Predicting Active Sites in Photocatalytic Degradation Process Using an Interpretable Molecular-Image Combined Convolutional Neural Network
Abstract
:1. Introduction
2. Results
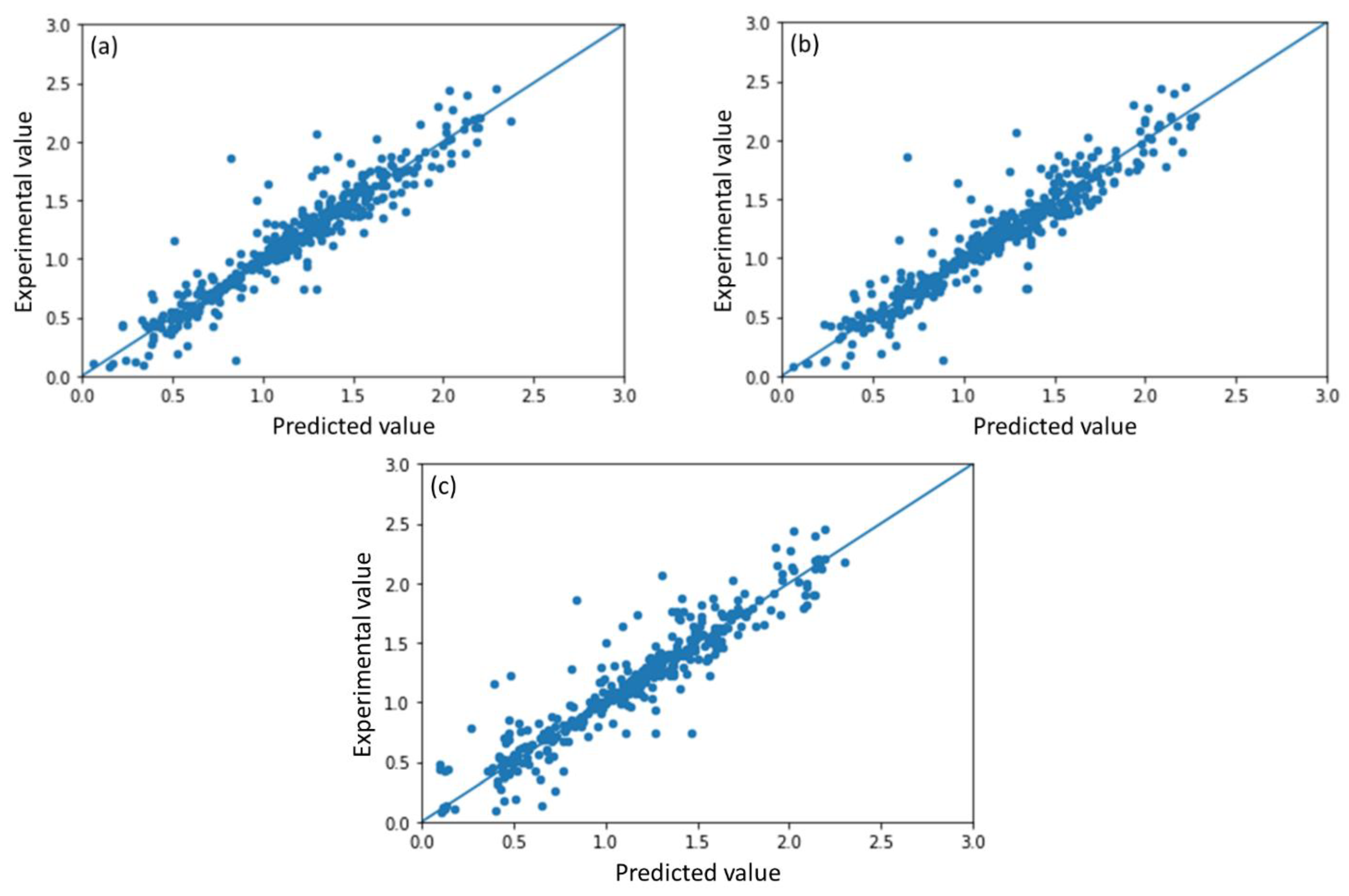
2.1. Model Performance and Comparison
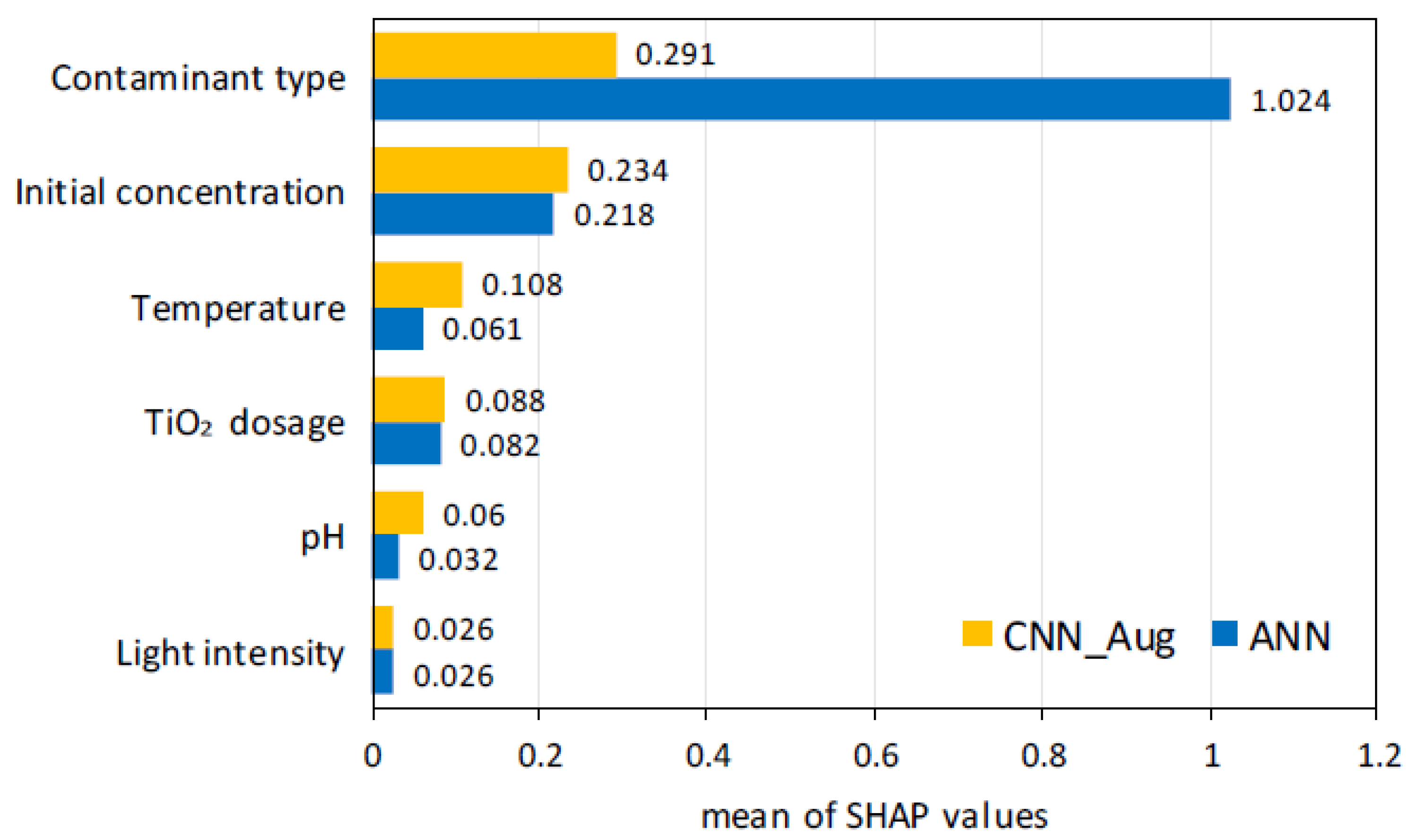
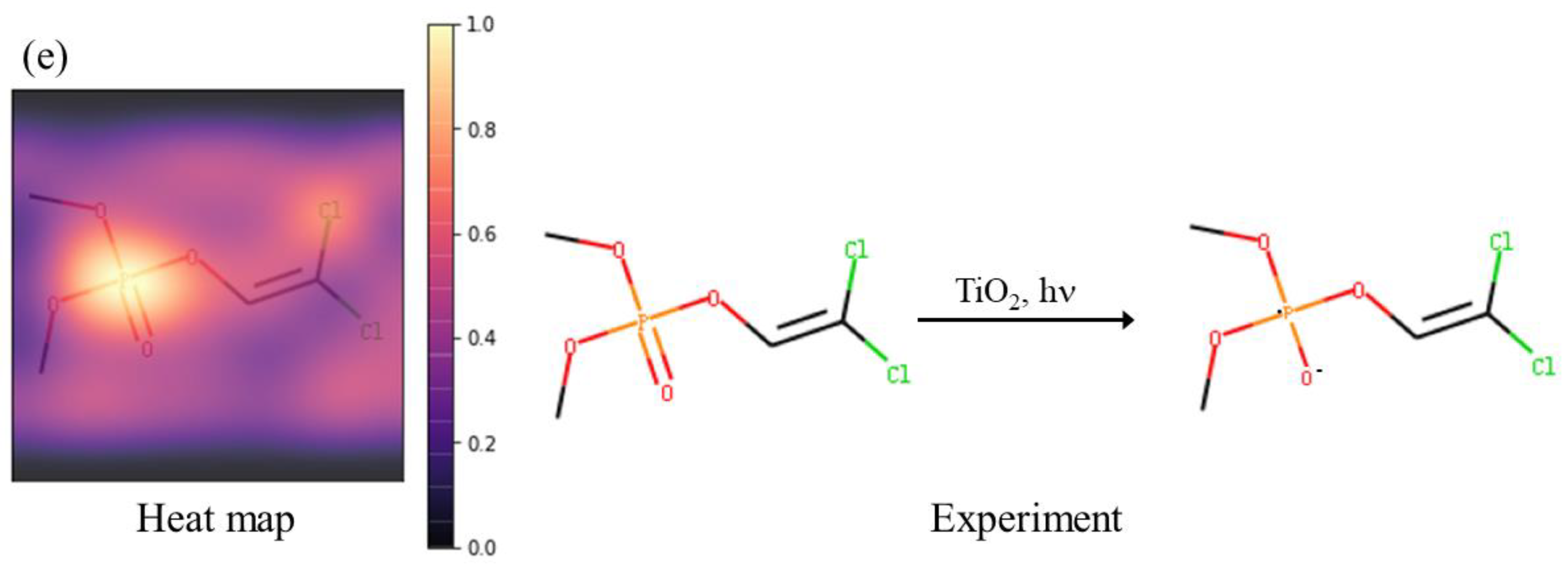
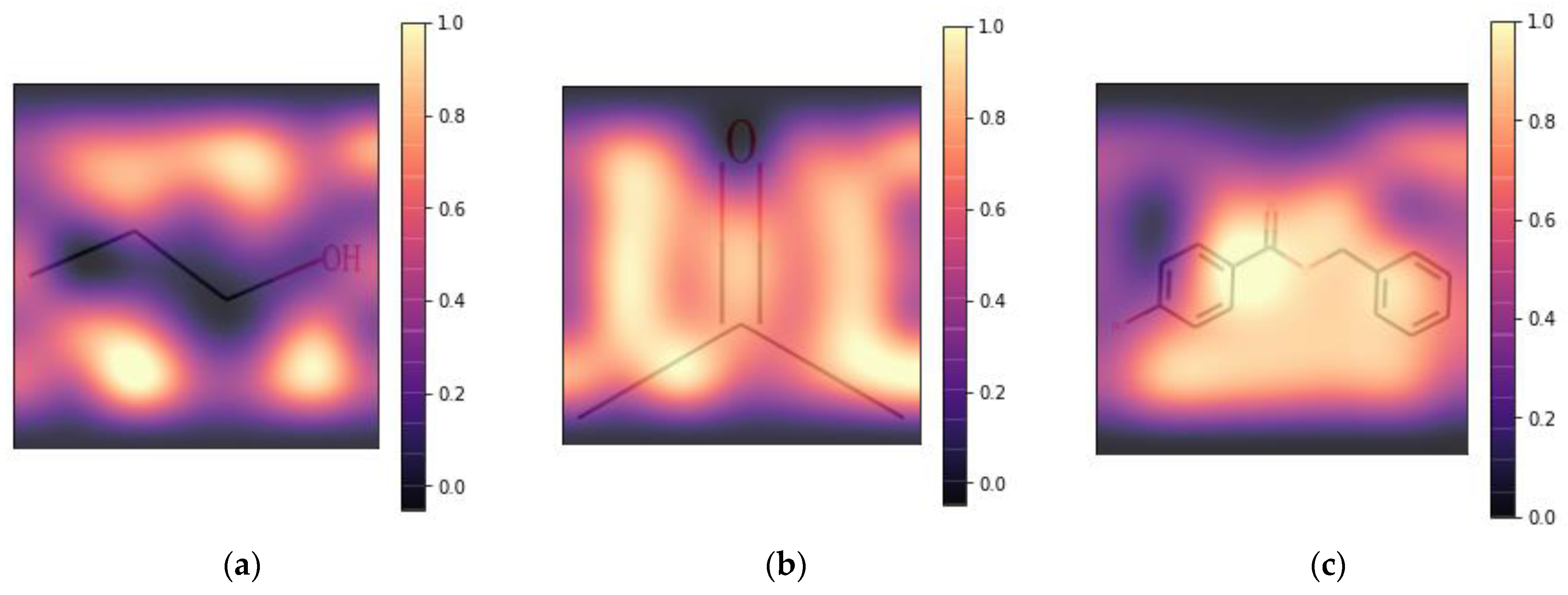
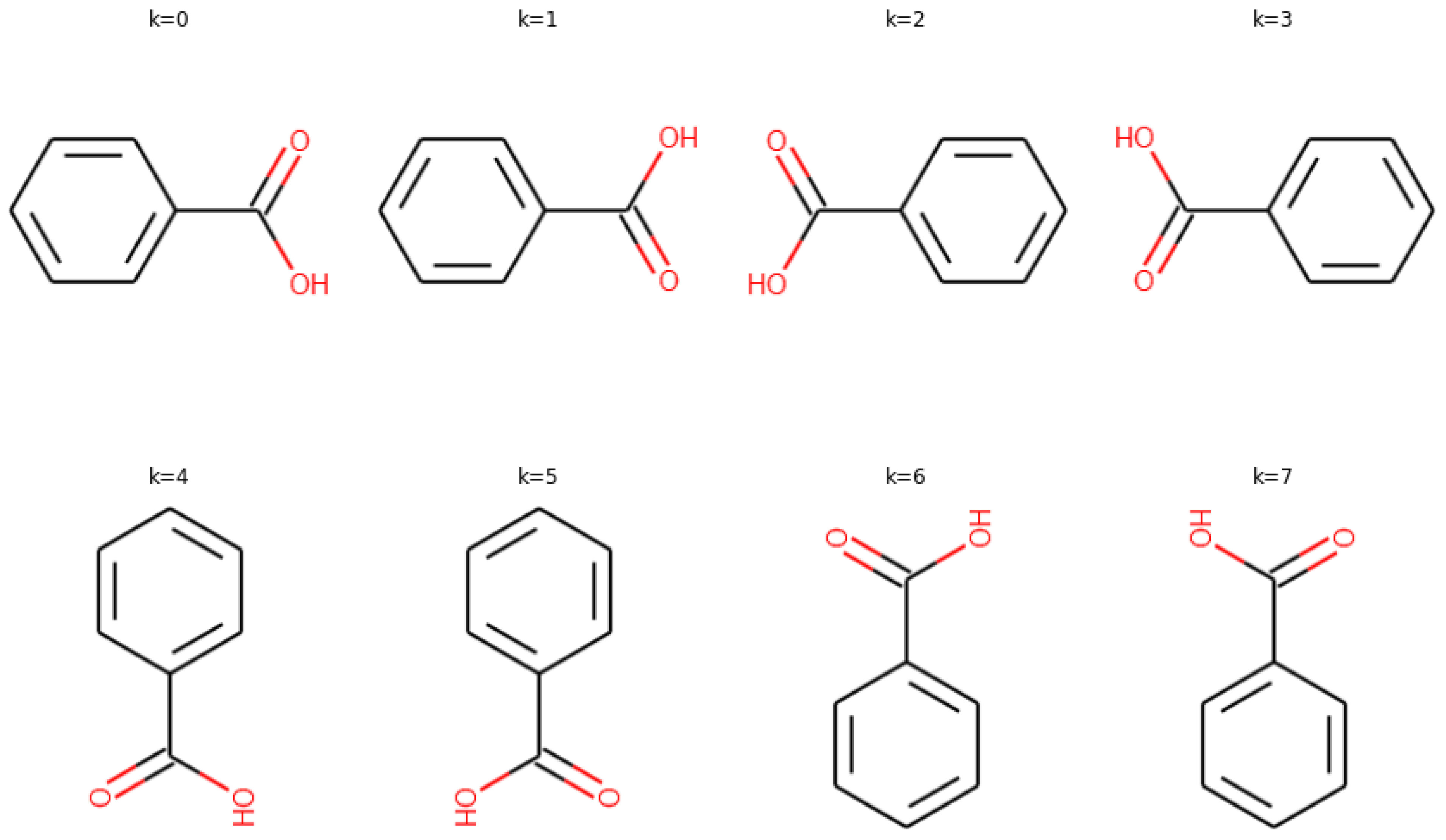
2.2. Feature Importance
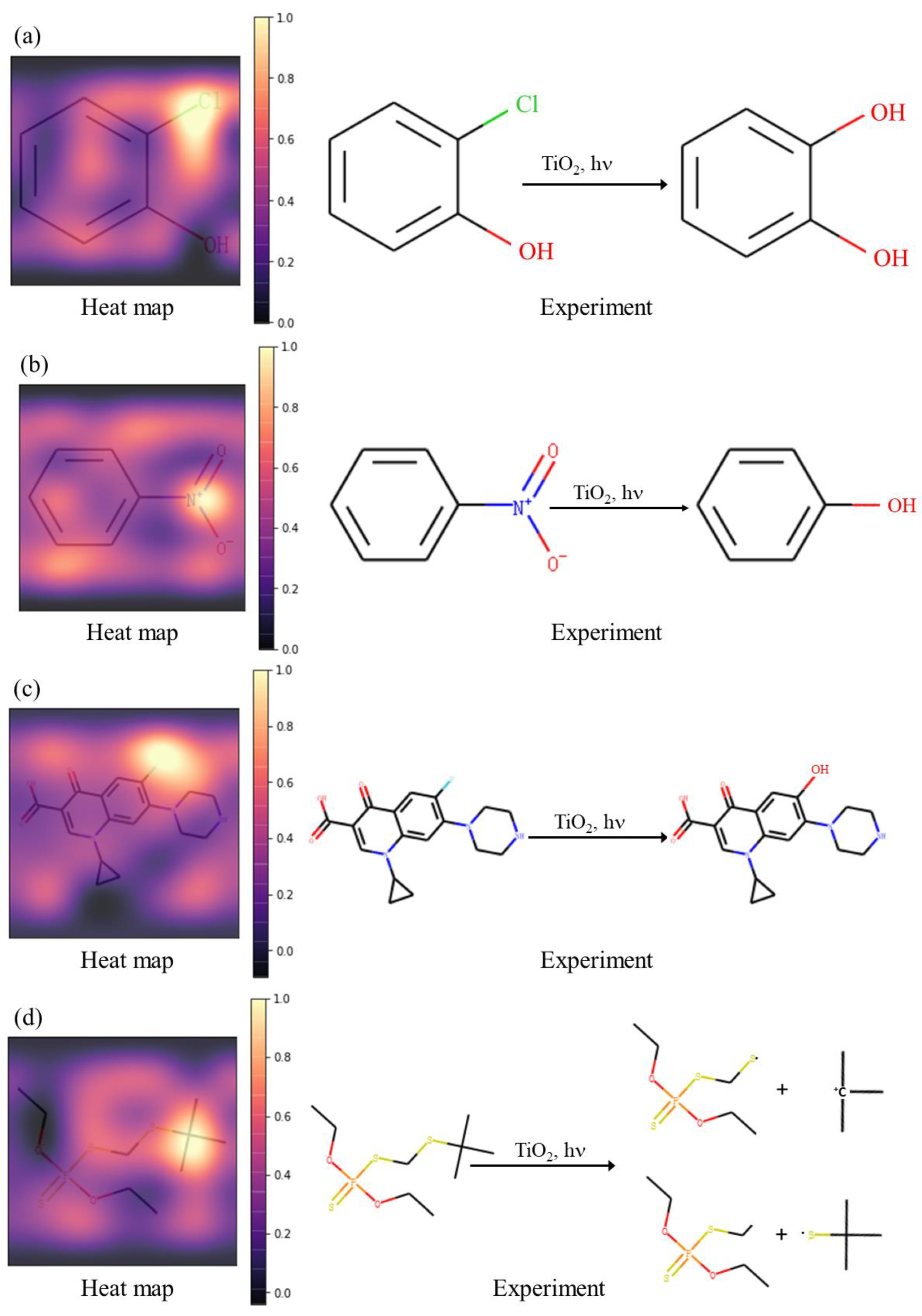
2.3. Interpretability of Active Site Prediction
3. Methods
3.1. Datasets
3.2. Data Augmentation
3.3. Model Structure
4. Conclusions
5. Discussions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flah, M.; Nunez, I.; Chaabene, W.B.; Nehdi, M.L. Machine learning algorithms in civil structural health monitoring: A systematic review. Arch. Comput. Methods Eng. 2020, 28, 2621–2643. [Google Scholar] [CrossRef]
- Ying, Y.; Garrett, J.H., Jr.; Oppenheim, I.J.; Soibelman, L.; Harley, J.B.; Shi, J.; Jin, Y. Toward data-driven structural health monitoring: Application of machine learning and signal processing to damage detection. J. Comput. Civ. Eng. 2013, 27, 667–680. [Google Scholar] [CrossRef]
- Granata, F.; Papirio, S.; Esposito, G.; Gargano, R.; De Marinis, G. Machine learning algorithms for the forecasting of wastewater quality indicators. Water 2017, 9, 105. [Google Scholar] [CrossRef] [Green Version]
- Miller, T.H.; Gallidabino, M.D.; MacRae, J.I.; Hogstrand, C.; Bury, N.R.; Barron, L.P.; Snape, J.R.; Owen, S.F. Machine learning for environmental toxicology: A call for integration and innovation. Environ. Sci. Technol. 2018, 52, 12953–12955. [Google Scholar] [CrossRef] [Green Version]
- Kitchin, J.R. Machine learning in catalysis. Nat. Catal. 2018, 1, 230–232. [Google Scholar] [CrossRef]
- Yang, W.; Fidelis, T.T.; Sun, W.H. Machine Learning in Catalysis, From Proposal to Practicing. ACS Omega 2019, 5, 83–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zang, Q.; Mansouri, K.; Williams, A.J.; Judson, R.S.; Allen, D.G.; Casey, W.M.; Kleinstreuer, N.C. In silico prediction of physicochemical properties of environmental chemicals using molecular fingerprints and machine learning. J. Chem. Inf. Model. 2017, 57, 36–49. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, G. Machine learning based toxicity prediction: From chemical structural description to transcriptome analysis. Int. J. Mol. Sci. 2018, 19, 2358. [Google Scholar] [CrossRef] [Green Version]
- Cipullo, S.; Snapir, B.; Prpich, G.; Campo, P.; Coulon, F. Prediction of bioavailability and toxicity of complex chemical mixtures through machine learning models. Chemosphere 2019, 215, 388–395. [Google Scholar] [CrossRef] [Green Version]
- Xi, X.; Wei, Z.; Xiaoguang, R.; Yijie, W.; Xinxin, B.; Wenjun, Y.; Jin, D. A comprehensive evaluation of air pollution prediction improvement by a machine learning method. In Proceedings of the 2015 IEEE International Conference on Service Operations and Logistics, And Informatics (SOLI), Yasmine Hammamet, Tunisia, 15–17 November 2015; pp. 176–181. [Google Scholar]
- Chowdhury, A.J.; Yang, W.; Walker, E.; Mamun, O.; Heyden, A.; Terejanu, G.A. Prediction of adsorption energies for chemical species on metal catalyst surfaces using machine learning. J. Phys. Chem. C 2018, 122, 28142–28150. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Xu, X. Machine learning band gaps of doped-TiO2 photocatalysts from structural and morphological parameters. ACS Omega 2020, 5, 15344–15352. [Google Scholar] [CrossRef] [PubMed]
- Rajan, A.C.; Mishra, A.; Satsangi, S.; Vaish, R.; Mizuseki, H.; Lee, K.R.; Singh, A.K. Machine-learning-assisted accurate band gap predictions of functionalized MXene. Chem. Mater. 2018, 30, 4031–4038. [Google Scholar] [CrossRef]
- Masood, H.; Toe, C.Y.; Teoh, W.Y.; Sethu, V.; Amal, R. Machine Learning for Accelerated Discovery of Solar Photocatalysts. ACS Catal. 2019, 9, 11774–11787. [Google Scholar] [CrossRef]
- Jiang, Z.; Hu, J.; Zhang, X.; Zhao, Y.; Fan, X.; Zhong, S.; Zhang, H.; Yu, X. A Generalized Predictive Model for TiO2–Catalyzed Photo-degradation Rate Constants of Water Contaminants through Artificial Neural Network. Environ. Res. 2020, 187, 109697. [Google Scholar] [CrossRef]
- Rudin, C.; Radin, J. Why are we using black box models in AI when we don’t need to? A lesson from an explainable AI competition. Harv. Data Sci. Rev. 2019, 1. [Google Scholar] [CrossRef]
- Mattessich, S.; Tassavor, M.; Swetter, S.M.; Grant-Kels, J.M. How I learned to stop worrying and love machine learning. Clin. Dermatol. 2018, 36, 777–778. [Google Scholar] [CrossRef] [PubMed]
- Pazzani, M.J.; Billsus, D. Content-based recommendation systems. In The Adaptive Web; Springer: Berlin/Heidelberg, Germany, 2007; pp. 325–341. [Google Scholar]
- Xie, X.; Wang, B. Web page recommendation via twofold clustering: Considering user behavior and topic relation. Neural Comput. Appl. 2018, 29, 235–243. [Google Scholar] [CrossRef]
- He, X.; Pan, J.; Jin, O.; Xu, T.; Liu, B.; Xu, T.; Shi, Y.; Atallah, A.; Herbrich, R.; Bowers, S.; et al. Practical lessons from predicting clicks on ads at facebook. In Proceedings of the Eighth International Workshop on Data Mining for Online Advertising, New York, NY, USA, 24–27 August 2014; pp. 1–9. [Google Scholar]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine learning in medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- Ackermann, K.; Walsh, J.; De Unánue, A.; Naveed, H.; Navarrete Rivera, A.; Lee, S.J.; Bennett, J.; Defoe, M.; Cody, C.; Haynes, L. Deploying machine learning models for public policy: A framework. In Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, New York, NY, USA, 19–23 August 2018; pp. 15–22. [Google Scholar]
- Burscher, B.; Vliegenthart, R.; De Vreese, C.H. Using supervised machine learning to code policy issues: Can classifiers generalize across contexts? Ann. Am. Acad. Political Soc. Sci. 2015, 659, 122–131. [Google Scholar] [CrossRef] [Green Version]
- Ciolacu, M.; Tehrani, A.F.; Beer, R.; Popp, H. Education 4.0—Fostering student’s performance with machine learning methods. In Proceedings of the 2017 IEEE 23rd International Symposium for Design and Technology in Electronic Packaging (SIITME), Constanta, Romania, 26–29 October 2017; pp. 438–443. [Google Scholar]
- Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef] [Green Version]
- Shwartz-Ziv, R.; Tishby, N. Opening the black box of deep neural networks via information. arXiv 2017, arXiv:1703.00810. [Google Scholar]
- Madras, D.; Pitassi, T.; Zemel, R. Predict responsibly: Improving fairness and accuracy by learning to defer. Adv. Neural Inf. Process. Syst. 2018, 31, 6147–6157. [Google Scholar]
- Zou, J.; Huss, M.; Abid, A.; Mohammadi, P.; Torkamani, A.; Telenti, A. A primer on deep learning in genomics. Nat. Genet. 2019, 51, 12–18. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017, 4765–4774. [Google Scholar] [CrossRef]
- Lin, Y.; Ferronato, C.; Deng, N.; Chovelon, J.M. Study of benzylparaben photocatalytic degradation by TiO2. Appl. Catal. B Environ. 2011, 104, 353–360. [Google Scholar] [CrossRef]
- Daneshvar, N.; Salari, D.; Niaei, A.; Khataee, A.R. Photocatalytic degradation of the herbicide erioglaucine in the presence of nanosized titanium dioxide: Comparison and modeling of reaction kinetics. J. Environ. Sci. Health Part B 2006, 41, 1273–1290. [Google Scholar] [CrossRef]
- Qamar, M.; Muneer, M.; Bahnemann, D. Heterogeneous photocatalysed degradation of two selected pesticide derivatives, triclopyr and daminozid in aqueous suspensions of titanium dioxide. J. Environ. Manag. 2006, 80, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Reza, K.M.; Kurny, A.S.W.; Gulshan, F. Parameters affecting the photocatalytic degradation of dyes using TiO2: A review. Appl. Water Sci. 2017, 7, 1569–1578. [Google Scholar] [CrossRef] [Green Version]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE international Conference on Computer Vision, Venice, Italy, 27–29 October 2017; pp. 618–626. [Google Scholar]
- D’Oliveira, J.C.; Al-Sayyed, G.; Pichat, P. Photodegradation of 2- and 3-chlorophenol in titanium dioxide aqueous suspensions. Environ. Sci. Technol. 1990, 24, 990–996. [Google Scholar] [CrossRef]
- Bertelli, M.; Selli, E. Reaction paths and efficiency of photocatalysis on TiO2 and of H2O2 photolysis in the degradation of 2-chlorophenol. J. Hazard. Mater. 2006, 138, 46–52. [Google Scholar] [CrossRef]
- Jeong, S.; Lee, H.; Park, H.; Jeon, K.J.; Park, Y.K.; Jung, S.C. Rapid photocatalytic degradation of nitrobenzene under the simultaneous illumination of UV and microwave radiation fields with a TiO2 ball catalyst. Catal. Today 2018, 307, 65–72. [Google Scholar] [CrossRef]
- Zeng, Y.; Chen, D.; Chen, T.; Cai, M.; Zhang, Q.; Xie, Z.; Li, R.; Xiao, Z.; Liu, G.; Lv, W. Study on heterogeneous photocatalytic ozonation degradation of ciprofloxacin by TiO2/carbon dots: Kinetic, mechanism and pathway investigation. Chemosphere 2019, 227, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.J.; Chen, C.C.; Chen, M.H.; Lu, C.S. Titanium dioxide-mediated heterogeneous photocatalytic degradation of terbufos: Parameter study and reaction pathways. J. Hazard. Mater. 2009, 162, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Muneer, M. Photocatalysed degradation of two selected pesticide derivatives, dichlorvos and phosphamidon, in aqueous suspensions of titanium dioxide. Desalination 2005, 181, 161–172. [Google Scholar] [CrossRef]
- RDKit, an Open-Source Cheminformatics Software. Available online: www.rdkit.org (accessed on 16 December 2020).
- Tan, M.; Le, Q.V. Efficientnet: Rethinking model scaling for convolutional neural networks. arXiv 2019, arXiv:1905.11946. [Google Scholar]
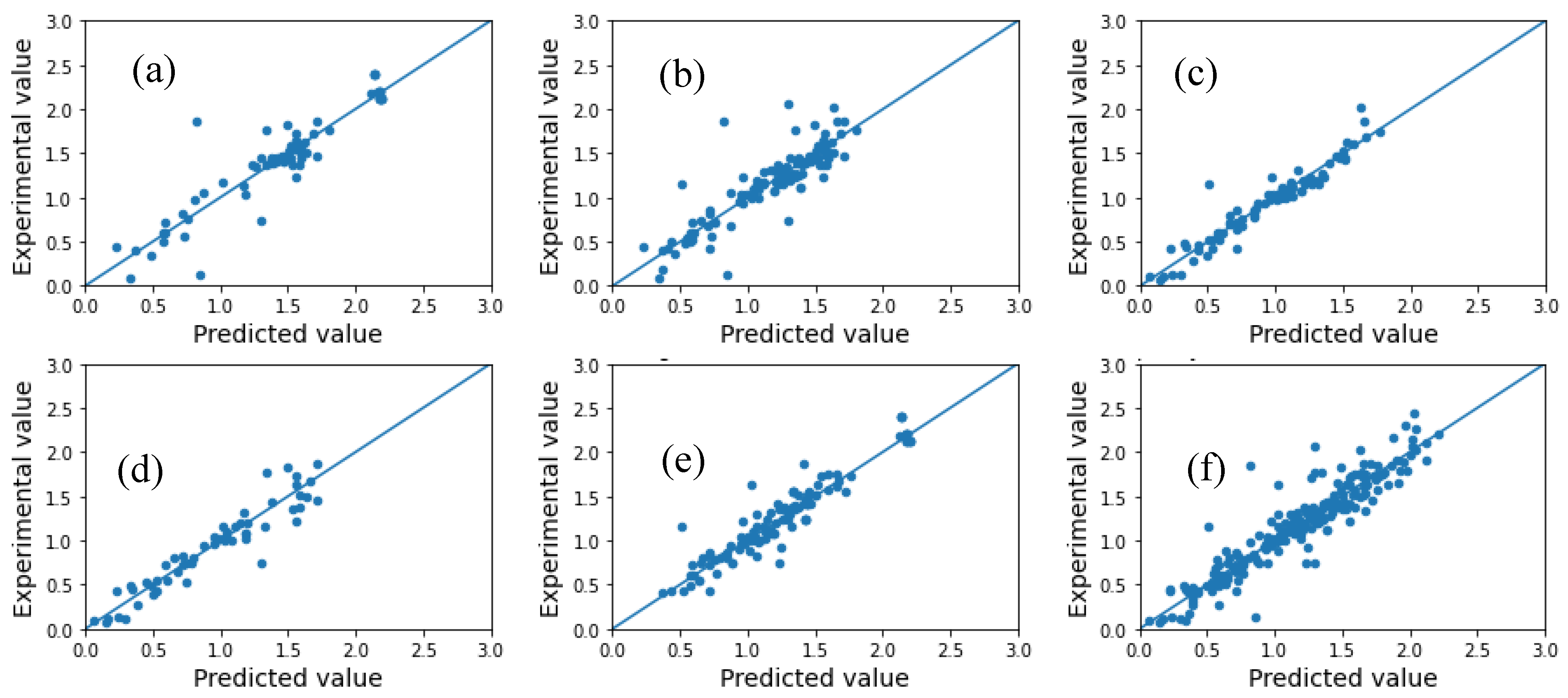




| Model | Metric | Subgroup 1 | Subgroup 2 | Subgroup 3 | Subgroup 4 | Subgroup 5 | Average |
|---|---|---|---|---|---|---|---|
| CNN_Aug | R2 | 0.909 | 0.925 | 0.842 | 0.923 | 0.884 | 0.897 |
| MAE | 0.102 | 0.088 | 0.116 | 0.085 | 0.102 | 0.099 | |
| RMSE | 0.149 | 0.141 | 0.197 | 0.125 | 0.161 | 0.156 | |
| CNN_Ori | R2 | 0.893 | 0.934 | 0.812 | 0.915 | 0.886 | 0.889 |
| MAE | 0.113 | 0.080 | 0.131 | 0.093 | 0.107 | 0.105 | |
| RMSE | 0.161 | 0.132 | 0.214 | 0.131 | 0.160 | 0.163 | |
| ANN | R2 | 0.882 | 0.902 | 0.799 | 0.919 | 0.868 | 0.873 |
| MAE | 0.110 | 0.101 | 0.134 | 0.084 | 0.111 | 0.108 | |
| RMSE | 0.170 | 0.162 | 0.222 | 0.128 | 0.172 | 0.173 |
| Metric | Amide | Amine | Carboxylic Acid | Ether | Halogen | Phenyl |
|---|---|---|---|---|---|---|
| R2 | 0.868 | 0.756 | 0.917 | 0.891 | 0.905 | 0.869 |
| MAE | 0.127 | 0.128 | 0.079 | 0.114 | 0.101 | 0.118 |
| RMSE | 0.210 | 0.209 | 0.124 | 0.156 | 0.151 | 0.184 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Z.; Hu, J.; Samia, A.; Yu, X. Predicting Active Sites in Photocatalytic Degradation Process Using an Interpretable Molecular-Image Combined Convolutional Neural Network. Catalysts 2022, 12, 746. https://doi.org/10.3390/catal12070746
Jiang Z, Hu J, Samia A, Yu X. Predicting Active Sites in Photocatalytic Degradation Process Using an Interpretable Molecular-Image Combined Convolutional Neural Network. Catalysts. 2022; 12(7):746. https://doi.org/10.3390/catal12070746
Chicago/Turabian StyleJiang, Zhuoying, Jiajie Hu, Anna Samia, and Xiong (Bill) Yu. 2022. "Predicting Active Sites in Photocatalytic Degradation Process Using an Interpretable Molecular-Image Combined Convolutional Neural Network" Catalysts 12, no. 7: 746. https://doi.org/10.3390/catal12070746
APA StyleJiang, Z., Hu, J., Samia, A., & Yu, X. (2022). Predicting Active Sites in Photocatalytic Degradation Process Using an Interpretable Molecular-Image Combined Convolutional Neural Network. Catalysts, 12(7), 746. https://doi.org/10.3390/catal12070746






