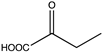
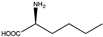
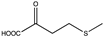
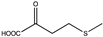
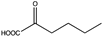
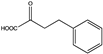
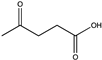
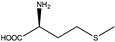
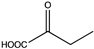
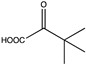
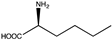
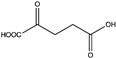
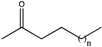
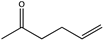
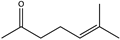
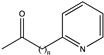
Abstract
Amino acid dehydrogenases (AADHs) are a group of enzymes that catalyze the reversible reductive amination of keto acids with ammonia to produce chiral amino acids using either nicotinamide adenine dinucleotide (NAD+) or nicotinamide adenine dinucleotide phosphate (NADP+) as cofactors. Among them, glutamate dehydrogenase, valine dehydrogenase, leucine dehydrogenase, phenylalanine dehydrogenase, and tryptophan dehydrogenase have been classified as a superfamily of amino acid dehydrogenases (s-AADHs) by previous researchers because of their conserved structures and catalytic mechanisms. Owing to their excellent stereoselectivity, high atom economy, and low environmental impact of the reaction pathway, these enzymes have been extensively engineered to break strict substrate specificities for the synthesis of high value-added chiral compounds (chiral amino acids, chiral amines, and chiral amino alcohols). Substrate specificity engineering of s-AADHs mainly focuses on recognition engineering of the substrate side chain R group and substrate backbone carboxyl group. This review summarizes the reported studies on substrate specificity engineering of s-AADHs and reports that this superfamily of enzymes shares substrate specificity engineering hotspots (the inside of the pocket, substrate backbone carboxyl anchor sites, substrate entrance tunnel, and hinge region), which sheds light on the substrate-specific tailoring of these enzymes.
1. Introduction
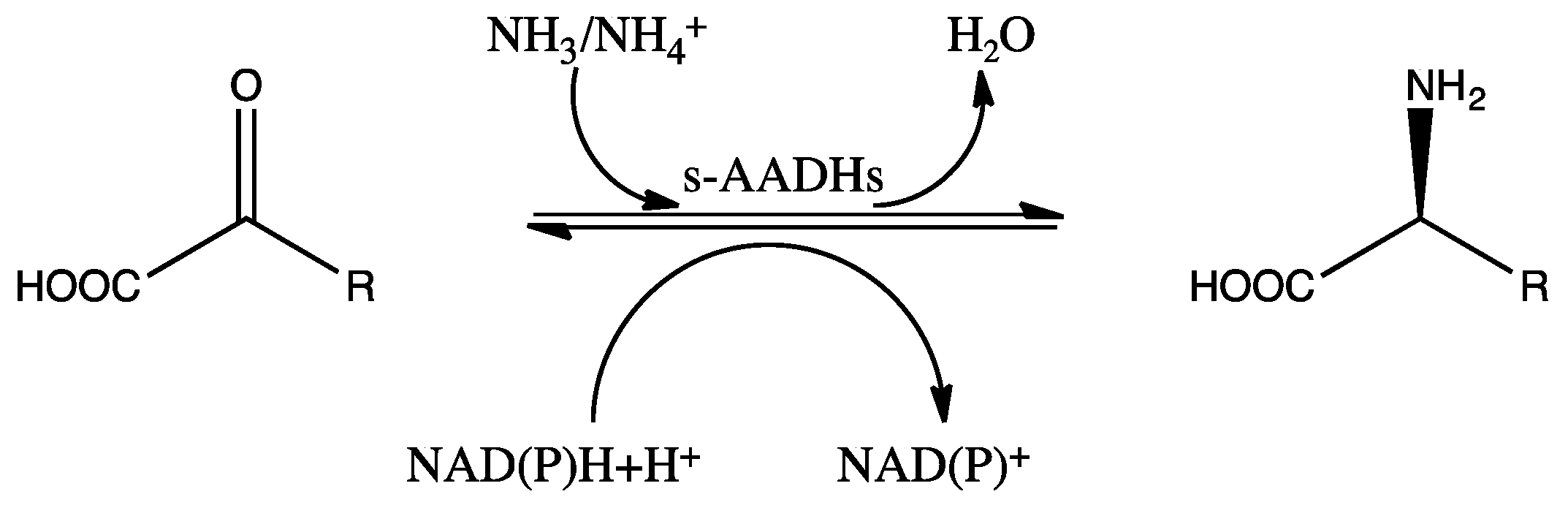
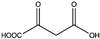
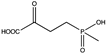
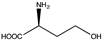
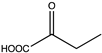
Amino acid dehydrogenases (AADHs, EC 1.4.1.X) catalyze the reversible nicotinamide adenine dinucleotide phosphate (NAD(P)+)-dependent oxidative deamination of L-amino acids to their corresponding α-keto acids and ammonia (Scheme 1), which have important physiological functions in organisms [1,2,3,4].
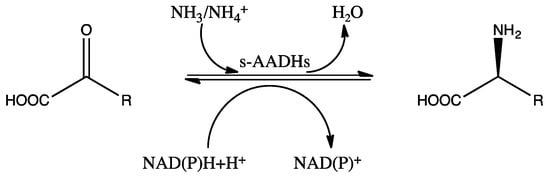
Scheme 1.
Schematic diagram of the reaction catalyzed by s-AADHs.
Among AADHs is a superfamily of amino acid dehydrogenases (s-AADHs) related by divergent evolution [5,6], which, to date, contains glutamate dehydrogenase (GluDH), valine dehydrogenase (ValDH) [7], leucine dehydrogenase (LeuDH), phenylalanine dehydrogenase (PheDH), and tryptophan dehydrogenase (TryDH) [8]. The equilibrium of reactions catalyzed by s-AADHs is preferred for reductive amination, with Keq values ranging from 1014 to 1018 [9]. Additionally, the reaction has high optical purity, high atom utilization, and environmentally friendly characteristics, which is very attractive for the synthesis of chiral amino acids [10,11]. s-AADHs usually exhibit distinct substrate specificity. GluDHs prefer glutamate as a substrate over all other amino acids [12,13], whereas ValDHs and LeuDHs only catalyze the oxidative deamination of short, aliphatic amino acids [14], and PheDH and especially TryDH have a marked preference for aromatic amino acids [8,15] (Table 1).

Table 1.
The superfamily of amino acid dehydrogenases.
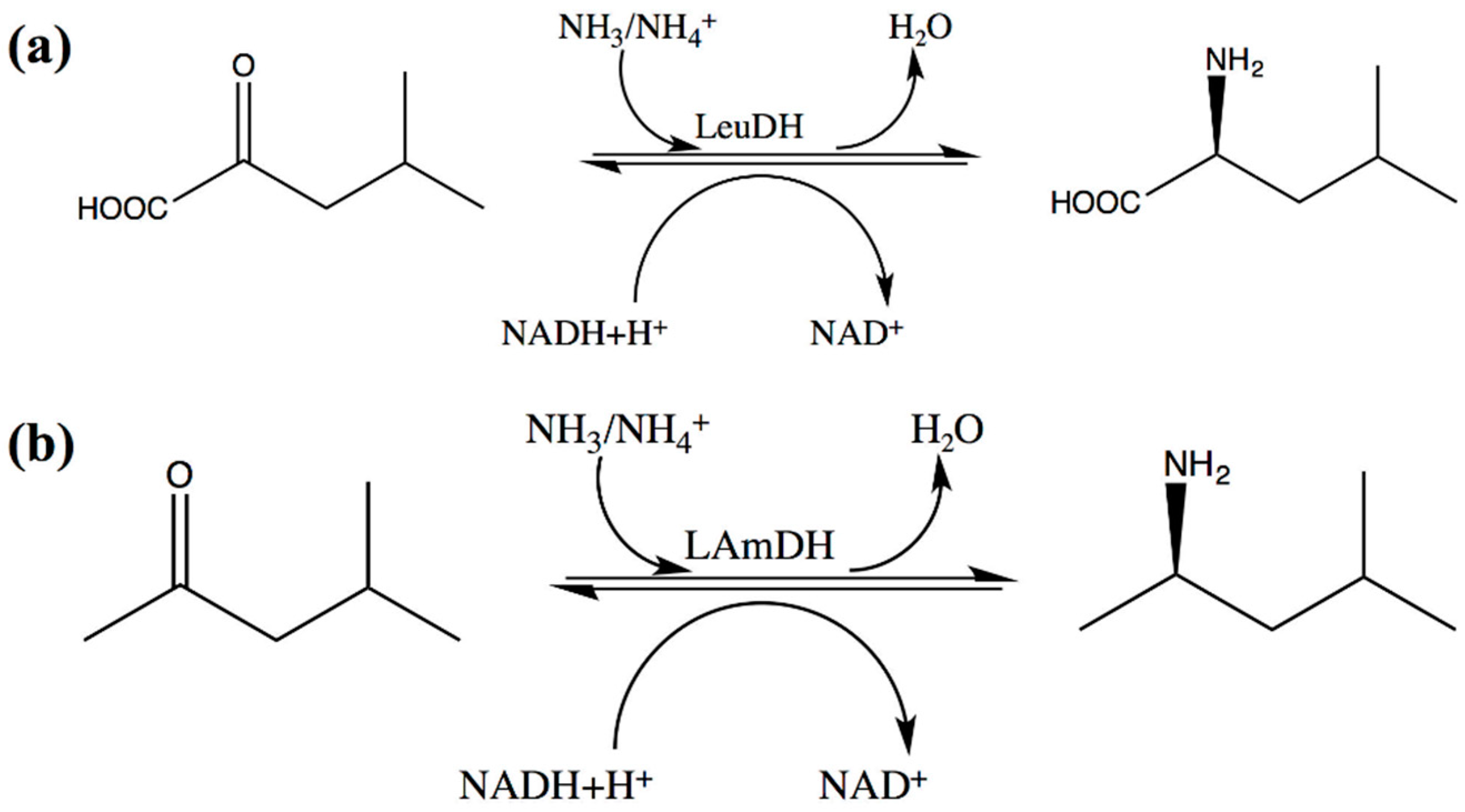
The strict substrate spectrum limits the application scope of s-AADHs. Accordingly, many studies have been conducted to expand the substrate specificity of s-AADHs [16,17,18]. The substrate-specific engineering of s-AADHs can be mainly divided into two aspects, substrate side chain R group recognition engineering and substrate backbone carboxyl recognition engineering. The former mainly enhances the ability of s-AADHs to synthesize optically pure non-canonical L-α-amino acids, including homoalanine [18,19], L-tert-leucine [20], para-bromo-L-phenylalanine [21], and L-homophenylalanine [22]. Mutations associated with this type of substrate-specific engineering are mainly concentrated inside the substrate pocket. In 2012, Abrahamson et al. [23,24,25,26] successfully converted LeuDH into an engineered amine dehydrogenase (LAmDH). In contrast to wild-type LeuDH, LAmDH catalyzes the asymmetric reductive amination of ketones, mainly owing to mutations in two residues anchored to the carboxyl group of the substrate (K68S/N261L) (Scheme 2). The anchor sites of the substrate carboxyl group of s-AADHs have become a new mutation hotspot [26,27,28]. The subsequently constructed engineered amine dehydrogenase can asymmetrically synthesize a series of chiral amines that play an important role in the pharmaceutical industry, such as (R)-amphetamine, (R)-1-methyl-3-phenylpropylamine [28], and (S)-2-amino-1-butanol [24]. In addition, other potential mutation hotspots (the substrate entrance tunnel and hinge) were found in the substrate-specific engineering of s-AADHs, and the mutation in these regions can effectively improve the catalytic activity of the engineered enzyme for non-natural substrates [29,30]. Owing to the similarity in structure and catalytic mechanism of s-AADHs, these mutational hotspots have certain universality for the substrate-specific engineering of these enzymes. This review summarizes the substrate-specific modification and important engineering hotspots of s-AADHs based on reported studies, which will shed light on the substrate-specific tailoring of this superfamily of enzymes.
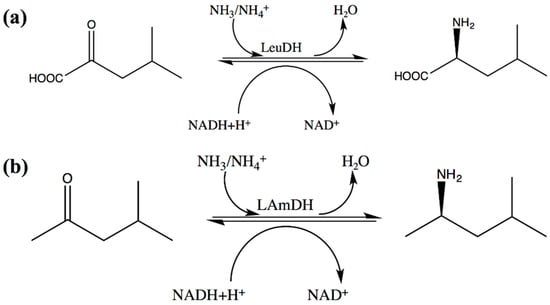
Scheme 2.
The reaction of LeuDH and its corresponding engineered amine dehydrogenase. (a) Wild-type LeuDH reaction; (b) LAmDH reaction.
2. Enzyme Structure and Catalytic Mechanism of s-AADHs
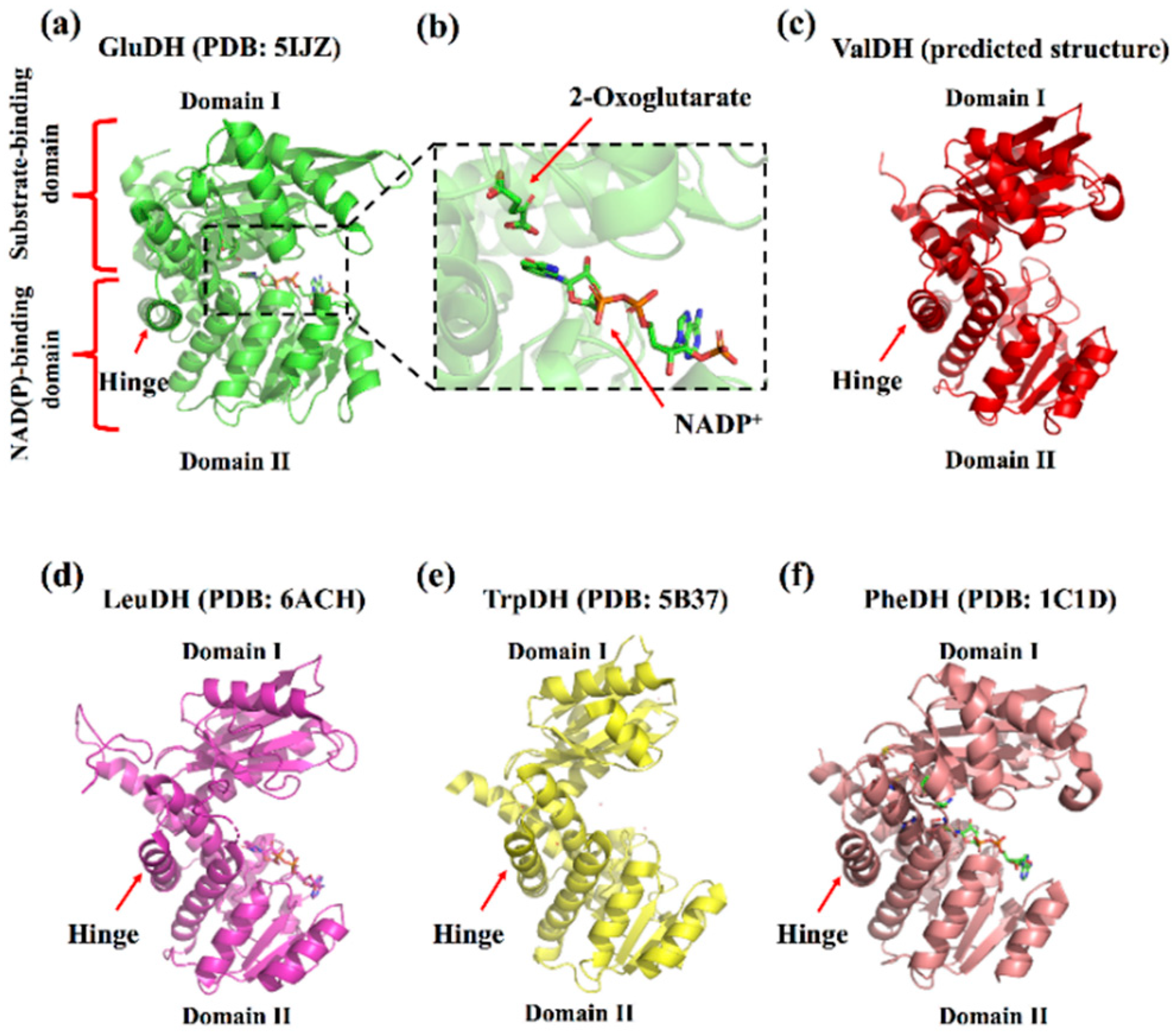
Although the five types of amino acid dehydrogenases in this superfamily have different optimal reaction substrates, they have high similarity in their crystal structures (no studies have reported the crystal structure of ValDH, but it has a high sequence identity with LeuDH) [6,8,14]. A single subunit of these enzymes primarily consists of two domains (domains I and II) that are separated by a deep cleft containing the active site. Domain I includes the substrate-binding site, whereas domain II contains a typical Rossmann fold for the binding of NAD(P)+/NAD(P)H. Additionally, there is “hinge” between the two domains, which plays an important role in catalysis [31] (Figure 1).
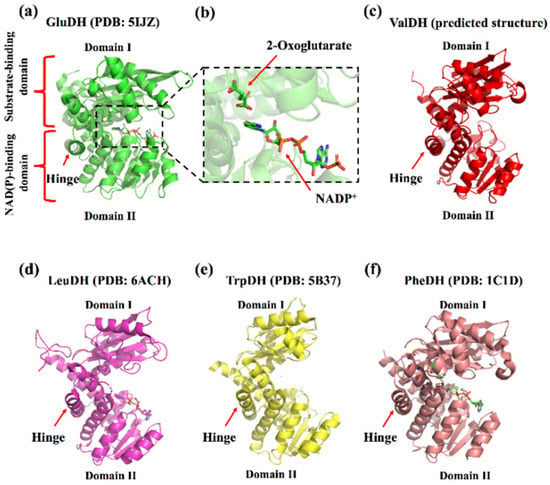
Figure 1.
Representative crystallography structures of s-AADHs subunit. (a) GluDH (PDB ID: 5IJZ). (b) Overview of active site of GluDH (PDB ID: 5IJZ). (c) ValDH (predicted by AlphaFold, https://alphafold.ebi.ac.uk/entry/Q06539 (accessed on 9 December 2021)). (d) LeuDH (PDB ID: 6ACH). (e) TrpDH (PDB: 5B37). (f) PheDH (PDB ID: 1C1D).
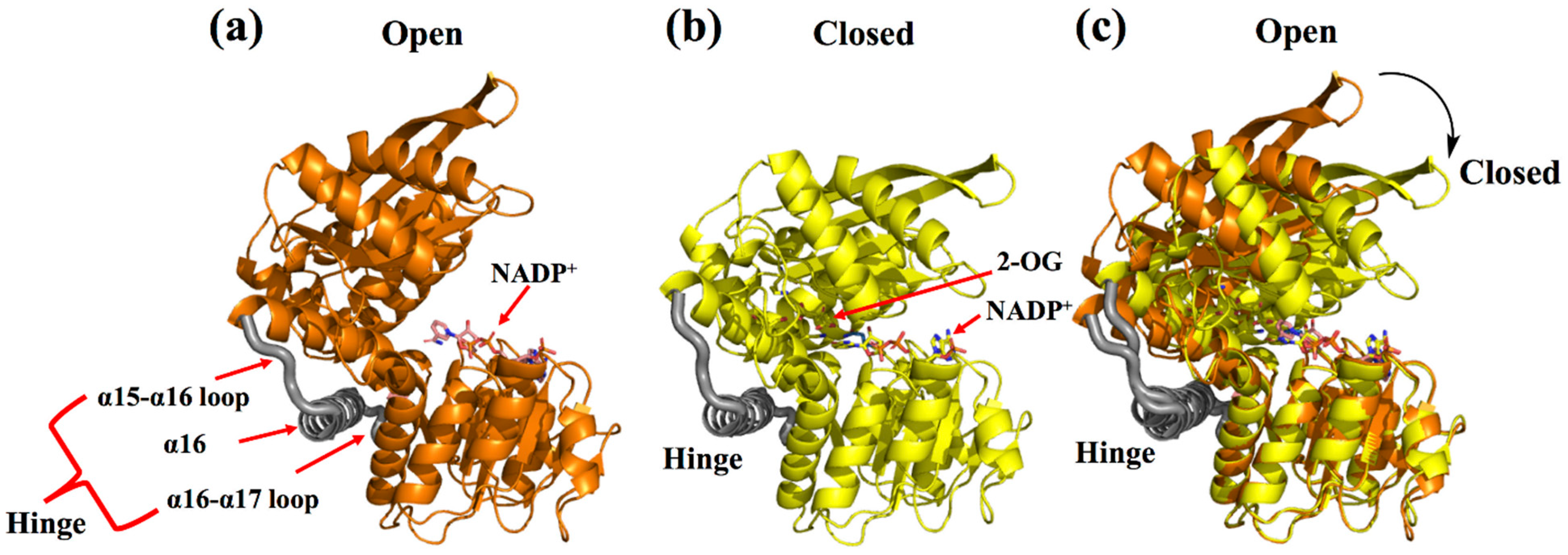
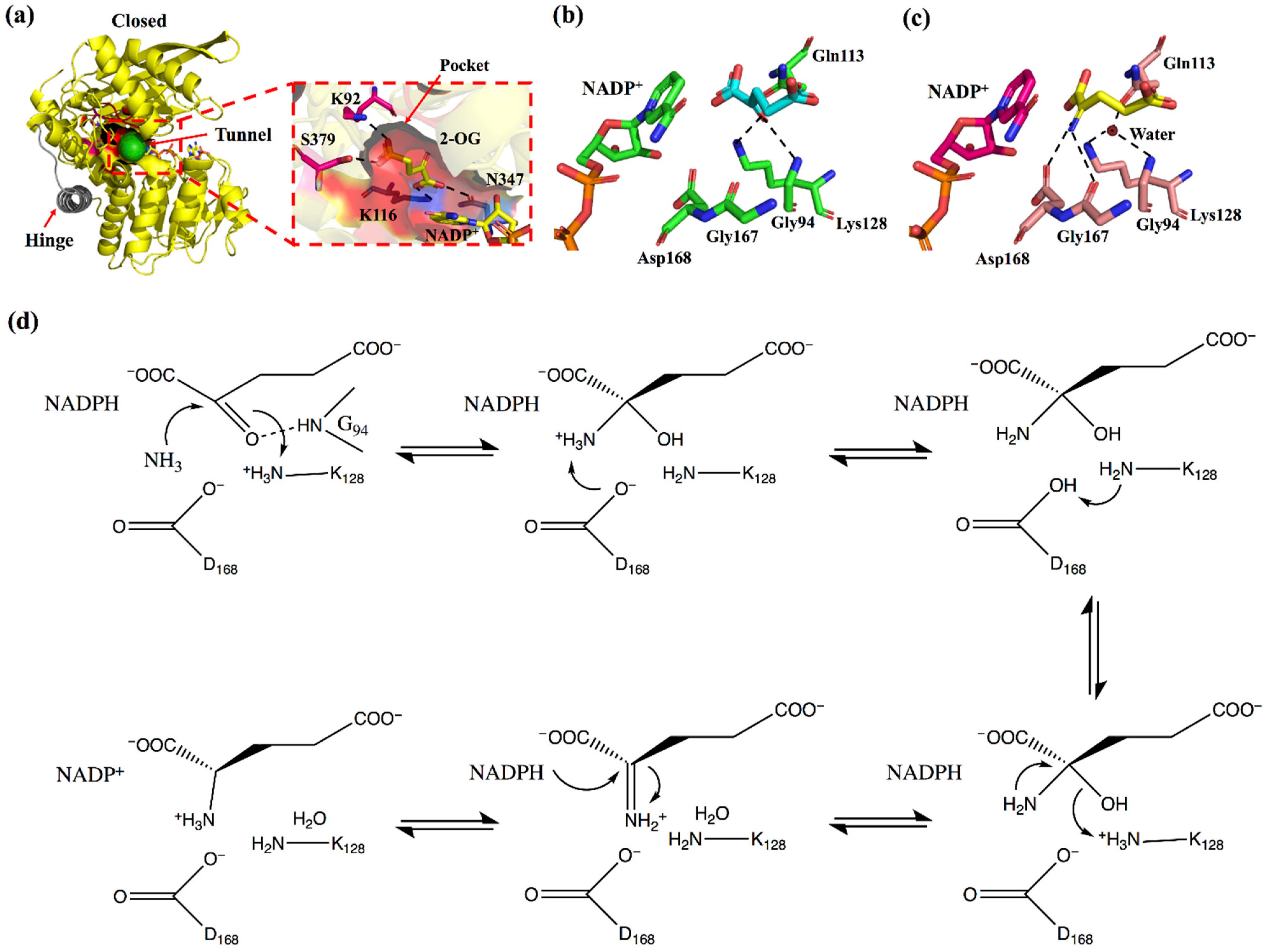
This review considers GluDH from Corynebacterium glutamicum (CgGluDH) as an example to describe the catalytic mechanism [32]. In the catalytic process, the “hinge” plays a key role [31]. As shown in Figure 2, the hinge region (in gray) is composed of two connecting loops (α15-α16 and α16-α17) and α16 helix. Domain I (bound to substrate) and Domain II (bound to coenzyme) of GluDH are linked together by the hinge. Before the hinge undergoes a conformational change, the distance between the Domain I and the Domain II is far away (Figure 2a), and the catalysis cannot take place. However, when the flexible loop on the hinge region undergoes a conformational change, Domain I and Domain II come close to each other, allowing the substrate and coenzyme to interact (Figure 2b). When 2-oxoglutarate (2-OG) enters the red substrate pocket through the green substrate entrance tunnel (calculated by CAVER plugin of PyMOL) [33] and binds to the enzyme, the protein in an open state undergoes a conformational change to a closed state, with α16 helix playing a role as an axis in the domain movement (Figure 2c and Figure 3a). The 1-carboxyl group and 5-carboxyl group of 2-OG are fixed by four residues (Lys116, Asn347, Lys92, and Ser379), in which Lys116 and Asn347, interacting with the 1-carboxyl group, occur outside the pocket, whereas Lys92 and Ser379, interacting with the 5-carboxyl group, are inside the pocket (Figure 3a). The 2-oxo group of 2-OG is recognized by the ε-amino group of Lys128 through an electrostatic interaction and by Gly94 through a hydrogen bond (Figure 3b). The space between 2-OG and NADPH appears to accept an ammonia molecule through hydrogen bonds with Gly167 and Asp168. Through an ammonia attack on the carbonyl-C atom of the ligand and deprotonation of Lys128, a carbinolamine intermediate is formed. Subsequently, the deprotonated Lys128 obtains the proton of Asp168 and is protonated again. The intermediate uses protonated Lys128 to eliminate water molecules (stabilized by a strong charge-assisted hydrogen bond interaction with Lys128 and by two additional weaker hydrogen bonds with Gly94 and Gln113 before elimination) and forms 2-iminoglutarate (2-IG), which is stabilized through hydrogen bonds with Asp168 and Gly167 (Figure 3c). During the process, the 1-carboxyl and 5-carboxyl groups of the ligand are fixed at similar positions, whereas the 2-imino group of 2-IG is twisted approximately 90° from the 2-oxo group of 2-OG. The newly formed 2-imino group of 2-IG stacks with the plane of the nicotinamide ring in NADPH, and their relative positions determine the configuration of the product. The pro-S hydrogen at the C-4 position of the nicotinamide moiety of NADPH is transferred to the 2-imino group of 2-IG and finally forms L-glutamic acid (Figure 3d).
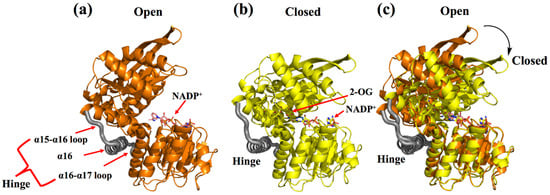
Figure 2.
Domain conformational change of CgGDH. (a) The open conformations of CgGDH binding NADP+ (the D chain of 5GUD with orange). (b) The closed conformations of CgGDH binding 2-OG and NADP+ (the A chain of 5IJZ in yellow). (c) Superposition of an open form of CgGluDH and a closed form of CgGluDH. The hinge (α15-α16 loop, α16-α17 loop, and α16 helix) is marked in gray.
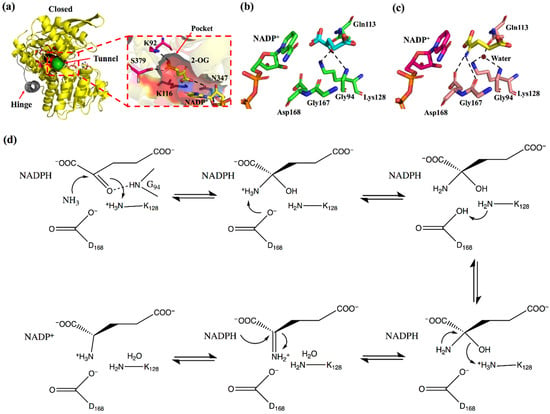
Figure 3.
The reaction mechanism of GluDH. (a) The closed conformations of CgGDH binding 2-OG and NADP+ (the A chain of 5IJZ in yellow), in which the inside of the binding pocket is marked in red, the substrate backbone carboxyl anchor sites (K116 and N347) outside the pocket are marked in blue, the substrate entrance tunnel is marked in green, and the hinge is marked in gray. (b) The CgGluDH·2-OG·NADP+ complex (the A chain of 5IJZ). (c) The CgGluDH·2-IG·NADP+ complex (the F chain of 5GUD). (d) A schematic model of the reaction for the production of glutamate from 2-OG.
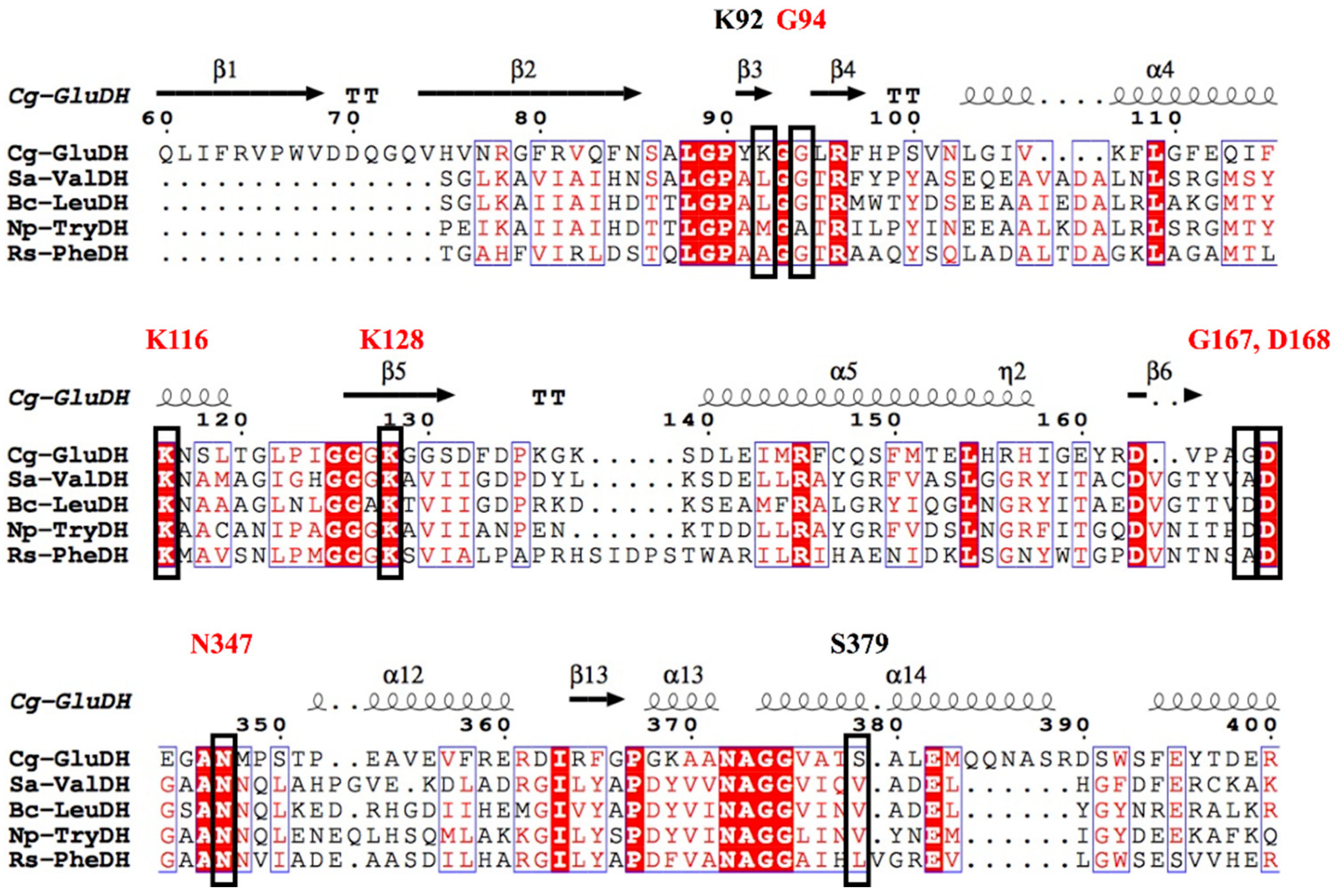
The residues that play key roles in the catalytic process are Gly94 and Lys128, which recognize the 2-oxo group; Asp168 and Gly167, which stabilize the 2-imino group; Lys116 and Asn347, which fix the 1-carboxyl group; and Lys92 and Ser379, which fix the 5-carboxyl group. The amino acid sequences of s-AADHs with different substrate specificities were compared, and Lys116, Asn347, Lys128, and Asp168 were found to be completely conserved, indicating that these residues might play the same role in the catalytic process (Figure 4). The catalytic mechanism of PheDH proposed by Norbert et al. [34] is the same as that of CgGluDH. The Gly167 residue in GluDH is replaced by Ala in PheDH, but both residues form a hydrogen bond with the amino group of the ligand through the main chain carbonyl-O atom. As shown in Figure 4, Lys92 and Ser379, which fix the 5-carboxyl group of the ligand during the catalysis of CgGluDH, are replaced with hydrophobic amino acids (A, V, L, and M) in other s-AADHs. These residues could be related to the recognition of substrates (such as Val, Leu, Phe, and Trp) [12,35]. Next, this review will separately describe the alterations to the substrate specificity of s-AADHs.
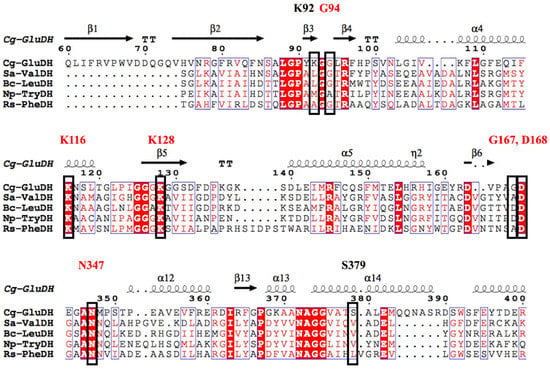
Figure 4.
Amino acid sequence alignment of s-AADHs and comparison of the active site. The black box indicates the key residues in the catalytic process of GluDH, of which the shared functional residues of s-AADHs are annotated in red.
3. Substrate-Specific Engineering of s-AADHs
To broaden the application scope of s-AADHs, many researchers have focused on the directed evolution of s-AADHs based on their activity on substrates. The modification of substrate specificity has mainly focused on adapting engineered enzymes to substrates with different R groups (charged, polar, aliphatic, aromatic, etc.) [18,22,36]. With the transformation of LeuDH into engineered amine dehydrogenase (AmDH) by researchers in 2012, the substrate scope of s-AADHs was expanded from keto acids to ketones without the carboxy group. Subsequently, engineered AmDHs with different substrate specificities were constructed [23,24,25,26]. In general, the substrate-specific engineering of s-AADHs can enable them to catalyze the synthesis of more diverse amino acids and chiral amines. In this section, substrate-specific engineering of s-AADHs is described in three parts (Section 3.1, Section 3.2 and Section 3.3), with a focus on the modification of mutation hotspot regions.
3.1. GluDH
GluDH is widely distributed among living organisms owing to its important role in the interconversion of nitrogen and carbon metabolism [32]. Based on their cofactor specificity, GluDH enzymes can be categorized into three sub-families, specifically NAD(H)-dependent (EC 1.4.1.2), NADP(H)-dependent (EC 1.4.1.4), and NAD(H)/NADP(H) dual-specific GluDHs (EC 1.4.1.3) [37]. GluDHs have strict substrate specificity for glutamate and 2-OG. The NAD+-dependent GluDH of Clostridium symbiosum (CsGluDH) is almost exclusively specific for glutamate in the detection of the catalytic activity of 21 amino acids [12]. Meanwhile, the NADP+-dependent GluDH from Pseudomonas putida (PpGluDH) and the dual cofactor-dependent GluDH from Acinetobacter tandoii (AtGluDH) have catalytic activity for aliphatic keto acid and aromatic keto acid, but their specific activity is less than 8% of the specific activity of 2-OG [18,36]. Only a few studies have reported the use of wild-type GluDH for the synthesis of other substances, such as L-6-hydroxynorleucine and L-phosphinothricin [38,39].
The strict substrate specificity of the wild-type GluDH limits its application. As shown in Table 2, researchers have engineered GluDHs to alter their substrate specificity for various amino acid synthesis.

Table 2.
Altered substrate specificity of engineered GluDHs and their synthesis of amino acids.
L-α-Amino acids with hydrophilic side chains. Some researchers have modified the substrate specificity of GluDH to make it more suitable for catalyzing other substrates with hydrophilic side chains (Entry 1–7 in Table 2). Khan et al. [13] changed the substrate specificity of GluDH from Bacillus subtilis (BsGluDH) from 2-OG to oxaloacetate (OAA). They found that the specificity of G82K and M101S for OAA was significantly increased, with kcat values of 3.45 and 5.68 s−1, respectively, which were 265 times and 473 times higher than that of 2-OG, respectively. Analysis of the docking results revealed that the replaced residues of the two variants fixed the side chain carboxyl group of OAA, which could play an important role in catalysis of the substrate, OAA. In the subsequent determination of the substrate spectrum, it was found that the two variants have increased activity for some aliphatic and aromatic 2-keto acids (such as pyruvate and p-hydroxyphenylpyruvate). Yin et al. [36] studied GluDH as a substrate for 2-oxo-4-[(hydroxy)(methyl)phosphinoyl]butyric acid (PPO), which is a structural analog of 2-OG. Through screening, Yin et al. found that eight NADP+-dependent GluDHs have activity on PPO. Among them, GluDH from P. putida (PpGluDH), with favorable soluble expression and high enzymatic activity (the specific activity for PPO is 0.002% of the specific activity of 2-OG), was used for further directed evolution. When residue I170 at the substrate entrance tunnel of PpGluDH was mutated, the obtained variant I170M had a 2.1-fold increase in PPO activity compared to that of the wild-type, but this still needs to be improved. Directed by modeling and docking simulations, Yin et al. [40] used site-directed mutagenesis to enlarge the pocket of PpGluDH so that it could accommodate the larger substrate, PPO. In this study, the researchers performed the same modification (A167G or V378A numbering according to PpGluDH) of GluDH from other sources and found that the activity of each variant on PPO and other macromolecular substrates was also improved. Even NAD+-dependent GluDH, which has no enzymatic activity for PPO, can have PPO activity after a mutation. This indicates that these two points have a certain universality in expanding the substrate pocket of GluDH to accommodate macromolecular substrates [36]. In further studies, Yin et al. [30] performed saturation mutagenesis on specific residues (Thr121, Leu123, Ala379, Leu383, Lys402, Ile406, and Ile410) in or near the hinge region of PpGluDH. A dozen positive variants with significantly improved PPO catalytic activities were obtained. The kcat/Km of T121N/L123Y was 97.9 times higher than that of the wild-type, which was mainly due to the improvement in the kcat. Molecular dynamics simulations revealed that the enhanced catalytic activity could be due to an increase in the conversion efficiency of the open/closed conformation of the protein. In addition, these advantageous variants, obtained by screening based on PPO as a substrate, also showed a universal increase in the catalytic activity for other non-polar keto acids (such as 2-ketobutyric acid and 2-oxo-phenylbutyric acid) [30]. Owing to the structural similarity of s-AADHs, the activity engineering of GluDHs in the hinge region might have implications for the substrate-specific modification of other s-AADHs. Geng et al. [41] modified the substrate specificity of GluDH from Escherichia coli (EcGluDH) using homoserine as the target substrate. The key site, K92, was identified by energy decomposition and structural analysis of the docking results of EcGluDH with homoserine or glutamate. Experimental validation showed that the variant K92V had the highest catalytic activity towards homoserine, which showed a 722% increase in specific activity (1328 ± 71 μU mg−1) compared to that of wild-type GluDH.
Aliphatic L-α-amino acids. Many studies have reported that GluDH can be engineered to catalyze the synthesis of aliphatic amino acids (Entry 8–13 in Table 2). Zhang et al. [19] used valine auxotrophic E. coli for the directed evolution of EcGluDH and obtained the variant K92V/T195S. Compared with that of wild-type EcGluDH, K92V/T195S had significantly enhanced catalytic activity with 2-ketobutyrate, with a 4-fold decrease in the Km and a 2-fold increase in the kcat. This might be caused by the mutation of lysine 92 to valine, which increases the hydrophobic properties of the pocket. Wang et al. [18] used site-directed mutagenesis to construct a hydrophobic network between the GluDH from A. tandoii (AtGluDH) and the terminal methyl group of the substrate 2-ketobutyric acid. The catalytic activity of the obtained double variant K76L/T180C for 2-ketobutyric acid was 17.2 times that of the wild-type. The kcat/Km of K76L/T180C was nearly 10 times compared with that of the wild-type. Through determination of the substrate spectrum, it was found that the variant has universal improvement in the aliphatic keto acid catalytic activity. Wang et al. [12] obtained the triple variant K89L/S380A/A163G of CsGluDH, which represents effective conversion of substrate specificity from glutamate to aliphatic amino acids (norvaline, methionine, norleucine). The triple variant showed no detectable GluDH activity. This team further relocated these three residues to GluDH from Halobacterium salinarum (HsGluDH) and obtained a variant (K89L/A163G/S367A), which was also active for aliphatic amino acids (norvaline = 1.75 U/mg, methionine = 3.1 U/mg, norleucine = 3.7 U/mg) [42].
Aromatic L-α-amino acids. There are also reports on the substrate specificity transformation of GluDH to aromatic substrates (Entry 14 in Table 2). Li et al. [43] developed a stepwise substrate walking strategy through two rounds of protein engineering, changing the substrate specificity of EcGluDH from 2-OG to 2-oxo-4-phenylbutyric acid (2-OPBA). Compared with that of wild-type GluDH, the kcat/Km of the obtained variant K92A/A166G/T195A/V377A/S380A was approximately 100 times higher for 2-OPBA.
γ-Amino acids. The substrate specific engineering for GluDH is not limited to the recognition of substrate side chain R groups, and the substrate specific engineering of GluDH can also occur in the recognition of substrate main chain carboxyl groups. Inspired by the work of Abrahamson et al. [23,27], Zhou et al. [26] developed an engineered amine dehydrogenase (K116Q/N348M) base on E. coli GluDH (Ec-GAmDH), which can transform levulinic acid into value-added (R)-4-aminopentanoic acid (Entry 15 in Table 2).
The GluDH substrate specific modifications reviewed in the first three items focused on the side chain R-group of the substrate. By introducing new hydrophilic residues or by adjusting the existing hydrophilic residues inside the substrate pocket, GluDH showed better catalytic activity towards other substrates with hydrophilic R-groups. By replacing the hydrophilic residues inside the substrate pocket with hydrophobic residues, GluDH exhibited significant catalytic activity towards aliphatic 2-keto acids. With the replacement of residues inside the pocket with smaller amino acids (Gly, Ala), GluDH became catalytically active against long-chain aliphatic 2-keto acids and even bulky aromatic 2-keto acids. This allows the engineered GluDH to catalyze the synthesis of more α-amino acids. However, the substrate-specific engineering in the last item occurs at the substrate backbone carboxyl anchor site. In previous studies, engineered amine dehydrogenases were developed using LeuDH or PheDH as scaffolds (a detailed summary will be provided in Section 3.2.2 and Section 3.3.2). Ec-GAmDH is the first reported engineered amine dehydrogenase constructed with GluDH as a scaffold.
3.2. ValDH and LeuDH
Compared to those of GluDH, the substrate specificities of ValDH and LeuDH are more promiscuous. They catalyze the reversible deamination of branched- and straight-chain amino acids (L-valine, L-leucine, L-isoleucine, L-norvaline, L-2-amino-butyrate, etc.) into the corresponding keto acids. ValDH catalyzes L-valine/2-oxoisovalerate conversion, whereas LeuDH preferentially catalyzes L-leucine/2-oxoisohexanoate reactions [14]. In a comparison of amino acid sequence of s-AADHs, it is found that ValDH and LeuDH have the highest similarity (approximately 50–55%), which explains the similar substrate spectrum of the two enzymes [44]. At present, most ValDH is found in Streptomyces species, and this enzyme has a higher activity with NAD+ compared to that with NADP+ [44]. This enzyme was first found in Streptomyces aureofaciens and is involved in the synthesis of the oligoketide antibiotic tylosin [45]. There are few reports on the substrate-specific modification of valine dehydrogenase. To the best of our knowledge, only Hyun et al. [16] have altered the substrate specificity of ValDH from Streptomyces albus (SaValDH). The variant A124G shows decreased activities toward most aliphatic amino acid substrates and dramatically increased catalytic activities toward L-phenylalanine, L-tyrosine, and L-methionine (Entry 1 in Table 3). Research on LeuDHs, thus far having only used NAD+ as a cofactor, has been relatively extensive. LeuDHs are found in a limited number of bacterial species, mainly in endospore-forming bacteria (such as Bacillus species) [46], and they can play a role in spore germination [1].

Table 3.
Selected examples of altered substrate specificity of engineered ValDH and LeuDH and their synthesis of L-α-amino acids.
3.2.1. The Substrate-Specific Engineering of LeuDHs and Their Synthesis of L-α-Amino Acids
Many studies have been reported on the use of engineered LeuDH to catalyze the synthesis of unnatural aliphatic L-α-amino acids (Entry 2–6 in Table 3). Zhou et al. [29] carried out the rational engineering of LeuDH from Bacillus cereus (BcLeuDH) and found that the replacement of residue E116 in the substrate entrance tunnel with valine can universally improve the catalytic efficiency of the enzyme for branched- and straight-chain amino acids and phenylglycine. The same mutation was also obtained in the high-throughput screening by Zhou et al. [20] to improve the catalytic efficiency of BcLeuDH for L-tert-leucine synthesis. Integrating protein structure information and amino acid sequence (LeuDH, PheDH, and GluDH) alignment, Kataoka and Tanizawa [17] pointed out important residues (A113, L40, and V294) that are assumed to be the determinants of substrate side-chain binding. The variant A113G has broader substrate specificities than the wild-type LeuDH from Bacillus stearothermophilus (BsLeuDH). The catalytic efficiency of A113G for L-norleucine was significantly improved, and the kcat/Km was 14.7 times that of the wild-type. At the same time, A113G is active toward phenylpyruvate. This indicates that A113 plays a critical role in discriminating the bulkiness of the side chain. In this study, L40 and V294 of LeuDH, corresponding to two sites in GluDH that interact with the γ-carboxyl group of the substrate L-glutamate, were mutated to L40K/V294S and L40D/V294S, respectively. Through determination of the substrate spectrum, it was found that L40K/V294S and L40D/V294S have catalytic activities for L-glutamate and L-lysine, respectively (Entry 7–8 in Table 3). These results suggest that substrate specific transformation of LeuDHs and GluDHs can be achieved by mutation of key residues within the pocket.
3.2.2. The Substrate-Specific Engineering of LeuDHs and Their Synthesis of Chiral Amines
By mutating the substrate carboxyl anchor site of LeuDH, the specificity of LeuDH can be expanded from the original 2-keto acid to ketones without carboxyl groups. Abrahamson et al. [23] first engineered LeuDH from Geobacillus stearothermophilus (GsLeuDH) into an amine dehydrogenase (Gs-LAmDH, K68S/N261L/E114V/V291C), providing a new dimension in the substrate-specific engineering of LeuDH and other s-AADHs. Instead of the natural substrate 2-oxoisohexanoate, the engineered amine dehydrogenase (AmDH) now accepts the analogous ketone (methyl isobutyl ketone), which is mainly due to the mutation of two residues (K68S/N261L) that interact with the carboxyl group of the substrate [23,27] (Entry 1 in Table 4). Since 2012, multiple groups have produced similarly engineered AmDHs based on LeuDH scaffolds and have conducted detailed studies on the substrate spectrum of these enzymes.
Aliphatic chiral amines. Chen et al. [47] constructed an engineered LeuDH (K77S/N270L) from Exiguobacterium sibiricum (Es-LAmDH), which exhibits broad catalytic activity towards acetophenone, short-chain secondary aliphatic ketones, and alkyl cyclic ketones (Entry 2–4 in Table 4). Inspired by Chen et al.’s [47] work, Löwe et al. [48] provided detailed insight into substrate scope of Es-LAmDH. They found that Es-LAmDH also has catalytic activity for a series of aromatic ketones, among which the activity for acetophenone was the most significant. To enable the engineered amine dehydrogenase to have catalytic activity for bulky aliphatic ketones, an engineered LeuDH (K68S/N261L) from Lysinibacillus fusiformis (Lf-LAmDH) with good thermal stability was tailored by Chen et al. [49] based on structural information. They found that A113 and T134 of Lf-LAmDH prevented the binding of bulky aliphatic ketones. When these two residues were mutated to smaller amino acids (G or A), the variants (A113G/T134A, A113G/T134G) exhibited higher activity toward 2-hexanone than Lf-LAmDH and exhibited activity against longer aliphatic ketones, the longest of which being 2-decanone (Entry 5–6 in Table 4). The effect of these two-site mutations was sufficiently adaptable to other engineered AmDHs (Es-LAmDH and Bs-LAmDH). Based on the studies by Abrahamson et al. [23,27] and Chen et al. [49], Franklin et al. [50] generated a series of mutations (D32A, L39A, F101S, A112G, C290V) in Gs-LAmDH, which were mentioned in previous studies. The specific activity of the obtained variant (K68S/N261L/E114V/V291C/D32A/L39A/F101S/A112G/C290V) for long-chain aliphatic ketones was further improved, and the catalytic activity of 2-decanone reached 140.5 mU/mg (Entry 7 in Table 4).
Chiral vicinal amino alcohols. Subsequent studies revealed that the substrate spectrum of LAmDHs is not limited to aliphatic ketones, but is also catalytically active for substrates with a hydroxymethyl backbone (Entry 8–11 in Table 4). Chen et al. [24] found that two-site combinatorial variants (sites K68 and N261 of Lf LeuDH) can catalyze new reactions that convert α-hydroxy-2-ketones to the corresponding chiral vicinal amino alcohols. Among these, K68T/N261L shows the highest activity toward 1-hydroxyl-2-butanone. Further mutations (A113G, A113G/T134A, and A113G/T134G) were introduced into the substrate-binding pocket of the variant (K68T/N261L) and were found to exhibit activity toward the bulkier 1-hydroxy-2-heptanone. Wang et al. [51] introduced two-site mutations (K68S/D261L, K70S/N263L) into LeuDH from other sources and also created engineered amine dehydrogenases capable of synthesizing chiral vicinal amino alcohols.
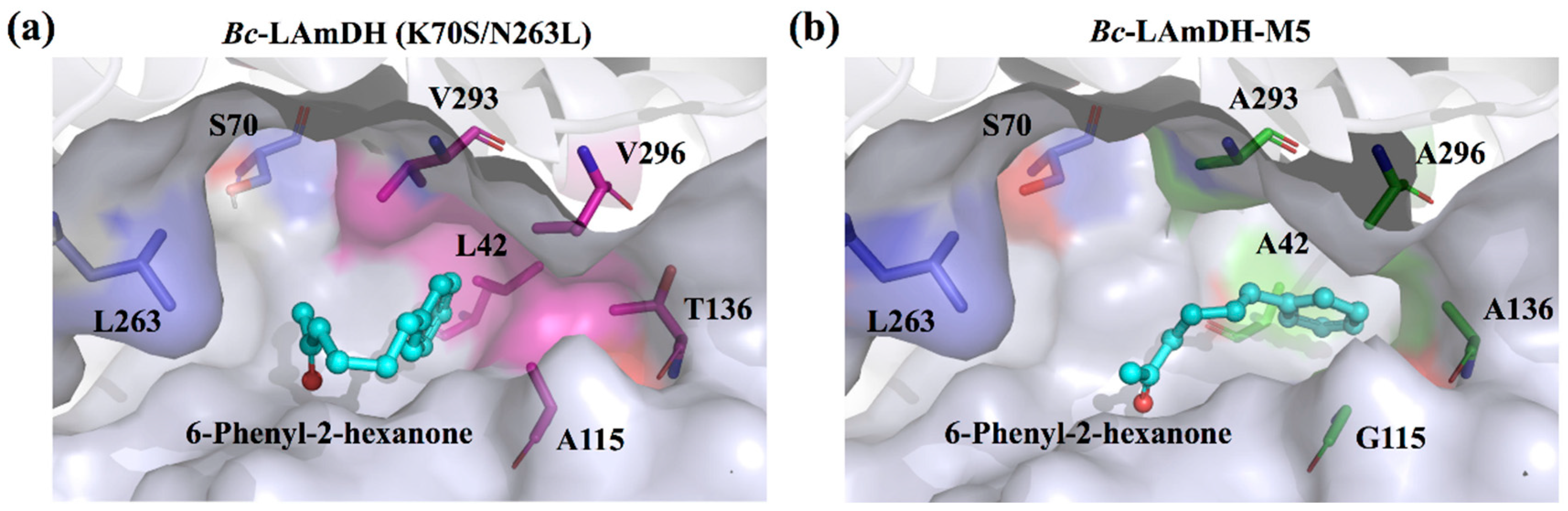
Aromatic chiral amines. The volume of the substrate-binding pocket of LAmDH is too small to accommodate the bulky aromatic ketones. However, by replacing sterically hindered residues inside the substrate pocket with smaller residues (Gly, Ala), LAmDH developed catalytic activity for bulky aromatic ketones (Entry 12–14 in Table 4). Using iterative alanine scanning mutagenesis, the volume of the substrate pocket of the engineered amine dehydrogenase (variant K70S/N263L of LeuDH from B. cereus, Bc-LAmDH) was gradually enlarged to accommodate aromatic ketones with longer chains. The eventual variant Bc-LAmDH-M5 was constructed by mutation of five residues (A115G/T136A/L42A/V296A/V293A), which successfully exhibited specificity toward the substrate 6-phenyl-2-hexanone [52] (Figure 5).
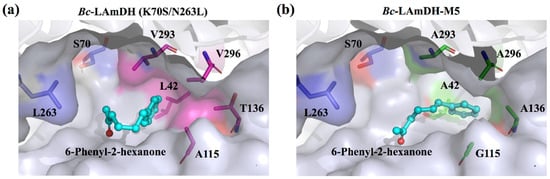
Figure 5.
Illustration of the substrate-binding cavities of Bc-LAmDH (K70S/N263L) (a) and Bc-LAmDH-M5 (K70S/N263L/A115G/T136A/L42A/V296A/V293A) (b) with docked substrate 6-phenyl-2-hexanone. Residues surrounding the substrate-binding pocket are shown in surface representation, and 6-phenyl-2-hexanone is shown as a ball and stick model. Residues 42, 115, 136, 293, and 296 responsible for steric hindrance are shown as magentas sticks in the Bc-LAmDH (K70S/N263L) and green sticks in Bc-LAmDH-M5 (K70S/N263L/A115G/T136A/L42A/V296A/V293A), respectively. The main two residues initially mutated to generate AmDH activity (S70 and L263) are shown as blue sticks.

Table 4.
Selected examples of altered substrate specificity of engineered LeuDH and their synthesis of chiral amines.
Table 4.
Selected examples of altered substrate specificity of engineered LeuDH and their synthesis of chiral amines.
| Entry No. | Enzyme | Variants a | Substrate | Conversion (%) | Product Configuration and ee (%) | Specific Activity (U/mg) | kcat (s−1), Km (mM) | Ref. |
|---|---|---|---|---|---|---|---|---|
| 1 | Gs-LAmDH | K68SO/N261LO/E114VT/V291CI | 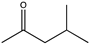 | 92.5 | (R), 99.8 | 0.690 | 0.46, 15.1 | [23] |
| 2 | Es-LAmDH | K77SO/N270LO | 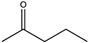 | 94 | (R), >99 | 0.796 | 0.84, 9.2 | [47,49] |
| 3 | 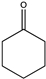 | 97 | N.D. b | 0.868 | N.D. | |||
| 4 | 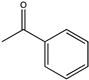 | 21 | (R), >99 | 0.0985 | N.D. | |||
| 5 | Lf-LAmDH | K68SO/N261LO | 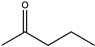 | >99 | (R), >99 | 0.930 | 1.7, 30.6 | [49] |
| 6 | K68SO/N261LO/A113GI/T134GI | 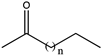 (n = 1, 2, 3, 4, 5) | 98, >99, >99, 80, 22 | (R), >99 | - c | N.D. | ||
| 7 | Gs-LAmDH | K68SO/N261LO/E114VT/V291CI/D32A/L39AI/F101S/A112GI/C290VI | 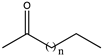 (n = 6) | N.D. | N.D. | 0.1405 | N.D. | [50] |
| 8 | Lf-LAmDH | K68TO/N261LO | 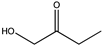 | 91 | (S), >99 | 0.233 | 0.227, 7.2 | [24] |
| 9 | K68TO/N261LO/A113GI/T134GI | 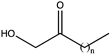 (n = 4) | 99 | (S), >99 | >2 | N.D. | ||
| 10 | Cs- LAmDH | K68SO/D261LO | 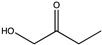 | >99 | (S), >99 | N.D. | 1.79, 2.53 | [51] |
| 11 | 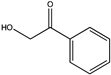 | 73 | (S), >99 | N.D. | N.D. | |||
| 12 | Bc-LAmDH | K70SO/N263LO/L42AI/A115GI/T136AI/V293AI/V296AI | 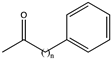 (n = 2) | >99 | (R), >99 | 0.0948 | 0.15, 10.5 | [52] |
| 13 | 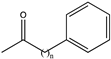 (n = 3) | N.D. | N.D. | 0.137 | 0.20, 6.7 | |||
| 14 | 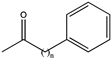 (n = 4) | 93 | (R), >99 | 0.0803 | 0.143, 11.6 |
a In this column, the mutated residues with a superscript “I” are inside the pocket of LeuDH, the mutated residues with a superscript “O” are the substrate backbone carboxyl anchor sites of LeuDH, and the mutated residues with a superscript “T” are at the substrate tunnel of LeuDH. The positions of all residues were determined by sequence alignment and structural comparison. b N.D. = not determined. c Specific activity is not shown here.
3.3. TrpDH and PheDH
TrpDH and PheDH recognize substrates with bulky aromatic rings, and their optimal catalytic substrates are L-tryptophan/3-indolepyruvate and phenylalanine/phenylpyruvate, respectively. TrpDH was first identified in several higher plants and was partially characterized in the 1980s [53,54]. There was then no further investigation of the enzyme until the first NAD+-dependent TrpDH was identified in Nostoc punctiforme ATCC 29133 in 2008, which was suggested to be involved in the biosynthesis of cyanobacterial sunscreen scytonemin [3]. TrpDH from N. punctiforme has strict substrate specificity, and they are catalytically active almost exclusively for L-tryptophan and 3-indolepyruvate [55,56]. To our knowledge, no studies have been reported in which the substrate specificity of TrpDH is altered, most likely owing to the fact that the cluster of hydrophobic residues around the active site of TrpDH is extremely sensitive to mutations [8]. Compared to TrpDH, PheDH has been much more extensively studied. PheDH is found in a limited number of Gram-positive aerobic bacteria [57]. In addition to reacting with aromatic amino acids, PheDHs show only slight activity towards aliphatic amino acids [58].
3.3.1. The Substrate-Specific Engineering of PheDHs and Their Synthesis of L-α-Amino Acids
L-α-Amino acids with bulky side chains. To date, many wild-type PheDHs have been studied for aromatic L-amino acid synthesis [10]. In terms of substrate specificity, PheDH from Bacillus sphaericus (BsPheDH) is unusual because its activities toward L-phenylalanine and L-tyrosine are similar [15]. However, the catalytic activity of PheDHs from other sources (BbPheDH from Bacillus badius, SuPheDH from Sporosarcina ureae, and RsPheDH from Rhodococcus sp. M4) for L-phenylalanine is 11–34 times higher than that of L-tyrosine. To find out the reason for the difference in the activity of PheDHs towards the substrates L-phenylalanine and L-tyrosine, Seah et al. [15] analyzed the amino acid sequences of PheDHs from different sources based on the existing amino acid dehydrogenase structures. They found that residue N145 of BsPheDH (the corresponding residue in BbPheDH and SuPheDH is valine) close to the terminus of the substrate side chain might provide relatively favorable possibilities for interactions with the hydroxyl group of the substrate tyrosine. Substitution of N145 in BsPheDH with aliphatic amino acids (A, V, I, and L) results in enzymes with greatly increased discrimination between the substrates L-phenylalanine and L-tyrosine, which is mainly due to the improved affinity for L-phenylalanine and the decreased affinity for L-tyrosine. The affinity of these variants for long chain aliphatic keto acids was improved compared with that of the wild-type (Entry 1–6 in Table 5). Yousefi et al. [59] mutated V144 of BbPheDH, which corresponds to position N145 in BsPheDH, and obtained variants that produced the same substrate-specific shift. They found that the variant V144N is more inclined to catalyze L-tyrosine than L-phenylalanine, whereas the variant V144L prefers to catalyze L-phenylalanine, which is due to the long chain of leucine residues enhancing the hydrophobicity of the enzyme pocket. In further studies, Engel’s group found that the variants (N145A, N145L, and N145V) of BsPheDH were more active than the wild-type enzyme for substrates with substituents (-CH3, -OCH3, and -CF3) at the 4-position on the phenyl ring, and the catalytic activity of a variant (N145A) based on Lysinibacillus sphaericus PheDH (LsPheDH) for para-bromo-L-phenylalanine was improved compared with that of the wild-type [21,60] (Entry 7–10 in Table 5). These results indicated that this residue (position 145 in BsPheDH) from different sources plays an important role in the recognition of the para-substituent of phenylpyruvate. In addition, the substitution of position 145 with a smaller amino acid allows PheDH to accommodate substrates with long side chains. Wu et al. [22] employed a modified steric hindrance engineering approach to create an enhanced biocatalyst toward L-homophenylalanine. The kcat and kcat/Km values of the optimal variant (V309G/L306V/V144G, V144 corresponding to position N145 of BsPheDH) were 12.7- and 12.9-fold higher than those of the wild-type, respectively (Entry 11 in Table 5).
L-α-Amino acids with small aliphatic side chains. The glycine residue at position 124 of BsPheDH, which is conserved in PheDHs, plays an important role in distinguishing between substrates with different steric hindrances. When this site was replaced by a residue with a larger side chain, the catalytic activity of PheDH towards smaller aliphatic amino acids was significantly improved (Entry 12–15 in Table 5). Seah et al. [57] found that glycine-124 and leucine-307 of BsPheDH are replaced by alanine and valine, respectively, both in LeuDH and ValDH, which leads to their differences in substrate specificity. The specific activity of the variant (G124A/L307V) of BsPheDH for L-phenylalanine and L-tyrosine is significantly reduced (5.7% and 0.56% of the wild-type specific activity, respectively). Furthermore, the specific activity of this variant for aliphatic amino acids (L-methionine, L-norleucine, L-norvaline, L-leucine, L-valine, and L-isoleucine) was found to be 1.3–22.6 times higher than that of the wild type, which is mainly caused by the greatly improved affinity of the variants for the substrates [58]. Replacement of glycine with serine at positions 123 and 124 of BbPheDH and BsPheDH, respectively, strikingly decrease the enzyme activity toward aromatic amino acids and result in an elevation in the relative activity toward aliphatic amino acids. The variant G123S of BbPheDH preferentially oxidizes aliphatic branched-chain amino acids (such as L-leucine and L-isoleucine), whereas the variant (G124S) of BsPheDH preferentially oxidizes straight-chain aliphatic amino acids (such as L-norvaline, L-methionine, and L-norleucine) [61]. Engel’s group engineered BsPheDH through directed evolution and obtained the optimal variant G124A/E313G, which shows improved activity and specificity towards L-propargylglycine [62].

Table 5.
Altered substrate specificity of engineered PheDHs and their synthesis of L-α-amino acids.
Table 5.
Altered substrate specificity of engineered PheDHs and their synthesis of L-α-amino acids.
| Entry No. | Enzyme | Variants a | Substrate | Conversion (%) | Product Configuration and ee (%) | Specific Activity (U/mg) c | kcat (s−1), Km (mM) | Ref. |
|---|---|---|---|---|---|---|---|---|
| 1 | BsPheDH | wild type | 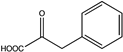 | N.D. b | N.D. | N.D. | 113, 0.37 | [15] |
| 2 | N145I | N.D. | N.D. | N.D. | 43, 0.012 | |||
| 3 | wild type | 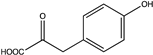 | N.D. | N.D. | N.D. | 90, 0.19 | ||
| 4 | N145I | N.D. | N.D. | N.D. | 80, 0.55 | |||
| 5 | wild type | 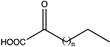 (n = 2) | N.D. | N.D. | N.D. | 51, 13 | ||
| 6 | N145A | N.D. | N.D. | N.D. | 66, 0.25 | |||
| 7 | N145L | 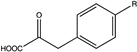 (R = -OCH3) | N.D. | N.D. | 114.8 (3.85) | N.D. | [60] | |
| 8 | N145A | 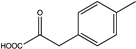 | N.D. | N.D. | 130.5 (4.47) | N.D. | ||
| 9 | 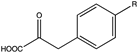 (R = -CF3) | N.D. | N.D. | 29.4 (16.33) | N.D. | |||
| 10 | LsPheDH | N145A | 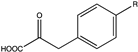 (R = -Br) | >99 | (S), >99 | >50 (≈1.67) | N.D. | [21] |
| 11 | BbPheDH | V309G/L306V/V144G | 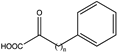 (n = 2) | > 99 | (S), >99 | N.D. | 83, 74 | [22] |
| 12 | BbPheDH | G123S | 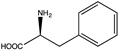 | N.D. | N.D. | 2.16 (0.03) | 1.55, 227 | [61] |
| 13 | 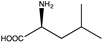 | N.D. | N.D. | 12 (5.59) | 1.54, 3.3 | |||
| 14 | 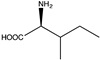 | N.D. | N.D. | 10.68 (74.6) | 1.48, 3.8 | |||
| 15 | BsPheDH | G124A/E313G | 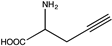 | N.D. | N.D. | N.D. | 0.92, 23.2 | [62] |
a In this column, all mutation sites are inside the pocket of PheDH. The positions of all residues were determined by sequence alignment and structural comparison. b N.D. = not determined. c In this column, the data in parentheses are the folds of the specific activity of the variant compared to the wild type.
3.3.2. The Substrate-Specific Engineering of PheDHs and Their Synthesis of Chiral Amines
Inspired by the work of Abrahamson et al. [23,27], many studies on the transformation of PheDH from different sources into engineered amine dehydrogenases (PAmDHs) and the substrate specificity of PAmDH have been intensively performed.
Construction of PAmDH and their substrate spectrum. Previous research on the evolution of an amine dehydrogenase from LeuDH allowed Abrahamson et al. [27] to rapidly improve an AmDH based on BbPheDH (Bb-PAmDH), which shares a reasonable sequence similarity with BsLeuDH (48% identity). The optimal variant (K77S/N276L) screened based on the model substrate p-fluorophenylacetone (PFPA) exhibits an impressive substrate scope, with considerable catalytic activity for aromatic ketones (PFPA, phenoxy-2-propanone), aliphatic ketones (2-hexanone, methyl isobutyl ketone), and cyclic ketones (3-methyl-2-butanone), among which the catalytic activity on PFPA is the most significant (Entry 1 in Table 6). The kcat value for PFPA was nearly 15-fold greater than the maximum observed kcat of 0.46 s−1 for the previously developed engineered AmDH based on LeuDH scaffolds. Ye et al. [28] conducted a similar directed evolution study based on RsPheDH using phenylacetone and benzylacetone as substrates and successfully obtained several engineered amine dehydrogenases (Rs-PAmDHs). The optimal variant, K66Q/N262C/S149G, had a kcat of 0.70 and 0.72 s−1 for the conversion of phenylacetone to (R)-amphetamine and for the conversion of 4-phenyl-2-butanone to (R)-1-methyl-3-phenylpropylamine, respectively (Entry 3–4 in Table 6).
In addition to obtaining PAmDH based on PheDH evolution, a new AmDH can also be constructed by domain shuffling. As mentioned in the introduction to the structure of s-AADHs, their subunits are constructed from the N-terminal substrate binding domains and the C-terminal coenzyme binding domains, which behave as an independently folding unit with different functions [63,64]. Based on the feature of these AADHs, Bommarius et al. [65] employed domain shuffling to generate a new chimeric amine dehydrogenase (cFL1-AmDH) consisting of an N-terminal substrate-binding domain of Bb-PAmDH and a C-terminal coenzyme-binding domain of Bs-LAmDH. Unlike previously reported chimeric AADHs [63,66], the substrate scope of cFL1-AmDH not only is an admixture of those of that of the two parent enzymes (such as p-fluorophenylacetone and acetophenone), but it also has substrates (such as 1-tetralone, adamantylmethylketone, and 3-methyl-1-phenylbutanone) not shared by the two parent enzymes in terms of catalytic activity (Entry 16–20 in Table 6). These differences are likely associated with conformational transitions involving domains I and II during the catalytic cycle [67]. The lower conversion of acetophenone by cFL1AmDH may be due to its substrate-binding domain derived from Bb-PAmDH, which has a larger substrate-binding pocket and is more suitable for catalyzing acetophenone and its derivatives (Entries 1, 3, 5, 6 in Table 6). However, the large substrate pocket makes it difficult for simple acetophenone to bind to the enzyme in the correct catalytic posture, resulting in low conversion rates.
Knaus et al. [68] detailed the substrate scope of three existing engineered amine dehydrogenases derived from PheDHs (cFL1-AmDH, Rs-PAmDH, Bb-PAmDH) through transformation experiments. The assayed substrates could be roughly divided into four categories, i.e., phenylacetone derivatives, alkyl methylketones, acetophenone derivatives, and bulky ketones. The results showed that cFL1-AmDH and Rs-PAmDH have broader substrate profiles than Bb-PAmDH. cFL1-AmDH is very active toward aliphatic ketones and acetophenone derivatives (Entry 16–26 in Table 6), whereas Rs-PAmDH preferentially catalyzes the reductive amination of phenylacetone derivatives and more sterically demanding ketones (Entry 5–15 in Table 6). Bb-PAmDH is more prone to the reductive amination of phenylacetone derivatives (Entry 1–2 in Table 6), which have similar substrate specificities for Ct-PAmDH (K68S/N266L) developed by the Ursula Schell group, based on Caldalkalibacillus thermarum PheDH [69].

Table 6.
Selected examples of the substrate spectrum of PAmDH.
Table 6.
Selected examples of the substrate spectrum of PAmDH.
| Entry No. | Enzyme | Variants a | Substrate | Conversion (%) | Product Configuration and ee (%) | Specific Activity (U/mg) | kcat (s−1), Km (mM) | Ref. |
|---|---|---|---|---|---|---|---|---|
| 1 | Bb-PAmDH | K77SO/N276LO | 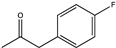 | >90 | (R), >99.8 | N.D.b | 6.85, 7.75 | [27] |
| 2 | Bb-PAmDH | K77SO/N276LO | 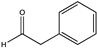 | 34 | n.a.c | N.D. | N.D. | [68] |
| 3 | Rs-PAmDH | K66QO/N262CO/S149GI | 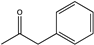 | N.D. | (R), >98 | 0.042 | 0.70, 4.0 | [28] |
| 4 | 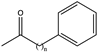 (n = 2) | 95 | (R), >98 | 0.043 | 0.72, 1.4 | |||
| 5 | 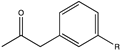 (R = -OCH3, -CF3) | 98, 98 | (R), >99 | N.D. | N.D. | [68] | ||
| 6 | 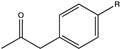 (R = -OCH3, -CH3) | >99, 98 | (R), >99 | N.D. | N.D. | |||
| 10 | 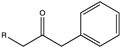 (R = -CH3, -CH2CH3) (R = -CH3, -CH2CH3) | >99, 71 | (R), >99 | N.D. | N.D. | |||
| 11 | 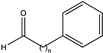 (n = 2) | 96 | n.a. | N.D. | N.D. | |||
| 12 | 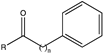 (n = 2; R = -CH3, -CH2CH3) | 99, 87 | (R), >99 | N.D. | N.D. | |||
| 13 | 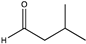 | 99 | n.a. | N.D. | N.D. | |||
| 14 | 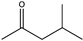 | 91 | (R), >99 | N.D. | N.D. | |||
| 15 | 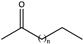 (n = 3, 4) | 99, 93 | (R), >99 | N.D. | N.D. | |||
| 16 | cFL1-AmDH | N-terminal domain of Bb-PAmDH and C-terminal domain of Bs-LAmDH | 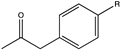 (R = -F) | 93 | (R), >99 | 1.725 | 1.24, 1.1 | [68] |
| 17 |  | 34 | (R), >99 | 0.301 | 0.24, 5.2 | |||
| 18 |  | N.D. | N.D. | 0.107 | N.D. | [65] | ||
| 19 |  | N.D. | N.D. | 0.069 | N.D. | |||
| 20 | 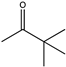 | N.D. | N.D. | 0.133 | N.D. | |||
| 21 | 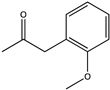 | >99 | (R), >99 | N.D. | N.D. | [68] | ||
| 22 | 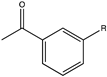 (R = -CH3, -F) | 39, 43 | (R), >99 | N.D. | N.D. | |||
| 23 | 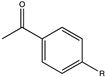 (R = -CH3, -F) | 9, 22 | (R), >99 | N.D. | N.D. | |||
| 24 | 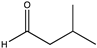 | >99 | n.a. | N.D. | N.D. | |||
| 25 | 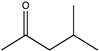 | 96 | (R), >99 | N.D. | N.D. | |||
| 26 | 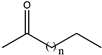 (n = 1, 2, 3, 4) | 75, 92, 98, 50 | (R), >99 | N.D. | N.D. |
a In this column, the mutated residues with a superscript “I” are inside the pocket of PheDH, and the mutated residues with a superscript “O” are the substrate backbone carboxyl anchor sites of PheDH. The positions of all residues were determined by sequence alignment and structural comparison. b N.D. = not determined. c n.a. = Not applicable.
Catalytic activity improvement and substrate spectrum expansion of PAmDH. The substrate specificity and catalytic activity of these engineered amine dehydrogenases still limit their application scope. Liu et al. [25] developed a Gk-PAmDH (K78S/N276L) that displays a broad substrate scope and good activity via the directed evolution of a Geobacillus kaustophilus PheDH. Gk-PAmDH exhibits considerable activity towards phenylacetone derivatives, alkyl methylketones, and benzylacetone and can even reduce aminate hydroxy ketones (such as 1-hydroxy-3-phenyl-2-propanone and 1-hydroxy-2-butanone) to the corresponding chiral vicinal amino alcohols (Entry 1–3 in Table 7). To develop a spectrum-extended AmDH that tolerates bulky ketones, Wang et al. [70] enlarged the binding pocket of Gk-PAmDH (K78S/N276L) with benzylacetone as the model substrate and obtained a triple variant V144A/V309A/A310G (Gk-PAmDH-M3) that displays a specific activity of 0.65 U mg−1 toward benzylacetone, 32-fold higher than that of Gk-PAmDH (Entry 4 in Table 7). Through error-prone PCR using the Gk-PAmDH-M3 gene as the template, Wang et al. [70] obtained a better variant (V144A/V309A/A310G/S156T/Q308A/C79N/F86M, Gk-PhAmDH-M8), which displayed a specific activity of 2.2 U mg−1 toward benzylacetone, an up to 110-fold increase compared with that of Gk-PAmDH (Entry 10 in Table 7). Experiments on the transformation of a series of benzylacetone derivatives showed that space-generating mutations (Gk-PAmDH-M3 and Gk-PAmDH-M8) are beneficial for expanding the substrate scope of Gk-PAmDH (Entry 4–9 in Table 7). To study the catalytic potential of these variants, a broader range of bulky substrates (aliphatic 2(or 3, 4)-ketones, methyl ketones with a remote functional group, and methyl ketones with a heterocycle) was examined. Variants M3 and M8 exhibited significantly improved reactivity toward all these challenging substrates (Entry 10–18 in Table 7). Interestingly, through a comparison with the structure of RsPheDH (PDB: 1C1D), the additional mutation sites on Gk-PAmDH-M8 were all found to be inside the pocket, with V144 and V309 coinciding with the previously selected sites of BbPheDH substrate-specific modification [22]. These sites can serve as important references in the subsequent substrate-specific engineering of PheDH.

Table 7.
Altered substrate specificity of Gk-PAmDHs and their synthesis of chiral amines.
4. Conclusions and Perspectives
This review summarizes research on the substrate-specific engineering of s-AADHs by combining the structure and catalytic mechanism. From the substrate-specific engineering studies summarized in Table 2, Table 3, Table 4, Table 5, Table 6 and Table 7, it can be seen that the mutation sites for substrate specificity engineering are mainly concentrated in four regions as follows: 1. inside the substrate pocket, 2. the carboxyl anchor site outside the substrate pocket, 3. the substrate entrance tunnel, and 4. the hinge region (the position in the structure is shown in Figure 3a).
Many mutation sites are inside the pocket, which allows s-AADHs to accommodate substrates with different R groups. Combinatorial mutation of the two carboxyl anchor sites outside the pocket can expand the substrate specificity of s-AADHs from α-keto acid to α-hydroxy-2-ketones, methyl ketones, ethyl ketone, and propyl ketone. Moreover, mutations at these two hotspots have additive effects, which further expands the substrate spectrum of engineered s-AADHs. Currently, the engineering of substrate carboxyl anchor sites is mainly focused on LeuDH, PheDH, and GluDH, of which there is only one report based on GluDH. The sequence alignment in Figure 4 shows that two residues at the carboxyl-anchoring site are conserved in s-AADHs, suggesting that similar substrate specificity modifications apply to ValDH and TrpDH. Given the wide distribution of GluDH in organisms, the construction of engineered amine dehydrogenases based on GluDH has great potential.
The other two hotspots (the substrate entrance tunnel and hinge region of s-AADHs) are the subject of relatively few reports. However, the catalytic activity of the engineered enzymes obtained by site mutations at the substrate entrance tunnel of GluDH and LeuDH has been improved for multiple substrates, which indicates that engineering at the tunnel site has great potential for the modification of the substrate spectrum of s-AADHs. An interesting case, domain shuffling, has been reported in substrate-specific engineering. The chimeric enzymes obtained by domain shuffling have the substrate specificity of the two parent enzymes and can even catalyze inactive substrates of the parent enzymes. In a study on the crystal structure of a chimeric GluDH, Oliveira et al. [67] found that the change in the angle between the two domains of the chimeric enzyme, caused by an interdomain flexibility change at a hinge region, promotes improvements in its catalytic efficiency. This study coincides with Xinjian Yin et al.’s [30] hinge engineering to improve the catalytic efficiency of enzymes, suggesting that domain shuffling might be a special form of hinge engineering. Domain shuffling and hinge engineering result in significant changes in the substrate specificity of s-AADHs, in which the variant (T121N/L123Y) obtained by hinge engineering shows a nearly 100-fold improvement in catalytic efficiency for unnatural substrates compared with that of the wild-type. As shown in Figure 1, s-AADHs all contain hinge structures, which means that domain shuffling and hinge engineering can be applied to the substrate-specific engineering of other s-AADHs.
s-AADHs catalyze the asymmetric reductive amination of keto acids or ketones using inexpensive ammonia as the reagent, with only water as a by-product, which is a highly desired catalyst in green and sustainable pharmaceutical manufacturing [71]. The rational combination of these mutation hotspots is expected to lead to the construction of more excellent engineered enzymes, thereby paving the way for the green industrial synthesis of chiral amino acids and chiral amines.
Author Contributions
Conceptualization, F.Z.; investigation, F.Z.; Writing—original draft preparation, F.Z.; writing—review and editing, F.Z., Y.N. and X.M.; funding acquisition, Y.N. and X.M.; Supervision, Y.X., Y.N. and X.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (Grant Numbers 2021YFC2100100), the National Natural Science Foundation of China (NSFC) (Grant Numbers 21336009 and 21176103), the Postgraduate Research and Practice Innovation Program of Jiangsu Province (KYCX19_1831), and the National First-Class Discipline Program of Light Industry Technology and Engineering (Grant Number LITE2018-09).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Tao Wu, the author of the article (DOI: 10.1002/cctc.202101558), for providing the original files of docking simulation in Figure 5.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
AADHs, amino acid dehydrogenases; s-AADHs, superfamily of amino acid dehydrogenases; NAD+, nicotinamide adenine dinucleotide; NADP+, nicotinamide adenine dinucleotide phosphate; GluDH, glutamate dehydrogenase; ValDH, valine dehydrogenase; LeuDH, leucine dehydrogenase; PheDH, phenylalanine dehydrogenase; TryDH, tryptophan dehydrogenase; LAmDH, engineered amine dehydrogenase based on LeuDH; 2-OG, 2-oxoglutarate; 2-IG, 2-iminoglutarate; OAA, oxaloacetate; PPO, 2-oxo-4-[(hydroxy)(methyl)phosphinoyl]butyric acid; PpGluDH, GluDH from Pseudomonas putida; EcGluDH, GluDH from E. coli; CsGluDH, GluDH from Clostridium symbiosum; 2-OPBA, 2-oxo-4-phenylbutyric acid; Ec-GAmDH, engineered amine dehydrogenase based on EcGluDH; BcLeuDH, LeuDH from Bacillus cereus; BsLeuDH, LeuDH from Bacillus stearothermophilus; Es-LAmDH, engineered amine dehydrogenase based on Exiguobacterium sibiricum LeuDH; Lf-LAmDH, engineered amine dehydrogenase based on Lysinibacillus fusiformis LeuDH; Gs-LAmDH, engineered amine dehydrogenase based on Geobacillus stearothermophilus LeuDH; Bc-LAmDH, engineered amine dehydrogenase BcLeuDH; BsPheDH, PheDH from Bacillus sphaericus; SuPheDH, PheDH from Sporosarcina ureae; RsPheDH, PheDH from Rhodococcus sp. M4; BbPheDH, PheDH from Bacillus badius; PFPA, phenoxy-2-propanone; Bb-PAmDH, engineered amine dehydrogenase based on BbPheDH; Rs-PAmDH, engineered amine dehydrogenase based on RsPheDH; cFL1-AmDH, a chimeric amine dehydrogenase with domain I of Bb-PAmDH and domain II of Bs-LAmDH; Gk-PAmDH, engineered amine dehydrogenase based on Geobacillus kaustophilus PheDH.
References
- Obermeier, N.; Poralla, K. Experiments on the role of leucine dehydrogenase in initiation of Bacillus subtilis spore germination. FEMS Microbiol. Lett. 1979, 5, 81–83. [Google Scholar] [CrossRef][Green Version]
- Tang, L.; Zhang, Y.X.; Hutchinson, C.R. Amino acid catabolism and antibiotic synthesis: Valine is a source of precursors for macrolide biosynthesis in Streptomyces ambofaciens and Streptomyces fradiae. J. Bacteriol. 1994, 176, 6107–6119. [Google Scholar] [CrossRef] [PubMed]
- Balskus, E.P.; Walsh, C.T. Investigating the initial steps in the biosynthesis of cyanobacterial sunscreen scytonemin. J. Am. Chem. Soc. 2008, 130, 15260–15261. [Google Scholar] [CrossRef] [PubMed]
- Prakash, P.; Punekar, N.S.; Hareshwar, P.V. Structural basis for the catalytic mechanism and α-ketoglutarate cooperativity of glutamate dehydrogenase. J. Biol. Chem. 2018, 293, 6241–6258. [Google Scholar] [CrossRef]
- Britton, K.L.; Baker, P.J.; Engel, P.C.; Rice, D.W.; Stillman, T.J. Evolution of substrate diversity in the superfamily of amino acid dehydrogenases. J. Mol. Biol. 1993, 234, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.J.; Waugh, M.L.; Wang, X.-G.; Stillman, T.J.; Turnbull, A.P.; Engel, P.C.; Rice, D.W. Determinants of substrate specificity in the superfamily of amino acid dehydrogenases. Biochemistry 1997, 36, 16109–16115. [Google Scholar] [CrossRef] [PubMed]
- Hyun, C.G.; Kim, S.S.; Park, K.H.; Suh, J.W. Valine dehydrogenase from Streptomyces albus: Gene cloning, heterologous expression and identification of active site by site-directed mutagenesis. FEMS Microbiol. Lett. 2000, 182, 29–34. [Google Scholar] [CrossRef][Green Version]
- Wakamatsu, T.; Sakuraba, H.; Kitamura, M.; Hakumai, Y.; Fukui, K.; Ohnishi, K.; Ashiuchi, M.; Ohshima, T. Structural insights into L-tryptophan dehydrogenase from a photoautotrophic cyanobacterium, Nostoc punctiforme. Appl. Environ. Microbiol. 2017, 83, e02710-16. [Google Scholar] [CrossRef]
- Ohshima, T.; Soda, K. Stereoselective biocatalysis: Amino acid dehydrogenases and their applications. In Stereoselective Biocatalysis; Patel, R.N., Ed.; Marcel Dekker Inc.: New York, NY, USA, 2000; pp. 877–902. [Google Scholar] [CrossRef]
- Xue, Y.-P.; Cao, C.-H.; Zheng, Y.-G. Enzymatic asymmetric synthesis of chiral amino acids. Chem. Soc. Rev. 2018, 47, 1516–1561. [Google Scholar] [CrossRef] [PubMed]
- Hummel, W.; Gröger, H. Reductive Amination of Keto Acids. In Enzyme Catalysis in Organic Synthesis; John Wiley & Sons, Ltd.: Weinheim, Germany, 2012; pp. 1165–1203. [Google Scholar] [CrossRef]
- Wang, X.G.; Britton, K.L.; Stillman, T.J.; Rice, D.W.; Engel, P.C. Conversion of a glutamate dehydrogenase into methionine/norleucine dehydrogenase by site-directed mutagenesis. Eur. J. Biochem. 2001, 268, 5791–5799. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.H.; Kim, H.; Ashida, H.; Ishikawa, T.; Shibata, H.; Sawa, Y. Altering the substrate specificity of glutamate dehydrogenase from Bacillus subtilis by site-directed mutagenesis. Biosci. Biotechnol. Biochem. 2005, 69, 1802–1805. [Google Scholar] [CrossRef][Green Version]
- Turnbull, A.P.; Baker, P.J.; Rice, D.W. Analysis of the quaternary structure, substrate specificity, and catalytic mechanism of valine dehydrogenase. J. Biol. Chem. 1997, 272, 25105–25111. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seah, S.Y.K.; Britton, K.L.; Rice, D.W.; Asano, Y.; Engel, P.C. Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its discrimination between phenylalanine and tyrosine substrates. Biochemistry 2002, 41, 11390–11397. [Google Scholar] [CrossRef]
- Hyun, C.-G.; Suk Kim, S.; Lee, I.H.; Suh, J.-W. Alteration of substrate specificity of valine dehydrogenase from Streptomyces albus. Antonie Van Leeuwenhoek 2000, 78, 237–242. [Google Scholar] [CrossRef]
- Kataoka, K.; Tanizawa, K. Alteration of substrate specificity of leucine dehydrogenase by site-directed mutagenesis. J. Mol. Catal. B Enzym. 2003, 23, 299–309. [Google Scholar] [CrossRef]
- Wang, L.; Diao, S.; Sun, Y.; Jiang, S.; Liu, Y.; Wang, H.; Wei, D. Rational engineering of Acinetobacter tandoii glutamate dehydrogenase for asymmetric synthesis of L-homoalanine through biocatalytic cascades. Catal. Sci. Technol. 2021, 11, 4208–4215. [Google Scholar] [CrossRef]
- Zhang, K.; Li, H.; Cho, K.M.; Liao, J.C. Expanding metabolism for total biosynthesis of the nonnatural amino acid L-homoalanine. Proc. Natl. Acad. Sci. USA 2010, 107, 6234–6239. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Mu, X.; Nie, Y.; Xu, Y. Enhanced catalytic efficiency and coenzyme affinity of leucine dehydrogenase by comprehensive screening strategy for L-tert-leucine synthesis. Appl. Microbiol. Biotechnol. 2021, 105, 3625–3634. [Google Scholar] [CrossRef]
- Khorsand, F.; Murphy, C.D.; Whitehead, A.J.; Engel, P.C. Biocatalytic stereoinversion of D-para-bromophenylalanine in a one-pot three-enzyme reaction. Green Chem. 2017, 19, 503–510. [Google Scholar] [CrossRef]
- Wu, T.; Mu, X.; Xue, Y.; Xu, Y.; Nie, Y. Structure-guided steric hindrance engineering of Bacillus badius phenylalanine dehydrogenase for efficient L-homophenylalanine synthesis. Biotechnol. Biofuels 2021, 14, 207–213. [Google Scholar] [CrossRef]
- Abrahamson, M.J.; Vázquez-Figueroa, E.; Woodall, N.B.; Moore, J.C.; Bommarius, A.S. Development of an amine dehydrogenase for synthesis of chiral amines. Angew. Chem. Int. Ed. Engl. 2012, 51, 3969–3972. [Google Scholar] [CrossRef]
- Chen, F.-F.; Cosgrove, S.C.; Birmingham, W.R.; Mangas-Sanchez, J.; Citoler, J.; Thompson, M.P.; Zheng, G.-W.; Xu, J.-H.; Turner, N.J. Enantioselective synthesis of chiral vicinal amino alcohols using amine dehydrogenases. ACS Catal. 2019, 9, 11813–11818. [Google Scholar] [CrossRef]
- Liu, L.; Wang, D.-H.; Chen, F.-F.; Zhang, Z.-J.; Chen, Q.; Xu, J.-H.; Wang, Z.-L.; Zheng, G.-W. Development of an engineered thermostable amine dehydrogenase for the synthesis of structurally diverse chiral amines. Catal. Sci. Technol. 2020, 10, 2353–2358. [Google Scholar] [CrossRef]
- Zhou, F.; Xu, Y.; Mu, X.; Nie, Y. A sustainable approach for synthesizing (R)-4-aminopentanoic acid from levulinic acid catalyzed by structure-guided tailored glutamate dehydrogenase. Front. Bioeng. Biotechnol. 2022, 9, 770302. [Google Scholar] [CrossRef]
- Abrahamson, M.J.; Wong, J.W.; Bommarius, A.S. The evolution of an amine dehydrogenase biocatalyst for the asymmetric production of chiral amines. Adv. Synth. Catal. 2013, 355, 1780–1786. [Google Scholar] [CrossRef]
- Ye, L.J.; Toh, H.H.; Yang, Y.; Adams, J.P.; Snajdrova, R.; Li, Z. Engineering of amine dehydrogenase for asymmetric reductive amination of ketone by evolving Rhodococcus phenylalanine dehydrogenase. ACS Catal. 2015, 5, 1119–1122. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, Y.; Chen, J.; Xu, M.; Yang, T.; Zheng, J.; Zhang, X.; Rao, Z. Rational engineering of Bacillus cereus leucine dehydrogenase towards α-keto acid reduction for improving unnatural amino acid production. Biotechnol. J. 2018, 14, 1800253. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Liu, Y.; Meng, L.; Zhou, H.; Wu, J.; Yang, L. Semi-rational hinge engineering: Modulating the conformational transformation of glutamate dehydrogenase for enhanced reductive amination activity towards non-natural substrates. Catal. Sci. Technol. 2020, 10, 3376–3386. [Google Scholar] [CrossRef]
- Son, H.F.; Kim, I.-K.; Kim, K.-J. Structural insights into domain movement and cofactor specificity of glutamate dehydrogenase from Corynebacterium glutamicum. Biochem. Biophys. Res. Commun. 2015, 459, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Tomita, T.; Yin, L.; Nakamura, S.; Kosono, S.; Kuzuyama, T.; Nishiyama, M. Crystal structure of the 2-iminoglutarate-bound complex of glutamate dehydrogenase from Corynebacterium glutamicum. FEBS Lett. 2017, 591, 1611–1622. [Google Scholar] [CrossRef]
- Chovancova, E.; Pavelka, A.; Benes, P.; Strnad, O.; Brezovsky, J.; Kozlikova, B.; Gora, A.; Sustr, V.; Klvana, M.; Medek, P.; et al. CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 2012, 8, e1002708. [Google Scholar] [CrossRef]
- Brunhuber, N.M.; Thoden, J.B.; Blanchard, J.S.; Vanhooke, J.L. Rhodococcus L-phenylalanine dehydrogenase: kinetics, mechanism, and structural basis for catalytic specifity. Biochemistry 2000, 39, 9174–9187. [Google Scholar] [CrossRef]
- Sekimoto, T.; Matsuyama, T.; Fukui, T.; Tanizawa, K. Evidence for lysine 80 as general base catalyst of leucine dehydrogenase. J. Biol. Chem. 1993, 268, 27039–27045. [Google Scholar] [CrossRef]
- Yin, X.; Liu, Y.; Meng, L.; Zhou, H.; Wu, J.; Yang, L. Rational molecular engineering of glutamate dehydrogenases for enhancing asymmetric reductive amination of bulky α-keto acids. Adv. Synth. Catal. 2018, 361, 803–812. [Google Scholar] [CrossRef]
- Engel, P.C. Glutamate dehydrogenases: The why and how of coenzyme specificity. Neurochem. Res. 2014, 39, 426–432. [Google Scholar] [CrossRef]
- Fang, J.-M.; Lin, C.-H.; Bradshaw, C.W.; Wong, C.-H. Enzymes in organic synthesis: Oxidoreductions. J. Chem. Soc. Perkin Trans. 1 1995, 967–978. [Google Scholar] [CrossRef]
- Hanson, R.L.; Schwinden, M.D.; Banerjee, A.; Brzozowski, D.B.; Chen, B.C.; Patel, B.P.; McNamee, C.G.; Kodersha, G.A.; Kronenthal, D.R.; Patel, R.N.; et al. Enzymatic synthesis of L-6-hydroxynorleucine. Bioorg. Med. Chem. 1999, 7, 2247–2252. [Google Scholar] [CrossRef]
- Yin, X.; Wu, J.; Yang, L. Efficient reductive amination process for enantioselective synthesis of L-phosphinothricin applying engineered glutamate dehydrogenase. Appl. Microbiol. Biotechnol. 2018, 102, 4425–4433. [Google Scholar] [CrossRef] [PubMed]
- Geng, F.; Ma, C.-W.; Zeng, A.-P. Reengineering substrate specificity of E. coli glutamate dehydrogenase using a position-based prediction method. Biotechnol. Lett. 2017, 39, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Munawar, N.; Engel, P.C. Prospects for robust biocatalysis: Engineering of novel specificity in a halophilic amino acid dehydrogenase. Extremophiles 2013, 17, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liao, J.C. Development of an NADPH-dependent homophenylalanine dehydrogenase by protein engineering. ACS Synth. Biol. 2014, 3, 13–20. [Google Scholar] [CrossRef]
- Oikawa, T.; Yamanaka, K.; Kazuoka, T.; Kanzawa, N.; Soda, K. Psychrophilic valine dehydrogenase of the antarctic psychrophile, Cytophaga sp. KUC-1. Eur. J. Biochem. 2001, 268, 4375–4383. [Google Scholar] [CrossRef] [PubMed]
- Vancurová, I.; Vancura, A.; Volc, J.; Neuzil, J.; Flieger, M.; Basarová, G.; Bĕhal, V. Isolation and characterization of valine dehydrogenase from Streptomyces aureofaciens. J. Bacteriol. 1988, 170, 5192–5196. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, T.; Misono, H.; Soda, K. Properties of crystalline leucine dehydrogenase from Bacillus sphaericus. J. Biol. Chem. 1978, 253, 5719–5725. [Google Scholar] [CrossRef]
- Chen, F.-F.; Liu, Y.-Y.; Zheng, G.-W.; Xu, J.-H. Asymmetric amination of secondary alcohols by using a redox-neutral two-enzyme Cascade. ChemCatChem 2015, 7, 3838–3841. [Google Scholar] [CrossRef]
- Löwe, J.; Ingram, A.A.; Gröger, H. Enantioselective synthesis of amines via reductive amination with a dehydrogenase mutant from Exigobacterium sibiricum: Substrate scope, co-solvent tolerance and biocatalyst immobilization. Bioorg. Med. Chem. 2017, 26, 1387–1392. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.-F.; Zheng, G.-W.; Liu, L.; Li, H.; Chen, Q.; Li, F.-L.; Li, C.-X.; Xu, J.-H. Reshaping the active pocket of amine dehydrogenases for asymmetric synthesis of bulky aliphatic amines. ACS Catal. 2018, 8, 2622–2628. [Google Scholar] [CrossRef]
- Franklin, R.D.; Mount, C.J.; Bommarius, B.R.; Bommarius, A.S. Separate sets of mutations enhance activity and substrate scope of amine dehydrogenase. ChemCatChem 2020, 12, 2436–2439. [Google Scholar] [CrossRef]
- Wang, H.; Qu, G.; Li, J.K.; Ma, J.A.; Guo, J.; Miao, Y.; Sun, Z. Data mining of amine dehydrogenases for the synthesis of enantiopure amino alcohols. Catal. Sci. Technol. 2020, 10, 5945–5952. [Google Scholar] [CrossRef]
- Mu, X.; Wu, T.; Mao, Y.; Zhao, Y.; Xu, Y.; Nie, Y. Iterative alanine scanning mutagenesis confers aromatic ketone specificity and activity of L-amine dehydrogenases. ChemCatChem 2021, 13, 5243–5253. [Google Scholar] [CrossRef]
- Minh, T.H.; Kutáček, M.; Šebánek, J. Growth-correlative effect of the root on the apical part of the epicotyl in pea seedlings regarding the IAA content and L-tryptophan aminotransferase and L-tryptophan dehydrogenase activities. Biol. Plant. 1984, 26, 342–348. [Google Scholar] [CrossRef]
- El Bahr, M.K.; Kutáček, M.; Opatrný, Z. L-Tryptophan aminotransferase and L-tryptophan dehydrogenase, enzymes of IAA synthesis, in normal and tumorous tobacco tissue cultures. Biochem. Physiol. Pflanz. 1987, 182, 213–222. [Google Scholar] [CrossRef]
- Ogura, R.; Wakamatsu, T.; Mutaguchi, Y.; Doi, K.; Ohshima, T. Biochemical characterization of an L-tryptophan dehydrogenase from the photoautotrophic cyanobacterium Nostoc punctiforme. Enzym. Microb. Technol. 2014, 60, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Matsui, D.; Okazaki, S.; Matsuda, M.; Asano, Y. Enhancement of stability of L-tryptophan dehydrogenase from Nostoc punctiforme ATCC29133 and its application to L-tryptophan assay. J. Biotechnol. 2015, 196–197, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Seah, S.Y.; Britton, K.L.; Baker, P.J.; Rice, D.W.; Asano, Y.; Engel, P.C. Alteration in relative activities of phenylalanine dehydrogenase towards different substrates by site-directed mutagenesis. FEBS Lett. 1995, 370, 93–96. [Google Scholar] [CrossRef]
- Seah, S.Y.K.; Britton, K.L.; Rice, D.W.; Asano, Y.; Engel, P.C. Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase activity with guidance from homology-based modelling. Eur. J. Biochem. 2003, 270, 4628–4634. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, F.; Ataei, F.; Arab, S.S.; Hosseinkhani, S. Increase of Bacillus badius phenylalanine dehydrogenase specificity towards phenylalanine substrate by site-directed mutagenesis. Arch. Biochem. Biophys. 2017, 635, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Busca, P.; Paradisi, F.; Moynihan, E.; Maguire, A.R.; Engel, P.C. Enantioselective synthesis of non-natural amino acids using phenylalanine dehydrogenases modified by site-directed mutagenesis. Org. Biomol. Chem. 2004, 2, 2684. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, S.; Kuwamori, Y.; Asano, Y. Discrimination of aliphatic substrates by a single amino acid substitution in Bacillus badius and Bacillus sphaericus phenylalanine dehydrogenases. Biosci. Biotechnol. Biochem. 2009, 73, 729–732. [Google Scholar] [CrossRef][Green Version]
- Chen, S.; Engel, P.C. Efficient screening for new amino acid dehydrogenase activity: Directed evolution of Bacillus sphaericus phenylalanine dehydrogenase towards activity with an unsaturated non-natural amino acid. J. Biotechnol. 2009, 142, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, K.; Takada, H.; Tanizawa, K.; Yoshimura, T.; Esaki, N.; Ohshima, T.; Soda, K. Construction and characterization of chimeric enzyme consisting of an amino-terminal domain of phenylalanine dehydrogenase and a carboxy-terminal domain of leucine dehydrogenase. J. Biochem. 1994, 116, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, T.; Kataoka, K.; Jin, Y.; Suzuki, S.; Soda, K. Fragmentary form of thermostable leucine dehydrogenase of Bacillus stearothermophilus: Its construction and reconstitution of active fragmentary enzyme. Biochem. Biophys. Res. Commun. 2001, 280, 1177–1182. [Google Scholar] [CrossRef]
- Bommarius, B.R.; Schürmann, M.; Bommarius, A.S. A novel chimeric amine dehydrogenase shows altered substrate specificity compared to its parent enzymes. Chem. Commun. 2014, 50, 14953–14955. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, M.A.; Engel, P.C. Modular coenzyme specificity: A domain-swopped chimera of glutamate dehydrogenase. Proteins 2009, 77, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, T.; Sharkey, M.A.; Engel, P.C.; Khan, A.R. Crystal structure of a chimaeric bacterial glutamate dehydrogenase. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2016, 72, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Knaus, T.; Böhmer, W.; Mutti, F.G. Amine dehydrogenases: Efficient biocatalysts for the reductive amination of carbonyl compounds. Green Chem. 2017, 19, 453–463. [Google Scholar] [CrossRef]
- Pushpanath, A.; Siirola, E.; Bornadel, A.; Woodlock, D.; Schell, U. Understanding and overcoming the limitations of Bacillus badius and Caldalkalibacillus thermarum amine dehydrogenases for biocatalytic reductive amination. ACS Catal. 2017, 7, 3204–3209. [Google Scholar] [CrossRef]
- Wang, D.-H.; Chen, Q.; Yin, S.-N.; Ding, X.-W.; Zheng, Y.-C.; Zhang, Z.; Zhang, Y.-H.; Chen, F.-F.; Xu, J.-H.; Zheng, G.-W. Asymmetric reductive amination of structurally diverse ketones with ammonia using a spectrum-extended amine dehydrogenase. ACS Catal. 2021, 11, 14274–14283. [Google Scholar] [CrossRef]
- Constable, D.J.C.; Dunn, P.J.; Hayler, J.D.; Humphrey, G.R.; Leazer, J.L., Jr.; Linderman, R.J.; Lorenz, K.; Manley, J.; Pearlman, B.A.; Wells, A.; et al. Key green chemistry research areas—A perspective from pharmaceutical manufacturers. Green Chem. 2007, 9, 411–420. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).