. Res. 2014, 23, 4631–4641. [Google Scholar] [CrossRef]
- Wiemann, J.; Heller, L.; Csuk, R. Targeting cancer cells with oleanolic and ursolic acid derived hydroxamates. Bioorg. Med. Chem. Lett. 2016, 26, 907–909. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.W.; Dou, T.Y.; Wang, P.; Lei, W.; Weng, Z.M.; Hou, J.; Wang, D.D.; Fan, Y.M.; Zhang, W.D.; Ge, G.B.; et al. Structure-activity relationships of pentacyclic triterpenoids as potent and selective inhibitors against human carboxylesterase 1. Front. Pharmacol. 2017, 8, 435. [Google Scholar] [CrossRef]
- Alho, D.P.S.; Salvador, J.A.R.; Cascante, M.; Marin, S. Synthesis and antiproliferative activity of novel heterocyclic glycyrrhetinic acid derivatives. Molecules 2019, 24, 766. [Google Scholar] [CrossRef]
- Capel, C.S.; de Souza, A.C.D.; de Carvalho, T.C.; de Sousa, J.P.B.; Ambrósio, S.R.; Martins, C.H.G.; Cunha, W.R.; Galán, R.H.; Furtado, N.A.J.C. Biotransformation using Mucor rouxii for the production of oleanolic acid derivatives and their antimicrobial activity against oral pathogens. J. Ind. Microbiol. Biotechnol. 2011, 38, 1493–1498. [Google Scholar] [CrossRef]
- Martinez, A.; Rivas, F.; Perojil, A.; Parra, A.; Garcia-Granados, A.; Fernandez-Vivas, A. Biotransformation of oleanolic and maslinic acids by Rhizomucor miehei. Phytochemistry 2013, 94, 229–237. [Google Scholar] [CrossRef]
- Gong, T.; Zheng, L.; Zhen, X.; He, H.X.; Zhu, H.X.; Zhu, P. Microbial transformation of oleanolic acid by Trichothecium roseum. J. Asian Nat. Prod. Res. 2014, 16, 383–386. [Google Scholar] [CrossRef]
- Ludwig, B.; Geib, D.; Haas, C.; Steingroewer, J.; Bley, T.; Muffler, K.; Ulber, R. Whole-cell biotransformation of oleanolic acid by free and immobilized cells of Nocardia iowensis: Characterization of new metabolites. Eng. Life Sci. 2015, 15, 108–115. [Google Scholar] [CrossRef]
- Xu, S.H.; Wang, W.W.; Zhang, C.; Liu, X.F.; Yu, B.Y.; Zhang, J. Site-selective oxidation of unactivated C–H sp3 bonds of oleanane triterpenes by Streptomyces griseus ATCC 13273. Tetrahedron 2017, 73, 3086–3092. [Google Scholar] [CrossRef]
- Xu, S.H.; Chen, H.L.; Fan, Y.; Xu, W.; Zhang, J. Application of tandem biotransformation for biosynthesis of new pentacyclic triterpenoid derivatives with neuroprotective effect. Bioorg. Med. Chem. Lett. 2020, 30, 126947. [Google Scholar] [CrossRef] [PubMed]
- Ivshina, I.B.; Kuyukina, M.S.; Krivoruchko, A.V. Hydrocarbon-oxidizing bacteria and their potential in eco-biotechnology and bioremediation. In Microbial Resources; Kurtboke, I., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 121–148. ISBN 978-0-12-804765-1. [Google Scholar]
- Grishko, V.V.; Tarasova, E.V.; Ivshina, I.B. Biotransformation of betulin to betulone by growing and resting cells of the actinobacterium Rhodococcus rhodochrous IEGM 66. Process. Biochem. 2013, 48, 1640–1644. [Google Scholar] [CrossRef]
- Corno, G.; Villiger, J.; Pernthaler, J. Coaggregation in a microbial predator–prey system affects competition and trophic transfer efficiency. Ecology 2013, 94, 870–881. [Google Scholar] [CrossRef]
- Belfiore, C.; Curia, M.V.; Farías, M.E. Characterization of Rhodococcus sp. A5wh isolated from a high altitude Andean lake to unravel the survival strategy under lithium stress. Rev. Argent. Microbiol. 2018, 50, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, H.M.; Mayer, F.; Frabritius, D.; Steinbüchel, A. Formation of intracytoplasmic lipid inclusions. Arch. Microbiol. 1996, 165, 377–386. [Google Scholar] [CrossRef]
- Hernández, M.A.; Mohn, W.W.; Martínez, E.; Rost, E.; Alvarez, A.F.; Alvarez, H.M. Biosynthesis of storage compounds by Rhodococcus jostii RHA1 and global identification of genes involved in their metabolism. BMC Genom. 2008, 9, 600. [Google Scholar] [CrossRef] [PubMed]
- Cheremnykh, K.M.; Luchnikova, N.A.; Grishko, V.V.; Ivshina, I.B. Bioconversion of ecotoxic dehydroabietic acid using Rhodococcus actinobacteria. J. Hazard. Mater. 2018, 346, 103–112. [Google Scholar] [CrossRef]
- Scalon Cunha, L.C.; Andrade e Silva, M.L.; Cardoso Furtado, N.A.J.; Vinhólis, A.H.C.; Gomes Martins, C.H.; da Silva Filho, A.A.; Cunha, W.R. Antibacterial activity of triterpene acids and semi-synthetic derivatives against oral pathogens. Z. Für Nat. C 2007, 62, 668–672. [Google Scholar] [CrossRef]
- Krasteva, I.; Yotova, M.; Yosifov, D.; Benbassat, N.; Jenett-Siems, K.; Konstantinov, S. Cytotoxicity of gypsogenic acid isolated from Gypsophila trichotoma. Pharmacogn. Mag. 2014, 10, 430. [Google Scholar] [CrossRef]
- Fukushima, E.O.; Seki, H.; Sawai, S.; Suzuki, M.; Ohyama, K.; Saito, K.; Muranaka, T. Combinatorial biosynthesis of legume natural and rare triterpenoids in engineered yeast. Plant Cell Physiol. 2013, 54, 740–749. [Google Scholar] [CrossRef]
- Cunha, W.R.; Martins, C.; da Silva Ferreira, D.; Miller Crotti, A.E.; Lopes, N.P.; Albuquerque, S. In vitro trypanocidal activity of triterpenes from Miconia species. Planta Med. 2003, 69, 470–472. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, E.O.; Seki, H.; Ohyama, K.; Ono, E.; Umemoto, N.; Mizutani, M.; Saito, K.; Muranaka, T. CYP716A subfamily members are multifunctional oxidases in triterpenoid biosynthesis. Plant Cell Physiol. 2011, 52, 2050–2061. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, D.; Zapp, J.; Bernhardt, R. Hydroxylation of the triterpenoid dipterocarpol with CYP106A2 from Bacillus megaterium. FEBS J. 2012, 279, 1663–1674. [Google Scholar] [CrossRef] [PubMed]
- Çelik, A.; Flitsch, S.L.; Turner, N.J. Efficient terpene hydroxylation catalysts based upon P450 enzymes derived from actinomycetes. Org. Biomol. Chem. 2005, 3, 2930–2934. [Google Scholar] [CrossRef] [PubMed]
- Catalogue of the Strains of Regional Specialised Collection of Alkanotrophic Microorganisms. Available online: http://www.iegmcol.ru/strains/index.html (accessed on 20 September 2022).
- Postgate, J.R. Differential media for sulphur bacteria. J. Sci. Food Agric. 1959, 10, 669–674. [Google Scholar] [CrossRef]
- Neumann, G.; Veeranagouda, Y.; Karegoudar, T.B.; Sahin, Ö; Mäusezahl, I.; Kabelitz,, N.; Kappelmeyer, U.; Heipieper, H.J. Cells of Pseudomonas putida and Enterobacter sp. adapt to toxic organic compounds by increasing their size. Extremophiles 2005, 9, 163–168. [Google Scholar] [CrossRef]
- Mrunalini, B.R.; Girisha, S.T. Screening and characterization of lipid inclusions in bacteria by fluorescence microscopy and mass spectrometry as a source for biofuel production. Indian J. Sci. Technol. 2017, 10, 104166501. [Google Scholar] [CrossRef]
- CrysAlisPro, Version 1.171.37.33 (release 27-03-2014 CrysAlis171.NET); Agilent Technologies: Yarnton, UK, 2014.
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Spek, A.L. PLATON SQUEEZE: A tool for the calculation of the disordered solvent contribution to the calculated structure factors. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Way2Drug Predictive Services. PASS Online. Available online: http://www.pharmaexpert.ru/passonline/index.php (accessed on 20 September 2022).
 ), abiotic control (
), abiotic control ( ). OD630 in the presence of OA (
). OD630 in the presence of OA ( ) and without it (
) and without it ( ). HPLC data of the post-cultural extracts are presented.
). HPLC data of the post-cultural extracts are presented.
 ), abiotic control (
), abiotic control ( ). OD630 in the presence of OA (
). OD630 in the presence of OA ( ) and without it (
) and without it ( ). HPLC data of the post-cultural extracts are presented.
). HPLC data of the post-cultural extracts are presented.
 ). Biotic (
). Biotic ( ) and abiotic (
) and abiotic ( ) controls.
) controls.
 ). Biotic (
). Biotic ( ) and abiotic (
) and abiotic ( ) controls.
) controls.
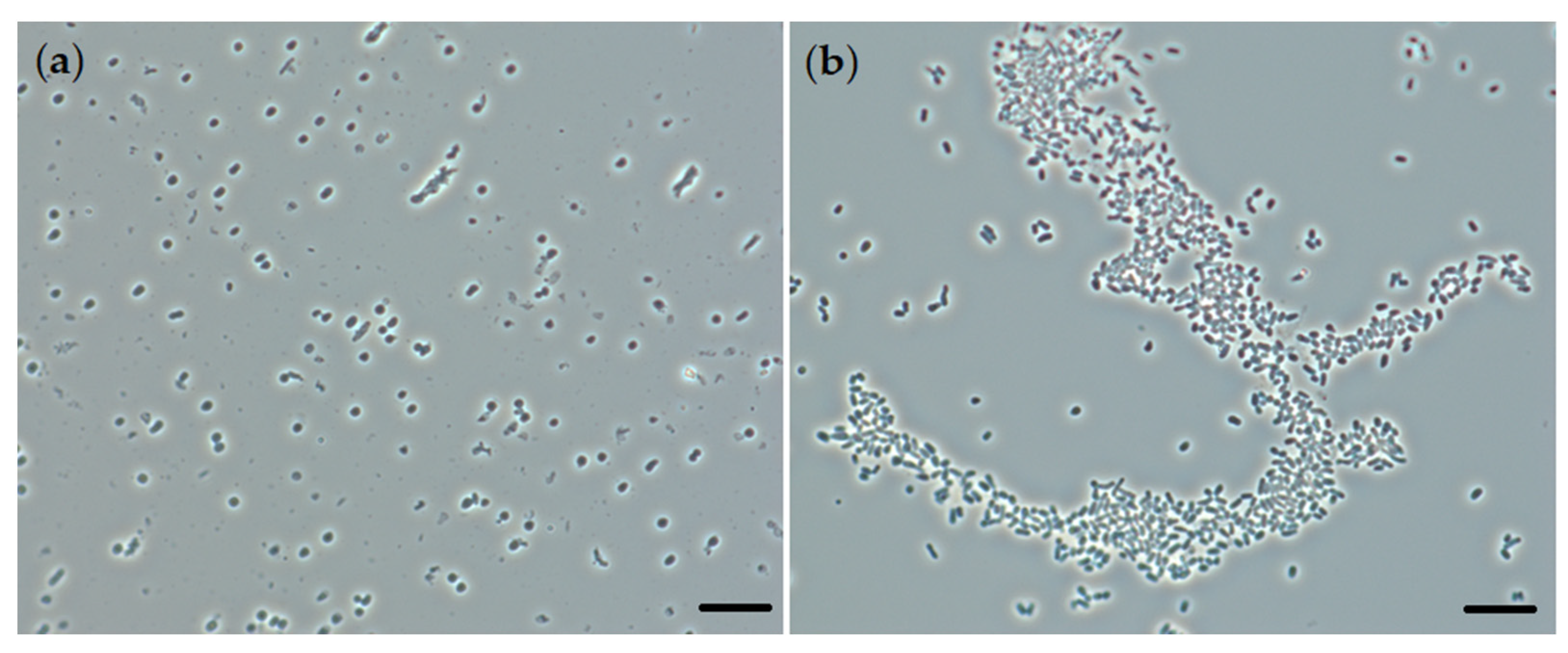
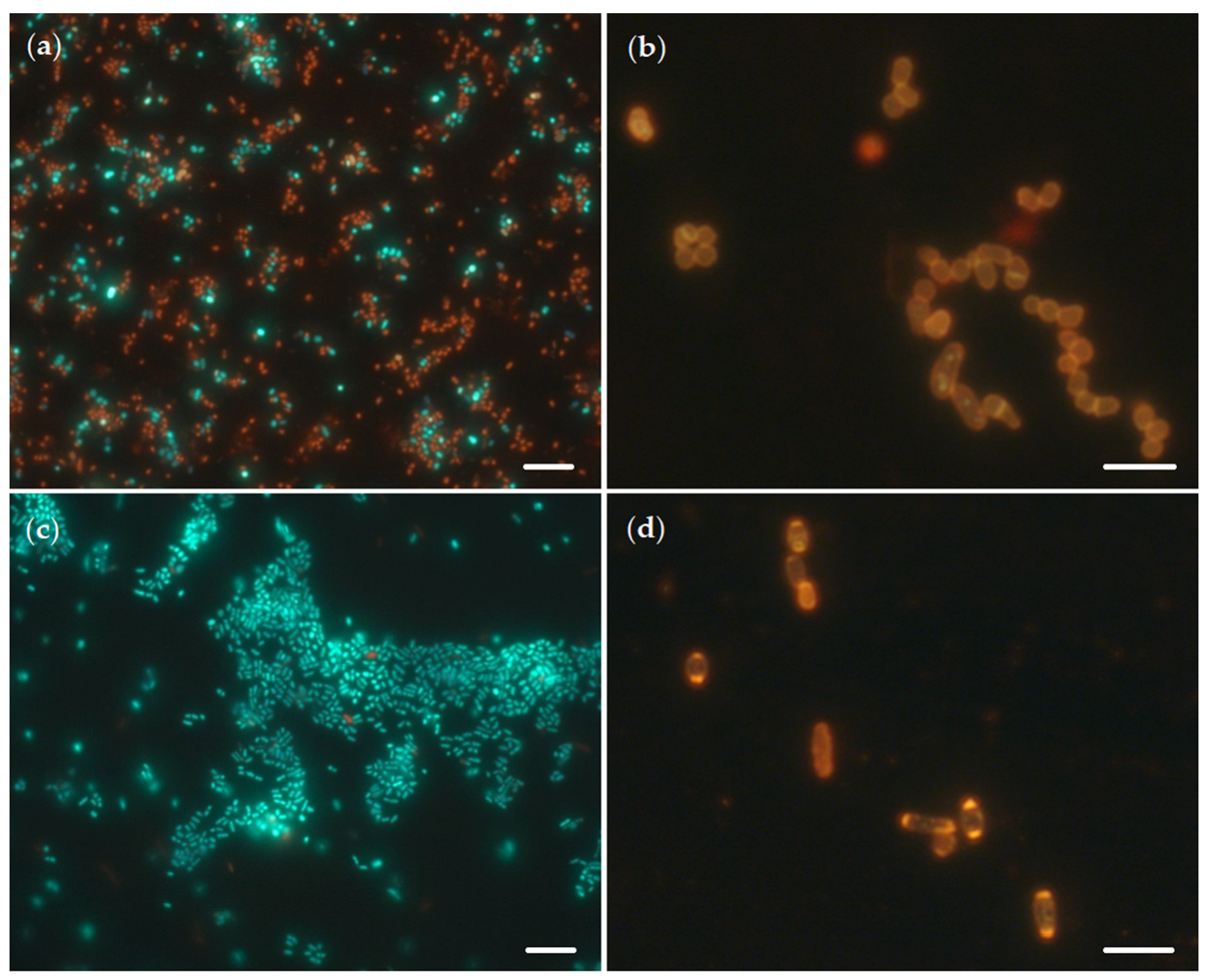

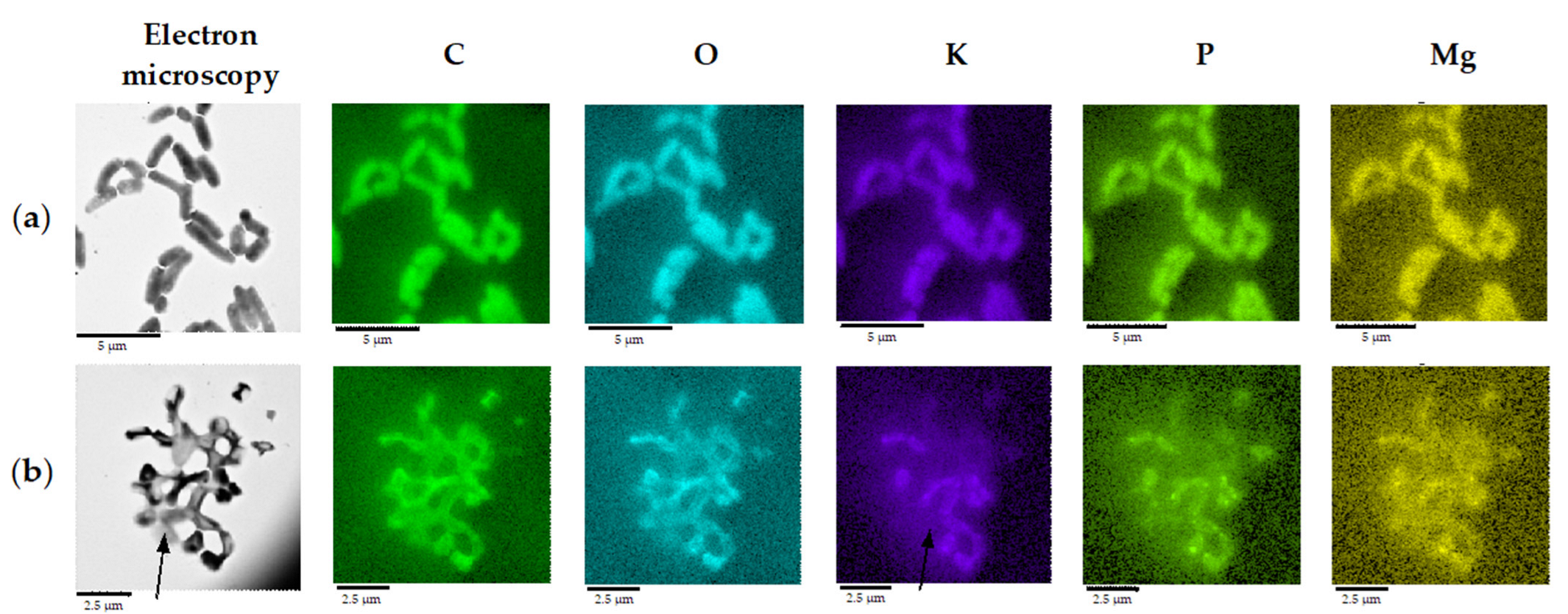
| Conditions | Length, µm | Width, µm | Area, µm2 | Volume, µm3 | Relative Area, µm−1 |
|---|---|---|---|---|---|
| Biotic control | 1.33 ± 0.09 | 1.01 ± 0.09 | 5.85 ± 0.71 | 1.08 ± 0.21 | 5.49 ± 0.35 |
| OA | 1.89 ± 0.33 | 1.00 ± 0.09 | 7.51 ± 1.36 | 1.49 ± 0.38 | 5.12 ± 0.46 |
| Test Organisms (Index, Exposure) | Concentration, mg/L | |
|---|---|---|
| OA | OA Metabolite | |
| Water solubility at 25 °C | 0.0018 | 1.391 |
| ECOSAR Class | Neutral Organics-acid | Neutral Organics-acid |
| Acute toxicity | ||
| Fish (LD50, 96 h) | 0.018 | 192.78 |
| Daphnia (LD50, 48 h) | 0.018 | 125.69 |
| Green algae (ED50, 96 h) | 0.127 | 165.80 |
| Chronic toxicity | ||
| Fish (ED50, 30 days) | 0.003 | 22.18 |
| Daphnia (ED50, 21 days) | 0.008 | 18.0 |
| Green algae (ED50, 16 days) | 0.112 | 59.07 |
| Estimated Activity | OA | OA Metabolite | ||
|---|---|---|---|---|
| Pa | Pi | Pa | Pi | |
| Apoptosis agonist | – | – | 0.908 | 0.004 |
| Insulin promoter | – | – | 0.906 | 0.003 |
| Chemoprophylactic | – | – | 0.892 | 0.002 |
| Analgesic | – | – | 0.778 | 0.002 |
| NFκB transcription factor stimulator | 0.908 | 0.001 | 0.946 | 0.001 |
| Antitumor | 0.810 | 0.010 | 0.823 | 0.009 |
| Hepatoprotective | 0.889 | 0.003 | 0.884 | 0.003 |
| Cell Fractions | Concentration, % * | |
|---|---|---|
| OA | OA Derivatives | |
| Supernatant with cytoplasmic enzymes (I) | 100.0 | 0.0 |
| Supernatant with extracted membrane-bound enzymes (II) | 35.5 | 0.0 |
| Resuspended cell precipitate with non-extractable enzymes (III) | 48.8 | 2.49 |
| Gene Number | Protein ID (Contig Number *) | Position | Size, bp |
|---|---|---|---|
| 1 | MCD2110228.1 (2) | c175904–177292 | 1389 |
| 2 | MCD2110230.1 (2) | c178067–179254 | 1188 |
| 3 | MCD2110232.1 (2) | c179967–181193 | 1227 |
| 4 | MCD2110413.1 (2) | c382010–383263 | 1254 |
| 5 | MCD2110571.1 (2) | 547960–549255 | 1296 |
| 6 | MCD2111197.1 (4) | 199296–200513 | 1208 |
| 7 | MCD2111485.1 (5) | c47424–48707 | 1284 |
| 8 | MCD2111502.1 (5) | c63058–64437 | 1380 |
| 9 | MCD2111603.1 (5) | 175155–176378 | 1214 |
| 10 | MCD2111604.1 (5) | 176468–177697 | 1210 |
| 11 | MCD2111618.1 (5) | 192090–193325 | 1236 |
| 12 | MCD2111684.1 (5) | c256103–257449 | 1347 |
| 13 | MCD2111707.1 (5) | c277043–278335 | 1293 |
| 14 | MCD2111739.1 (5) | c307984–309330 | 1337 |
| 15 | MCD2111762.1 (5) | 334358–335692 | 1335 |
| 16 | MCD2111873.1 (5) | c459464–>460666 | 1203 |
| 17 | MCD2112909.1 (9) | c117094–118344 | 1251 |
| 18 | MCD2113293.1 (11) | c76311–77486 | 1176 |
| 19 | MCD2113487.1 (12) | c78090–79403 | 1314 |
| 20 | MCD2113587.1 (12) | c190616–191827 | 1212 |
| 21 | MCD2114161.1 (17) | c40856–42100 | 1245 |
| 22 | MCD2114185.1 (17) | 64958–66229 | 1272 |
| 23 | MCD2114186.1 (17) | c66305–67549 | 1245 |
| 24 | MCD2114191.1 (17) | c71586–72848 | 1263 |
| 25 | MCD2114309.1 (19) | 19116–20480 | 1365 |
| 26 | MCD2114346.1 (20) | c1724–3109 | 1386 |
| 27 | MCD2114626.1 (25) | c8950–10212 | 1263 |
| 28 | MCD2114645.1 (25) | c30579–31841 | 1263 |
| 29 | MCD2114721.1 (27) | 27270–28601 | 1332 |
| 30 | MCD2115052.1 (44) | 4294–6654 | 2361 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luchnikova, N.A.; Grishko, V.V.; Kostrikina, N.A.; Sorokin, V.V.; Mulyukin, A.L.; Ivshina, I.B. Biotransformation of Oleanolic Acid Using Rhodococcus rhodochrous IEGM 757. Catalysts 2022, 12, 1352. https://doi.org/10.3390/catal12111352
Luchnikova NA, Grishko VV, Kostrikina NA, Sorokin VV, Mulyukin AL, Ivshina IB. Biotransformation of Oleanolic Acid Using Rhodococcus rhodochrous IEGM 757. Catalysts. 2022; 12(11):1352. https://doi.org/10.3390/catal12111352
Chicago/Turabian StyleLuchnikova, Natalia A., Victoria V. Grishko, Nadezhda A. Kostrikina, Vladimir V. Sorokin, Andrey L. Mulyukin, and Irina B. Ivshina. 2022. "Biotransformation of Oleanolic Acid Using Rhodococcus rhodochrous IEGM 757" Catalysts 12, no. 11: 1352. https://doi.org/10.3390/catal12111352
APA StyleLuchnikova, N. A., Grishko, V. V., Kostrikina, N. A., Sorokin, V. V., Mulyukin, A. L., & Ivshina, I. B. (2022). Biotransformation of Oleanolic Acid Using Rhodococcus rhodochrous IEGM 757. Catalysts, 12(11), 1352. https://doi.org/10.3390/catal12111352








