Abstract
High-molecular-weight glucansucrase (GS) URE 13-300 with two catalytic domains (CDs) synthesizes insoluble branched α-glucan. In the present work, we explore the role of the amino acid glycine 449 (G449) located in domain B of CD1 on the enzyme properties and polysaccharide structure. Glycine was substituted with lysine via site-directed mutagenesis and the mutant DNA was expressed in recombinant Escherichia coli BL21 (DE3). The obtained mutant glucansucrase U13M1 had a shifted optimum pH, from 5.3 to 6.5, and a decreased optimal temperature, from 30 to 20 °C. The modified glucan, synthesized using U13M1, retained the water-insoluble nature of the URE 13-300 glucan and also has altered linkage composition, with about 30% fewer α-(1 → 3) linked glucose residues in the main chain. This is the first mutagenesis study on glucansucrase with two catalytic domains in a non-truncated form.
1. Introduction
The use of bio-degradable polymers has reached a peak in the recent years, with various applications and advantages. Lactic acid bacteria (LAB) are well known to produce diverse sucrose-derived glucans that are widely used in the medicine, food, cosmetic and pharmaceutical industries [1,2]. Bacteria from the Streptococcus, Leuconostoc, Lactobacillus or Weissella genera produce enzymes from the glycoside hydrolase family 70 (GH70) that synthesize α-D-glucans with various types of linkages and degrees of branching. There are two types of activities differentiated in the family by their preferred substrate: the α-4,6 and α-4,3 glucanotransferasesm which use amylose or maltodextrines, and the glucansucrases (GSs), which solely utilize sucrose as the D-glucopyranosyl donor to synthesize α-D-glucans with a very high molar mass with the attendant release of D-fructose. These are also referred to as glucosyltransferases (GTFs) [3]. Depending on linkage specificity in the synthesized glucan polymers, GSs are classified as dextransucrases (EC 2.4.1.5) synthesizing dextran (>95% α-(1 → 6) linkages between glucose units), alternansucrases (EC 2.4.1.140), synthesizing alternan (alternating α-(1 → 6) and α-(1 → 3) linkages in the main chain), mutansucrases (EC 2.4.1.5) synthesizing mutan (mostly α-(1 → 3) linked glucose units) and reuteransucrases (EC 2.4.1.5) synthesizing reuteran (with α-(1 → 4) and α-(1 → 6) linked glucose units in the polymer chain) [4]. In recent years, a subfamily of GH70 enzymes was identified, called branching sucrases (BRS). These are able to add single α-(1→ 2) or α-(1→ 3) branched residue onto the non-reducing end of linear dextran as an acceptor, resulting in highly branched polysaccharides with a comb-like structure [5]. The degree of branching, the ratio between the different types of linkages and their organization in these polymers result in variations in their functional properties, such as solubility, flow and viscosity or conformation in solution [6].
The majority of glucans mainly consisting of α-(1 → 3) linkages are water-insoluble polymers, whereas glucans consisting of a high level of α-(1 → 6) linkages are presumed to be soluble in water [7,8]. It is known that α-(1 → 6)-linked glucose units form a flexible chain which readily hydrates and dissolves in water, in contrast to sequences of α-(1 → 3)-linked glucose units that tend to form ribbon-like helices which self-associate and are water-insoluble, similar to cellulose [9,10]. The water-insoluble glucans produced by Streptococcus strains are structured with two types of regions of dextran-like α-(1 → 6) linkages and α-(1 → 3) linkages that result in graft- or block-type copolymers. This block or graft copolymer structure is quite different from the highly water-soluble alternan, which has similar proportions of α-(1 → 3) and α-(1 → 6) linkages arranged in a regular, alternating fashion with no extended sequences of either linkage type [11].
The structural organization of the catalytic domain of GSs was confirmed by resolving the 3D structure of dextransucrase GTF180, produced by Lactobacillus reuteri 180, and revealed an organization in five distinct domains: the domains A, B and C, structurally close to those of the GH13 family enzymes, and the domains IV and V, unique to GH70 enzymes [12,13]. With the exception of domain C, which forms the U-shaped fold of the domain, the other four domains are built as a non-contiguous chain [13]. There are five 3D structures of GSs and one of branching sucrase (BRS) available; however, they all correspond to N- or C-terminal truncated ends. It remains a difficult task to crystallize whole enzymes because of their large size (>150 kDa) and the mobility of their domain V [14]. A calcium ion presumed to be involved in enzyme stability is found at the same position in all structures at the interface between domains A and B [15].
The mechanism of dextransucrase action that controls the type of glycosidic linkage in the synthesized polysaccharides is still unclear [16]. In the last two decades, fairly large amounts of data have been generated through mutagenesis studies, in order to further elucidate the structure–function relationships of the GSs and their specificity. Most of these studies are focused on amino acids in the conserved motifs (II, III, IV), comprising the catalytic core, and especially those in proximity to the catalytic triad, situated in domain A [17,18,19,20]. For example, a mutation close to the transition state stabilizer (TSS) resulted in a double mutant R624G:V430I of dextransucrase GTFR from Streptococcus oralis that was able to synthesize insoluble polymers with a majority of α-1,3 linkages [21]. Recently, the location of acceptor subsites −1, +1, +2 and +3 was confirmed in dextransucrase DSR-M from L. citreum NRRL B-1299. Two tryptophan residues in loop B2 (domain B) that provide stacking platforms to the growing glucan chain were also identified [19]. Despite the numerous reports on this topic, mutations in full-length GSs are still limited [14].
Among all of the characterized glucansucrases of the GH70 family, only three of them are reported to have two catalytic domains. The first to be studied in detail was dsrE, secreted from Leuconostoc mesenteroides NRRL B-1299. Sequence analysis revealed two catalytic domains, CD1 and CD2, separated by a glucan-binding domain (GBD). CD1 and CD2, which share 45% identity and 65% similarity between one another, were both classified into family GH70. Both contain the highly conserved amino acids involved in the formation of the glucosyl enzyme intermediate [22]. Further biochemical characterization of two recombinant truncated forms (CD1-GBD and GBD-CD2) showed that CD1-GBD acts as a polymerase, synthesizing glucan with predominantly α-(1 → 6) linkages. The second form (GBD-CD2) was found to be exclusively responsible for the synthesis of α-(1 → 2) linkages, resulting in a highly branched polymer [23,24]. The other recently reported enzyme with two catalytic domains was GtfZ from the fructophilic bacterium Lactobacillus kunkeei DSM 12,361, responsible for the synthesis of highly α-(1 → 3) branched α-glucans [25].
The gene encoding the third glucansucrase URE 13-300 with two catalytic domains, secreted from L. mesenteroides URE 13-300 strain, was successfully cloned and expressed in E. coli BL21. Its ability to synthesize oligosaccharides with prebiotic properties was studied in detail [26]. The enzyme catalyzes the formation of polymer with α-1,6 linkages and α-1,3 linkages (16%) in the main chain and 8% α-1,3 linkages in the branching points [27]. Obtaining a single-point mutant of full-length glucansucrase with two catalytic domains allows us to explore the role of certain amino acid substitutions in enzyme linkage specificity and subsequently acquiring glucan with modified properties. Additionally, we could gain insights about the interaction between the domains for synthesizing a branched polymer.
The aim of the current work is to study the effect of amino acid substitution, located in domain B of CD1 of glucansucrase URE 13-300, on the degree of branching in the synthesized polymer and the relationship with the enzymatic reaction. A single-point mutation was performed, followed by determination of the kinetic properties of the mutated enzyme and its ability to be activated by metal ions. The structure of the glucan, synthesized by the mutated glucansucrase, was analyzed via NMR spectroscopy and the rheological properties of the modified glucan were also studied.
2. Results
2.1. Design of the Site-Directed Mutagenesis and Expression of the Mutated Enzyme U13M1
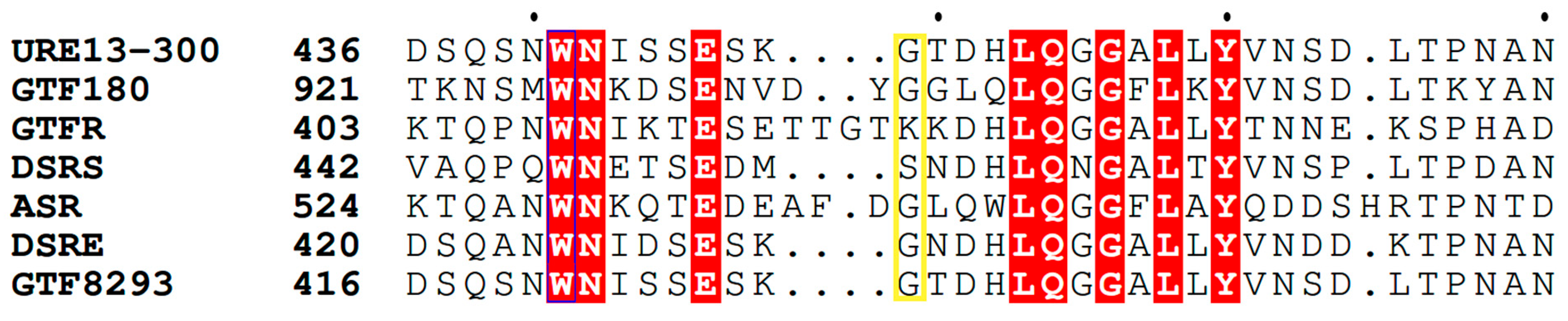
The gene encoding glucansucrase URE 13-300 containing two catalytic domains has been characterized previously [26]. With the aim of finding an important position for amino acid substitution, we performed a multiple sequence alignment with several dextransucrases, one mutansucrase (GtfI) and one alternansucrase (ASR) from GH70 family (Figure 1). Glycine, situated at the 449th position in domain B, is not highly conserved. However, it is the most common. In mutansucrase GtfI, the corresponding amino acid is tyrosine, in GTFR, it is lysine, and in DSRS, it is serine. Mutagenesis studies were performed at a few other positions in this site, comprising loop B1 [28,29,30].

Figure 1.
Sequence alignment of part of domain B from several GH70 glucansucrases and the position of the substituted amino acid (marked in the yellow rectangle): glucansucrase URE13-300 (AXI68673.1) from Leuconostoc mesenteroides URE 13-300; GTF180 (AY697430.1) from Lactobacillus reuteri 180; GTFR (AB025228.1) from S. oralis ATCC 10557; DSRS (AAD10952.1) from L. mesenteroides NRRL B-512; ASR (AJ250173.2) from L. mesenteroides NRRL B-1355, DSRE from L. mesenteroides NRRL B-1299; GTF8293 (ABJ61965) from L. mesenteroides ATCC 8293 and GTFI (AB025228.1) from Streptococcus downei. The strictly conserved amino acids are marked in red. Visualization of the performed alignment is used ESPript 3.0 [31].
The mutation was introduced by the designed primers, encoding the changed respective codon and conducting the PCR reaction for the synthesis of mutated plasmid DNA. The efficiency of the mutation was assessed as more than 95%. The substitution was confirmed by sequencing the plasmid, containing the mutated glucansucrase gene. In situ analysis showed that the molecular weight of the mutant enzyme U13M1 was about 300 kDa. This result is consistent with our previous results and the theoretical molecular weight, 311.85 kDa [26,27]. The expression of the gene encoding U13M1 was also optimized in regard to the concentration of the added inductor and the time of induction, as well as the duration of the fermentation.
2.2. Biochemical Characterization of the Mutant Enzyme U13M1
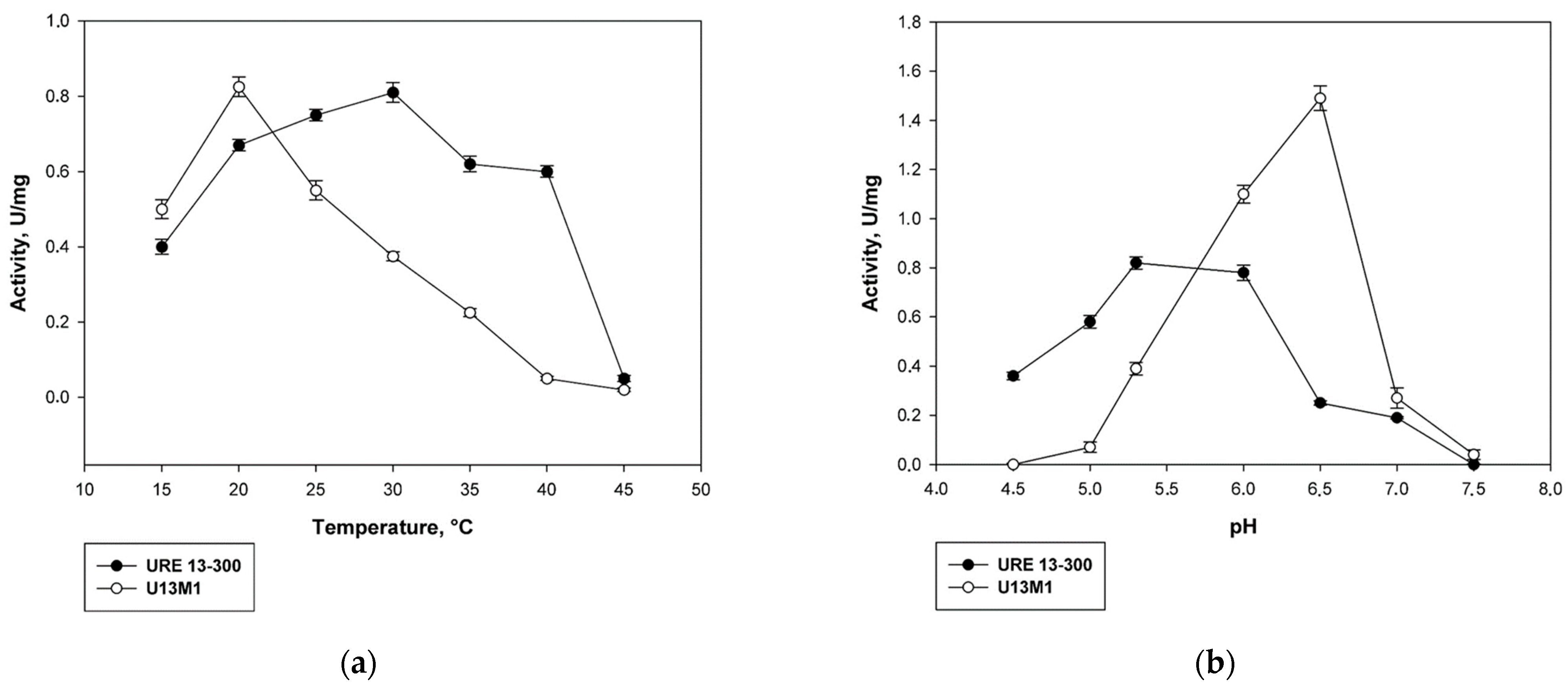
In order to evaluate the influence of the single-point mutation on the enzyme properties, we determined the optimal pH and temperature of action of the mutated variant U13M1 for conducting the enzyme reaction. The optimum temperature was estimated to be 20 °C, which is significantly lower than that of the wild-type glucansucrase—30 °C (Figure 2a). Increasing the temperature by 5 °C led to a 33% decrease in the enzyme activity. At 30 °C, the activity of U13M1 is more than 50% lower that at 20 °C. An enzyme reaction at 40 °C results in almost complete loss of activity. In contrast, at the same temperature, the wild-type variant URE 13-300 retains about 80% of its activity (Figure 2a).
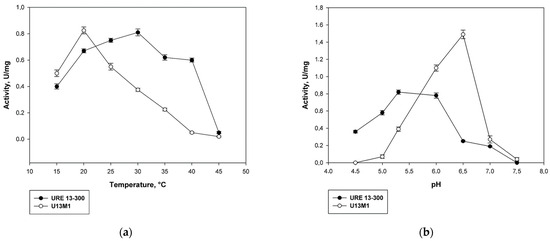
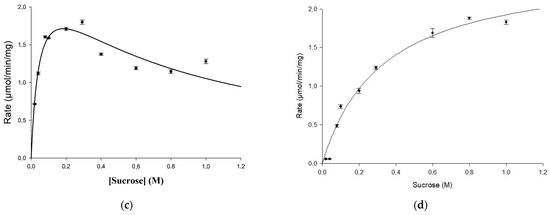
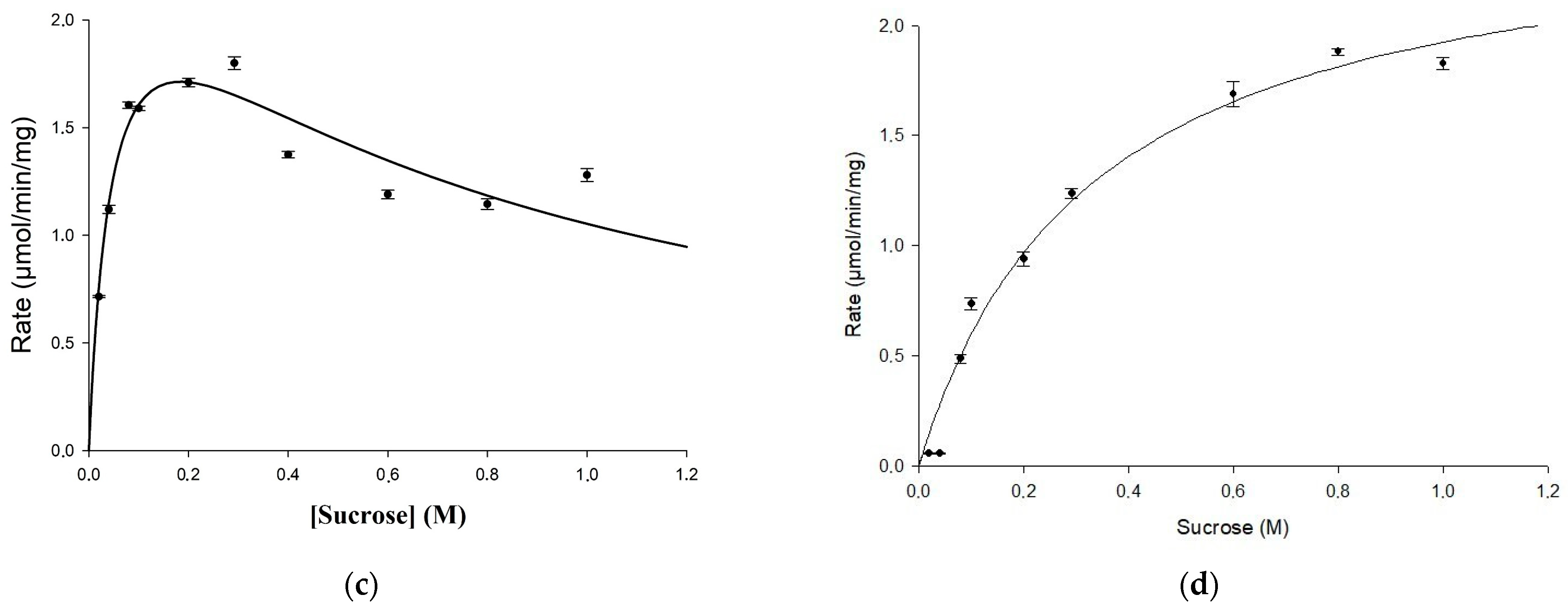
Figure 2.
Comparison between biochemical properties of the mutant U13M1 and the wild-type glucansucrase URE 13-300: (a) temperature; (b) pH; (c) kinetics of URE 13-300 with sucrose; (d) kinetics of U13M1 with sucrose.
The optimal pH conditions for the enzyme reaction of the mutant variant also showed very different values compared to the wild-type enzyme. The highest activity was reached at pH 6.5. In comparison, at pH 6.0, the activity decreased by 27% (Figure 2b). More alkaline buffers caused a drastic decrease in the enzyme activity—by more than 80% at pH of 7.0 and almost complete loss of activity at pH 7.5. The activity of U13M1 in acidic buffers was significantly decreased as well—about 70% lower at pH 5.5, as opposed to the optimal pH value, 6.5. All activity was lost at pH 4.5. In contrast, the optimal pH for the action of wild-type enzyme was 5.3. Moreover, URE 13-300 activity was diminished by 72% at pH 6.5, which was optimal for the U13M1 variant (Figure 2b).
Kinetic parameters for the mutated variant were examined and compared with the wild-type enzyme as well. Using the Michaelis–Menten equation and the non-linear regression approach, the value of Km = 0.048 M for the sucrose concentration, and the maximum reaction rate of Vmax = 2.6 U/mg were estimated for the wild-type URE 13-300 glucansucrase (Figure 2c). The value of Km = 0.33 M for the sucrose concentration, obtained for the mutant enzyme U13M1, is seven times higher than that for the wild-type enzyme. However, the enzyme reaction reached a maximum velocity at the same rate—Vmax = 2.6 U/mg (Figure 2d). Therefore, the affinity between the substrate and enzyme is much lower in the mutant enzyme reaction, while the reaction rate remains unaffected. Substrate inhibition of the mutant enzyme was observed at a sucrose concentration of 1.6 M, whereas the Ki of the wild type was 0.69 M. In Figure 2c,d, the kinetic curves are presented.
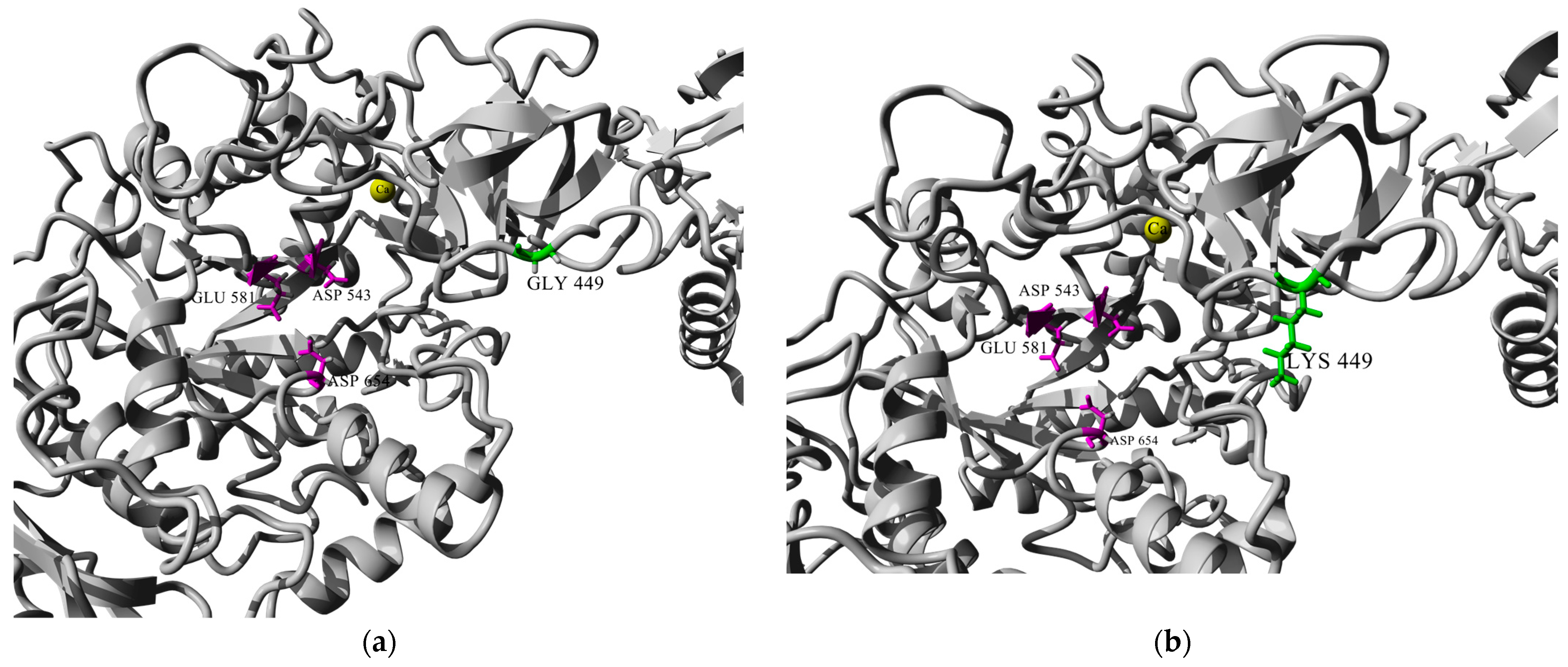
Resolving the 3D structures of the truncated forms of several glucansucrases, it was found that they include calcium ions in the same position close to the catalytic domains A and B [15]. According to Vujičić-Žagar et al., the removal of Ca2+ ions from the active site led to decrease in enzyme activity by about 40%. This effect might be due to the compromised binding of the substrate/acceptor at the site [13]. In Figure 3, we present a homology model of URE 13-300–CD1, based on the crystal structure of glucansucrase GtfB from Streptococcus mutans GS5. Comparison between the two models before and after the amino acid substitution shows that the calcium ion does not change its position at the active site.
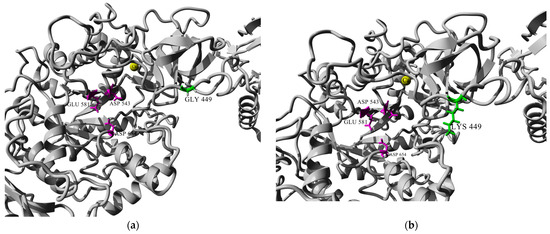
Figure 3.
Homology modeled structure of URE 13-300–CD1: (a) conformation of the catalytic site groove of CD1 before amino acid substitution with glycine (Gly); (b) conformation of the catalytic site groove of CD1 after amino acid substitution with lysine (Lys). Note: residues of the catalytic triad are colored in magenta; the substituted amino acid position is colored in green; and Ca2+ ions are colored in yellow.
We examined the influence of exogenous Ca2+ ions on the enzyme activity of the wild-type glucansucrase URE 13-300 and its mutated variant (Table 1). As reported for other glucansucrases, incubation of the enzymes with a chelating agent had a negative influence on enzyme activities, which is reversed with the addition of calcium ions [32]. Both enzymes were activated by the Ca2+ ions at a final concentration of 5 mM; however, the activation effect on the wild-type enzyme was two times higher, at 52%, as opposed to 17% higher enzyme activity for the mutant enzyme. An investigation of the increased concentration of calcium ions revealed that at the highest concentration tested, 25 mM, the wild-type enzyme reached 78% increased activity compared to the control reaction without Ca2+ ions present. In contrast, U13M1 activity was elevated with about 30% at a 20 mM Ca2+ concentration as opposed to the respective control reaction.

Table 1.
Influence of increasing concentrations of Ca2+ ions on the enzyme activity of the glucansucrases URE 13-300 and U13M1.
The four other tested ions, Mg2+, Fe2+, Ba2+, and Mn2+, all had positive influence on the parental enzyme. However, all showed an inhibitory effect on the mutated variant (Table 2). Notably, the Fe2+ ions had a similar activation effect to that of Ca2+ ions on the wild-type glucansucrase.

Table 2.
Influence of different metal ions on the enzyme activity of the glucansucrases URE 13-300 and U13M1.
Overall, incorporating the G449K single-point mutation decreased the activation effect of metal ions on U13M1 enzymes.
2.3. Structure and Linkage Specificity of URE 13-300 and U13M1 Polysaccharides
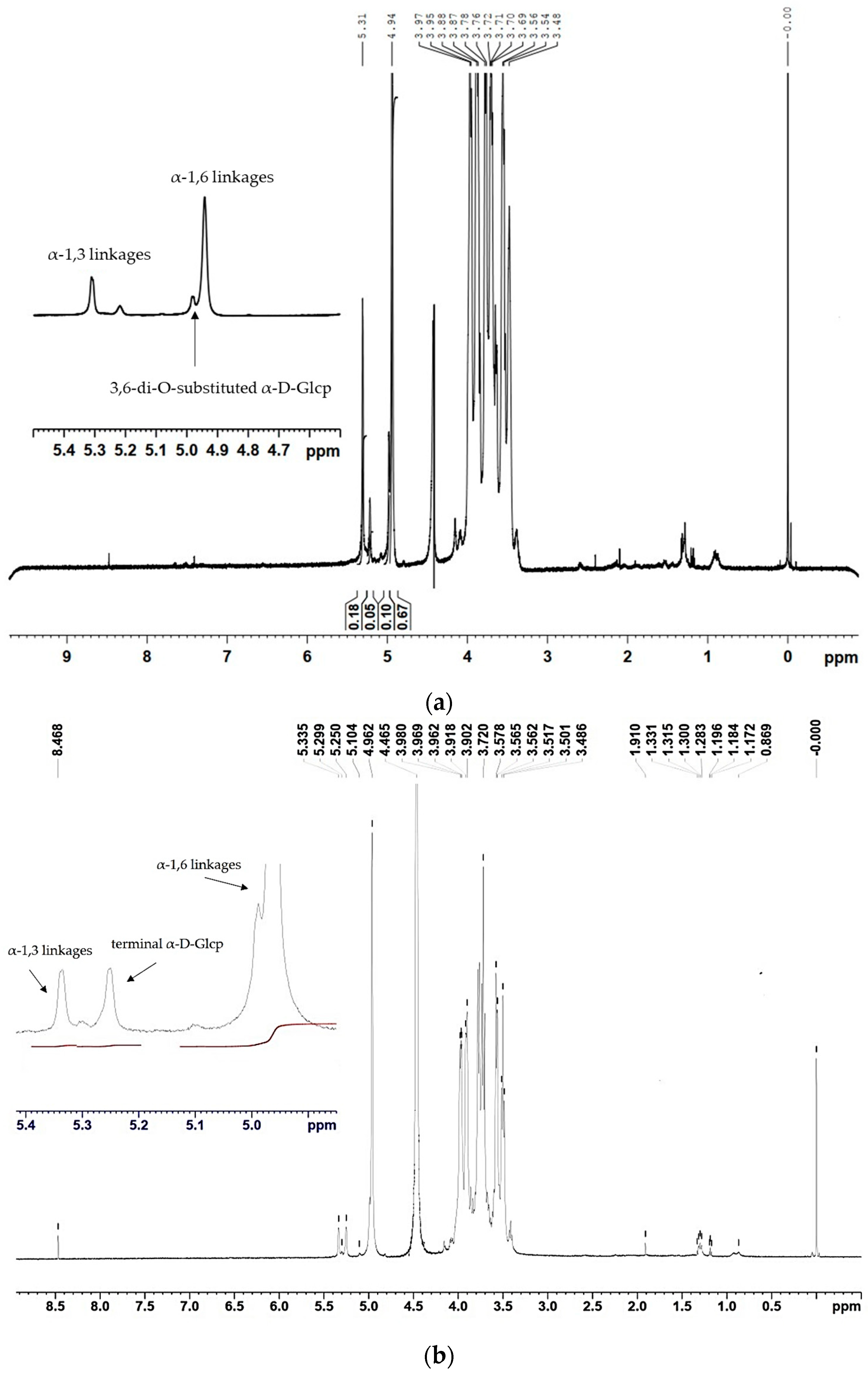
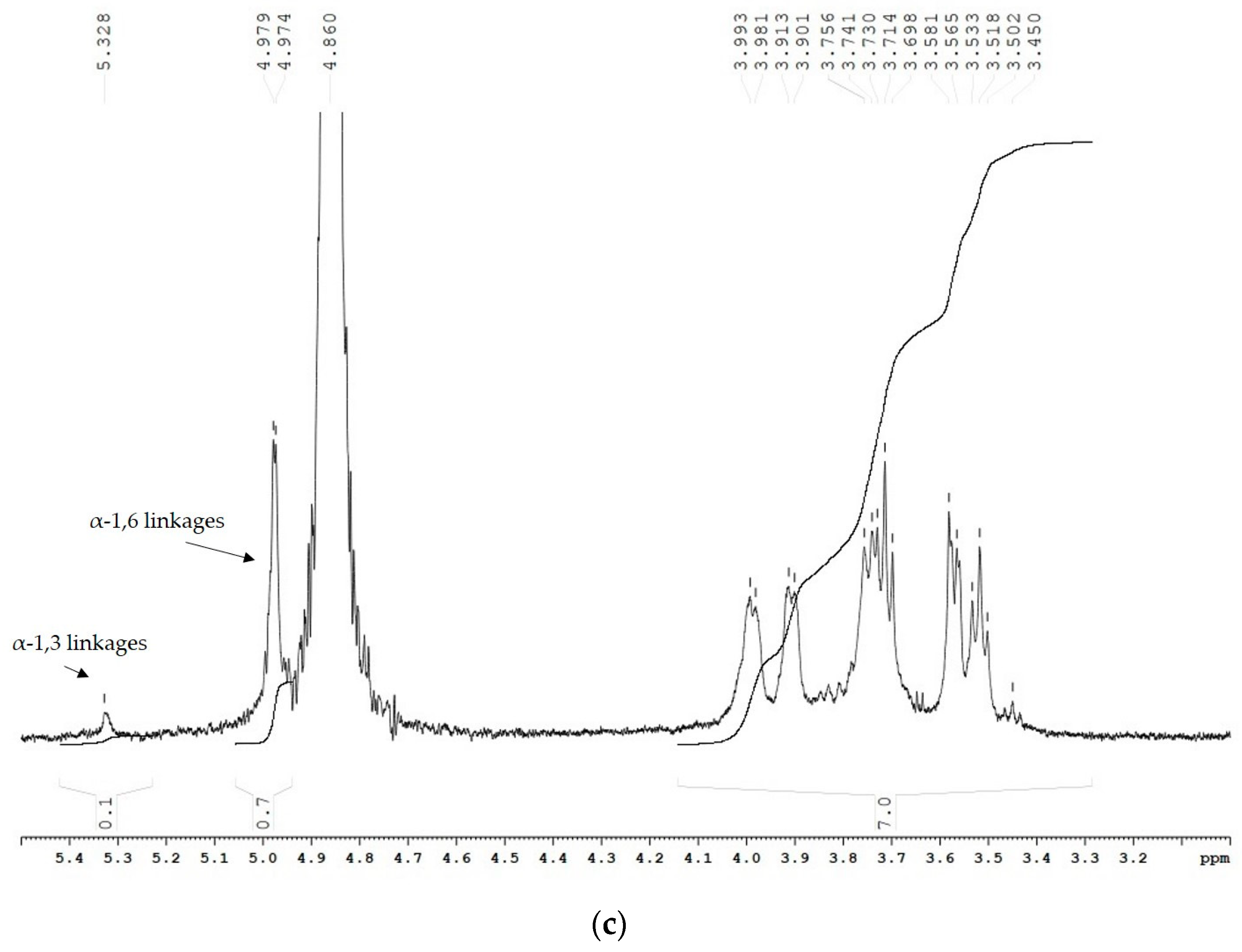
Nuclear magnetic resonance (NMR) results obtained from the 1H spectra of the two α-glucans showed that the ratio between the α-(1 → 6) and α-(1 → 3) linkages changed due to the incorporated mutation (Figure 4). The polymer, synthesized by the wild-type enzyme, consists of 67.2% α-(1 → 6) linkages and 16.2% α-(1 → 3) linkages, corresponding to D-Glcp residues with chemical shifts at 4.94 ppm and 5.31 ppm, respectively (Figure 4a). Moreover, the existing 1H NMR signal at 4.98 ppm indicated the presence of 3,6-di-O-substituted α-D-Glcp residues and the branched structure of the analyzed glucans estimated at 8.3% [27]. The 1H NMR spectra of the U13M1 polysaccharide, synthesized with a 600 mM initial sucrose concentration, revealed two anomeric protons with chemical shifts at 4.96 ppm, representing the presence of 90.6% α-(1 → 6) linkages, and those at 5.33 ppm, corresponding to the presence of 4.2% α-(1 → 3) linkages (Figure 4b). These results show the highly reduced amount of α-(1 → 3) linkages present in the U13M1 polymer structure. The peak at δH-1~4.96 is a typical structural reporter for (-)α-D-Glcp-(1 → 6) units (δ-range between 4.99 and 4.95 ppm) [33]. In both glucans synthesized by the wild-type and mutant enzyme, there is a signal at 5.22 and 5.25 ppm, respectively, that indicates terminal α-D-Glcp units [34]. There is also a distinguishable signal at the base of 4.96 ppm, indicating a small amount of 3,6-di-O-substituted α-D-Glcp residues, although not as a separate signal (Figure 4b). We performed a series of experiments at different concentrations of sucrose as well. The 1H NMR analysis of the polymer, synthesized at a 292 mM initial sucrose concentration, is presented in Figure 4c. Two peaks at δH-1∼4.97 ppm, corresponding to the presence of 87.5% α-(1 → 6) linkages, and-δH-1∼5.32 ppm, corresponding to the presence of 12.5% α-(1 → 3) linkages, were detected.
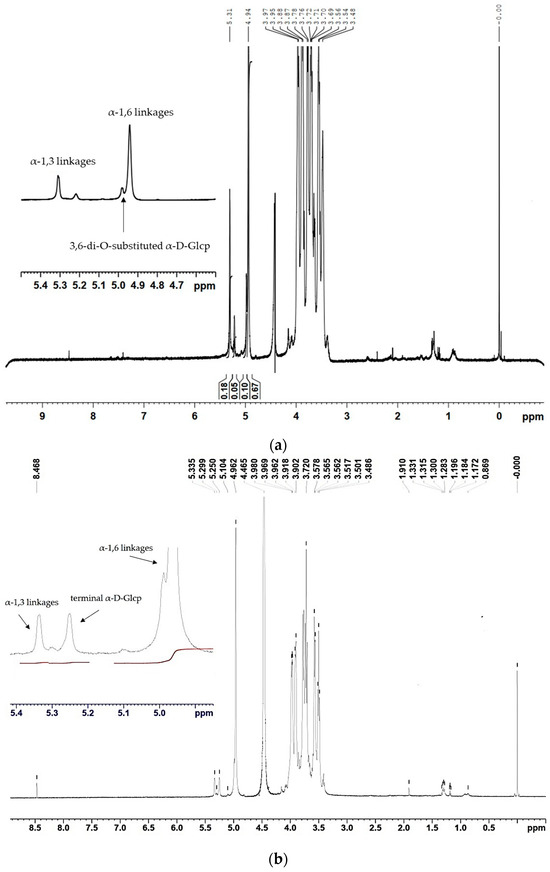
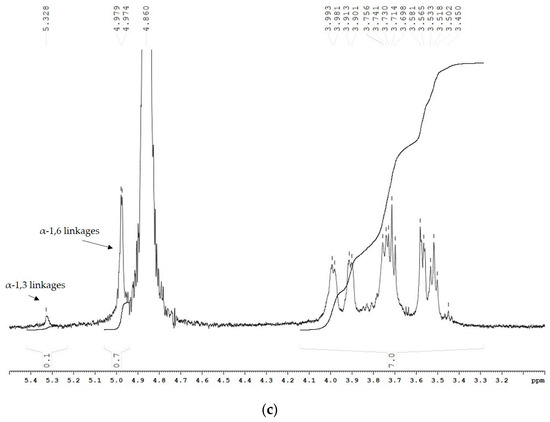
Figure 4.
1H NMR spectra of the α-glucans, synthesized by (a) the wild-type glucansucrase URE 13-300 with 292 mM sucrose; (b) the mutant variant U13M1 with 600 mM sucrose; (c) the mutant variant U13M1 with 292 mM sucrose.
2.4. Physical Properties of URE 13-300 and U13M1 Glucans
The water swelling behavior of both glucans was also assessed. The water holding capacity of U13M1 glucan was calculated to 1125% ± 7.6, which is more than 1.5 times higher than that of the wild-type polysaccharide, 715% ± 6.0. Polymer samples gradually absorbed the added water, and at the end of the experiment, they settled at the bottom of the cylinder, leaving the redundant water volume above. The insoluble polysaccharide, synthesized by U13M1, formed a more uniform suspension. However, it was not in the form of a hydrogel.
3. Materials and Methods
3.1. Bacterial Strains, Plasmid Template and Media
The gene, encoding glucansucrase URE 13-300, was previously cloned and expressed in recombinant strain Escherichia coli BL21 URE 13-300 as described [26]. The strain was cultivated in Luria–Bertani broth (LB) (Sigma, St. Louis, MO, USA) supplemented with 30 μg/mL kanamycin (Merck) overnight at 37 °C and 250 rpm. Pelleted cells were treated with QIAprep® Miniprep (Qiagen, Hilden, Germany) for isolation and purification of the plasmid vector pETite N-His SUMO/URE 13-300 according to the instructions provided by the kit manufacturer.
3.2. Site-Directed Mutagenesis
Site-directed mutagenesis was performed with QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA). The following primers were used to perform the amino acid substitution (alternated codon is underlined): ForG449K 5′CATTAGTAGTGAGTCTAAAAAAACAGATCATTTGCAAGGTGGTGCGCTC 3′; RevG449K 5′GAGCGCACCACCTTGCAAATGATCTGTTTTTTTAGACTCACTACTAATG 3′ (Bioneer, Daejeon, Republic of Korea). Primers were designed according to the kit instructions and checked via OligoEvaluator software (http://www.oligoevaluator.com (accessed on 14 November 2023)). PCR cycling parameters were set according to the kit instructions as well. Samples were treated with the 1U DpnI endonuclease enzyme to digest the methylated (parental) DNA template for 1 h at 37 °C. Chemically ultracompetent E. coli XL10-Gold cells (provided with the kit) were used in transformation with the plasmids harboring the mutated gene. Mutation efficiency was checked through a control reaction with the mutation in a pWhitescript plasmid for color screening, along with a pUC18 plasmid, used for control of the transformation.
Mutated plasmid DNA was subsequently purified and transformed into E. coli BL21 (DE3) for protein expression. Initially, the cells were grown on LB broth, containing 30 µg/mL kanamycin at 37 °C and at 250 rpm for 18 h. The clones containing the full gene were then cultivated in Terrific Broth medium (TB) (12.0 g/L tryptone (Sigma), 24.0 g/L yeast extract (Sigma), 9.4 g/L K2HPO4 (Sigma), and 2.2 g/L KH2PO4 (Sigma) supplemented with 30 μg/mL kanamycin on a rotary shaker (250 rpm) at 37 °C.
The screening for glucansucrase activity was performed on TB agar media supplemented with 20% sucrose and 1 mM of the inducer isopropyl β-D-thiogalactopyranoside (IPTG, Sigma). Positive clones forming glucan on the plates were sent for sequencing (Macrogen, Amsterdam, The Netherlands). After confirmation of correct amino acid substitution, one of the clones was used for the optimization of the expression conditions and production of mutated glucansucrase URE 13-300 M1 (U13M1).
3.3. Obtaining of Glucansucrase URE 13–300 Preparations
Crude cell-free extracts of the wild-type glucansucrase URE 13-300 were obtained as reported previously [26]. Cultivation of recombinant E. coli BL21 U13M1 was performed at 37 °C in a rotary shaker at 250 rpm until the optical density (OD) at 600 nm reached 1.6–1.8. Then, the inducer IPTG was added at a final concentration of 0.7 mM and glucansucrase gene expression was carried out at 16 °C for 13 h. Cultures were centrifugated at 8000× g at 4 °C, for 15 min and the collected cells were resuspended in 20 mM sodium acetate buffer, with a pH of 5.3. Subsequently, the cell pellet was disrupted in Nano DeBEE High Pressure Homogenizer (BEE International, Billerica, MA, USA). The cell debris was removed by means of centrifugation under the same conditions and the supernatant was used as a source of glucansucrase. The enzyme solution was not subjected to further purification, as it is known that the recombinant strain E. coli BL21 does not produce any other glucansucrase, hence it would not interfere with the obtained results of the following analyses [35].
3.4. SDS-PAGE Analysis
SDS-PAGE (with 3% concentrating and 5% separating acrylamide gels) was performed according to the method of Laemmli [36]. Glucansucrase in situ activity detection was performed by means of the incubation of the gel in 100 g/L sucrose (Sigma) overnight, followed by staining of the synthesized polysaccharide according to a periodic acid-Schiff’s procedure (PAS) [37]. The used protein standards (High Molecular Weight Calibration Kit for SDS Electrophoresis, Amersham, UK) were stained with Coomassie Brilliant Blue R 250 (Sigma–Aldrich, St. Louis, MO, USA).
3.5. Enzyme Activity Assay
Determination of the activity of the wild-type enzyme and its mutated variant was performed as described previously [26] via the DNS method [38]. One unit of glucansucrase activity was defined as the release of 1 μmol fructose per minute at 30 °C in 20 mM sodium acetate buffer, pH 5.3; 0.05 g/L CaCl2 (Sigma) and 100 g/L sucrose. The concentration of the released fructose was measured spectrophotometrically at 540 nm against the appropriate standard curve. The protein concentration was assayed using the Bradford method [39]. All measurements of enzyme activity were performed at least in triplicate, and the results from different experiments are presented with the standard deviations (±SD).
3.6. Kinetic Studies
The optimal temperature for the activity of both the wild-type and mutated enzyme was evaluated by means of enzyme incubation for 20 min at temperatures varying from 15 °C to 40 °C (at 5 °C steps) with constant conditions—20 mM sodium acetate buffer with a pH of 5.3 and a 292 mM sucrose concentration. Samples were taken every 5 min and fructose release was measured again via the DNS method.
Optimal pH values were evaluated by measuring the enzyme activities in consequent 20 mM buffers: sodium citrate buffer with pH 4.0 and 4.5, sodium acetate buffers with pH 5.0 to 6.5 (at increment of 0.5) and potassium phosphate buffers with pH 7.0 and 7.5. Reaction temperatures were set to 30 °C and 20 °C for the wild-type and mutant variant, respectively; the substrate concentration was set to 292 mM.
The determination of kinetic parameters for the wild-type and mutant enzyme was performed by measuring the enzyme activity in increasing sucrose concentrations as follows: 20 mM, 40 mM, 80 mM, 100 mM, 200 mM, 292 mM, 400 mM, 600 mM, 800 mM and 1 M. The constant conditions were 30 °C and pH 5.3 in the URE 13-300 enzyme reactions and 20 °C and sodium acetate buffer with pH 6.5 in the U13M1 reactions. Kinetic parameters such as the Michaelis constant (Km) and maximum velocity (Vmax) were determined using the Michaelis–Menten equation and the non-linear regression approach. Uniform enzyme preparations of 0.3 mL were used for each enzyme variant. All reactions were performed in triplicate, and the results are presented with the standard deviations (±SD).
3.7. Effect of Metal Ions on Enzyme Activity
The impact of several metal ions on enzyme activity of the wild-type glucansucrase URE 13-300 and its mutant variant U13M1 was assessed as well. Enzyme preparations were initially treated with 1 mM ethylene diamine tetraacetic acid (EDTA) as previously described [35]. The metal ions Ca2+, Mg2+, Fe2 +, Ba2+, and Mn2+ were added to the enzyme reaction in the form of salts at a final concentration of 5 mM. The presence of calcium ions was further tested with elevated concentrations from 5 to 25 mM. Reactions lacking metal ions were used as controls and the obtained activity values were set to 100%. Activity was measured at the optimal conditions for each of the enzymes—at 30 °C in a 20 mM sodium acetate buffer with pH 5.3 and a 292 mM sucrose substrate for the wild-type enzyme and at 20 °C in a 20 mM sodium acetate buffer with pH 6.5 and 600 mM sucrose for the mutant enzyme. All measurements of enzyme activity were performed at least in triplicate, and the results from different experiments are presented with the standard deviations (±SD).
3.8. In Vitro Glucan Synthesis
Glucan synthesis by glucansucrase URE 13-300 was carried out at 30 °C with 0.5 U/mL enzyme in a 20 mM sodium acetate buffer with a pH of 5.3 and 100 g/L sucrose. Glucansucrase U13M1 glucan was synthesized under the following conditions: 20 °C with 0.5 U/mL enzyme in a 20 mM sodium acetate buffer with a pH of 6.5 and 200 g/L sucrose. An additional reaction with a 100 g/L initial sucrose concentration was performed as well. All reactions were supplemented with 0.1% (w/v) sodium azide for the prevention of bacterial growth, and were performed in a rotary shaker for 24 h (until depletion of sucrose).
The obtained α-glucan fractions were separated by means of precipitation with two volumes of 96% ethanol. They were purified via triple washing with dH2O, followed by centrifugation and lyophilization (Labconco FreeZone 4.5; Labconco Corporation, Kansas City, MO, USA).
3.9. NMR Analysis
The linkages in the purified polysaccharides synthesized by the wild-type and mutant enzymes were determined by means of nuclear magnetic resonance (NMR) spectroscopy on Avance II (Bruker, Billerica, MA, USA), as described previously [6]. Samples (5 mg) were dissolved in 0.6 mL of 0.5 M NaOH diluted in D2O. Spectra were recorded at 600 MHz for 1H NMR analyses, and the temperature of the analysis was 353 K. 1H chemical shifts (δ) were expressed in ppm by reference to the TSPA standard (δ = 0.0 ppm). The data acquisition and processing were performed using TopSpin 4.2. software. The percentages of the linkages in glucans were calculated on the basis of the relative intensities of the anomeric protons.
3.10. Water Holding Capacity
The water holding capacity (WHC) of the α-glucans synthesized by U13M1 and URE 13-300 was evaluated as described in previous studies [40]. Sample preparation was performed in larger volumes as follows: we dissolved a 4.0 g sample in 150 mL of MilliQ water and stirred at room temperature for uniform dispersion. The dispersed sample was centrifuged at 8000× g for 30 min and the supernatant fractions were discarded. The settled polymers were analytically weighed.
The percentage of WHC was calculated using the following equation:
3.11. Sequence Analysis and Homology Modeling
Alignments of the protein sequence of URE 13-300 with other glucansucrases were carried out using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 31 May 2023) and ESPript 3.0. (https://espript.ibcp.fr/ESPript/ESPript/, accessed on 31 May 2023). The homology model of CD1 of URE 13-300 enzyme was created via the Swiss-Model web server (https://swissmodel.expasy.org, accessed on 7 July 2023) based on the crystal structure of glucansucrase GtfB from S. mutans GS5 (PDB ID: 8FJ9 [41], sharing 56.5% sequence identity) [42]. The model was optimized through energy minimization in the program YASARA, v22.5. The amino acid substitution was introduced in the model by swapping glycine amino acid code with lysine in the menu Edit > Swap > Residue, and it was again subjected to energy minimization using the default settings. Pictures of the models were obtained with YASARA.
3.12. Statistical Analysis
In all cases, the programmable scientific calculator “CASIO” fx-991ES Plus, statistical software package SigmaPlot v12.0 (Systat Software, Inc., Palo Alto, CA, USA) and Microsoft Excel were used for data analysis and graphical representation.
4. Discussion
Glucansucrases with two catalytic domains are an interesting subject for single-point mutations. The amino acid substitution that we investigated in this study is located in domain B of CD1 of the glucansucrase URE 13-300 at the 449th position (Figure 3). The effect of this substitution concerns both the enzyme kinetic parameters and the linkage composition of the synthesized polysaccharide.
The obtained mutant enzyme, U13M1, has noteworthy differences in its kinetic parameters compared to wild-type glucansucrase URE 13-300. The Km values for U13M1 glucansucrase are almost 7-fold higher than those of the wild-type enzyme, while retaining the same maximum velocity. The diminished affinity for the substrate that we observed may be due to change in the shape of the binding groove of the mutated enzyme [13]. In accordance with our results, mutants of the glucansucrase GTF180-∆N showed an increased Km for sucrose [43]. Mutations with cysteine, alanine, serine, glutamate and tryptophan showed a 4- to 5-fold increase in Km. Kinetic analysis of L938A and L938F revealed a decrease in activity. However, their Km values for sucrose were hardly affected (L938A) or showed a slight decrease (L938F) [43].
Along with the increased Km, in the current work we report a significantly altered optimal enzyme pH and temperature. The optimal pH of U13M1 increased by one unit compared to the wild-type enzyme, to 6.5. According to previous findings, the optimum pH of different glucansucrases is reported to lie between pH 4.7 and 7.4, but is often between pH 5.0 and 6.0 [32,44]. It is also suggested that a relatively high optimum pH can be important in different food environments, since it can result in increased glucan production [45,46]. The optimal temperature of most glucansucrases is between 30 °C and 40 °C. After the amino acid substitution with lysine, the optimal temperature of U13M1 shifted to 20 °C. Corresponding to our results, dextransucrase from L. mesenteroides 0326 has an optimal temperature of 25 °C, and its double mutant variant P473S/P856S retains almost its maximum activity at 20 °C [47].
As far as we know, such differences in the optimal conditions between these wild-type and mutated enzymes have not been previously reported. Moreover, the enzyme activity of U13M1 is less inhibited by the increase in sucrose concentrations. Further examinations of acceptor reactions with U13M1 are in progress to evaluate the ability of the mutant to synthesize larger yields of gluco-oligosaccharides and determine their degree of polymerization. Indeed, in the industrial process, higher temperatures are more favorable. However, in the current work, the aim was not to produce a more thermostable enzyme. Even so, these changes could be useful in the realization of unspecific transferase reactions. It is widely known that glucansucrases can use maltose, isomaltose, lactose and other oligosaccharides in acceptor reactions, and a wide range of non-carbohydrate acceptors are glycosylated as well.
Glucansucrase URE 13-300 is activated by Ca2+ and other metal ions, similarly to other glucansucrases [48,49]. However, this ability is abolished by the mutation. Furthermore, the increase in the Ca2+ ion concentration leads to more pronounced activation on the enzyme URE 13-300. Again, in contrast, this ability is diminished in U13M1. The reason for this may lie in the fact that the mutation site is in proximity to the Ca2+ ions’ binding site.
The mutant glucansucrase U13M1 synthesizes α-glucan with a 23% increase in the α-(1 → 6) linkages content and a respective decrease in the content of α-(1 → 3) linkages in the main chain and in the branches as well. The 1H NMR spectra of the polysaccharide showed no clear signal, detecting the 3,6-di-O-substituted α-D-Glcp residues. However, they are not completely absent, as they are visible in the 4.96 ppm signal. The shift in the ratio between α-(1 → 6) and α-(1 → 3) linkages corresponds to the results obtained by Funane et al. [29]. The single substitution of T350 or S445 with lysine in glucansucrase DSRS (corresponding to K344 and G449 in GS URE 13-300) resulted in mutant variants, synthesizing water-soluble glucans with an increased percentage of α-(1 → 6) linkages, from 70% for the wild-type enzyme to 84% and 86%, respectively. Both residues are positioned away from the catalytic triad; however, their double mutation resulted in increase in the number of α-(1 → 3) linkages and even acquired the ability to incorporate 4% α-(1→2) branching points.
Glucansucrase GTF180-∆N from Lb. reuteri 180 synthesizes an α-glucan with 69% α-(1 → 6) and 31% α-(1 → 3) linkages in the main chain and in branch points, corresponding to the type and ratio of linkages, comprising the URE 13-300 glucan [27,43,50]. Two positions in GTF180-∆N, L938 and L940, are reported to be involved in shaping the substrate/acceptor binding sites. The substitution of L938 with alanine, serine and lysine (L938A/L938S/L938K) led to increased amounts of α-(1 → 6) linkages from 67% in the wild-type enzyme to 78%, 76%, and 90%, respectively. Leucine at the 940th position (GTF180 numbering) is highly conserved in the GH70 family, and it is shown to be critical for linkage specificity [28]. Moreover, when amino acids with an aromatic side-chain were introduced, the shift towards α-(1 → 6) was almost complete with phenylalanine (93%), and substitution with tryptophane lead to the complete elimination of α-(1 → 3) linkage synthesis. In comparison, our study results show that the substitution of the small residue glycine with the much larger lysine G449 (corresponding to G936 in GTF180-∆N) leads to significant alterations in the ratio between α-(1 → 6) and α-(1 → 3) linkages. A possible reason for this may be that lysine could hinder the incorporation of α-(1,3) linkages into the main chain and/or into branches. Homology models are a good starting point for undertaking future mutations in the catalytic domains of URE 13-300 glucansucrase. Nevertheless, the model of just CD1 is lacking in terms of its incapability to show the relationship with CD2. We plan to further investigate the structure of the active sites of both domains in order to better understand the interplay between them.
The degree of branching in the glucan modified by U13M1 was severely impaired by the mutation. Nonetheless, the polymer retained the insoluble characteristics of the URE 13-300 glucan. In the comparative NMR analyses, we found that there were significant differences in the amount of α-(1 → 3)-linked glucose residues in the main chain of the glucan at the analyzed concentrations. The percentage of α-(1 → 3) linkages diminished by about 20%, where 292 mM sucrose was used, and this is comparable with the wild-type glucansucrase URE 13-300. When the initial sucrose concentration was 600 mM, nearly 70% fewer α-(1 → 3) linkages were detected. In previous studies, it was suggested that a higher percentage of alpha 1,3 linkages resulted in less water-soluble polymers. However, alternan, which has a large amount of α-(1 → 3) linkages, is very soluble in water [51]. In agreement with our results, an exopolysaccharide synthesized by Leuconostoc mesenteroides B–1355, containing only 5% α-(1 → 3) branched linkages, is less soluble than a polymer, synthesized by S. mutans 6715, which contains a relatively high percentage (35%) of α-(1 → 3) branched linkages [51,52]. Our results corroborate the fact that solubility of the different types of glucans is dependent on the distribution of the α-(1 → 3) linkages in the polymer structures. Complementary assays are underway to more accurately determine the determinants conditioning glucan solubility.
In addition, both glucans have pronounced water-holding capacity. High water-holding capacity is usually accompanied by a high water solubility due to the absorptive structure of the polymer caused by the formation of hydrogen bonds [53]. In contrast to our report, the water-soluble exopolysaccharides from L. lactis KC117496 and Lactobacillus kefiranofaciens ZW3 have water-holding capacity values of 117% and 496%, respectively [40,53]. The physical and rheological properties of the URE 13-300 glucan and the modified U13M1 glucan will be further investigated.
5. Conclusions
As a result of the performed single-point mutation at CD1 of glucansucrase URE 13-300, we report a mutant enzyme with a significant shift in the optimal pH and temperature conditions and also an altered substrate affinity without changing the maximum velocity. We hypothesize that these alterations in the enzyme properties could shed light on possible better conditions for the transferase reaction with the utilization of specific acceptors, even though the diminished catalytic potential of the mutant enzyme is usually undesirable in industrial applications. Moreover, the polysaccharide produced by the mutant enzyme U13M1 has a reduced ratio of α-(1 → 3)/α-(1 → 6) linkages by about 30% in the main chain compared to the wild-type glucan. The α-(1 → 3) branching points are nearly absent as well. The U13M1 polymer has retained a water-insoluble nature, which is interesting in light of its linkage composition. This successful mutation of full-length glucansucrase is a necessary base for studying the interplay between the two catalytic domains, acting like dextransucrase and branching sucrase, respectively. Therefore, novel polysaccharides with improved physical properties and applications could be designed.
Author Contributions
All authors designed and contributed to this study. Conceptualization, S.A. and I.I.; methodology, S.A., T.V., V.B. and I.I.; software, S.A.; validation, T.V., V.B. and I.I.; formal analysis, S.A., T.V. and V.B.; investigation, S.A. and I.I.; resources, S.A.; data curation I.I., T.V. and V.B.; writing—original draft preparation, S.A.; writing—review and editing, T.V., V.B. and I.I.; visualization, S.A.; supervision, I.I. and T.V.; project administration, I.I.; funding acquisition, I.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the operational program “Science and education for smart growth” 2014–2020, grant number BG05M2OP001-1.002-0005-C01, Personalized Innovative Medicine Competence Center (PERIMED).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data presented in this study are available from the corresponding author upon reasonable request.
Acknowledgments
We would like to express our gratitude to Svetlana Simova for her help in performing the NMR experiments and in the interpretation of the results.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Leemhuis, H.; Pijning, T.; Dobruchowska, J.M.; van Leeuwen, S.S.; Kralj, S.; Dijkstra, B.W.; Dijkhuizen, L. Glucansucrases: Three-dimensional structures, reactions, mechanism, α-glucan analysis and their implications in biotechnology and food applications. J. Biotechnol. 2013, 163, 250–272. [Google Scholar] [CrossRef]
- Gangoiti, J.; Pijning, T.; Dijkhuizen, L. Biotechnological potential of novel glycoside hydrolase family 70 enzymes synthesizing α-glucans from starch and sucrose. Biotechnol. Adv. 2018, 36, 196–207. [Google Scholar] [CrossRef]
- Faucard, P.; Grimaud, F.; Lourdin, D.; Maigret, J.E.; Moulis, C.; Remaud-Siméon, M.; Putaux, J.L.; Potocki-Véronèse, G.; Rolland-Sabaté, A. Macromolecular structure and film properties of enzymatically-engineered high molar mass dextrans. Carbohydr. Polym. 2018, 181, 337–344. [Google Scholar] [CrossRef]
- van Hijum, S.A.F.T.; Kralj, S.; Ozimek, L.K.; Dijkhuizen, L. Structure-function relationships of glucansucrase and fructansucrase enzymes from lactic acid bacteria. Microbiol. Mol. Biol. R. 2006, 70, 157–176. [Google Scholar] [CrossRef]
- Moulis, C.; André, I.; Remaud-Simeon, M. GH13 amylosucrases and GH70 branching sucrases, atypical enzymes in their respective families. Cell. Mol. Life Sci. 2016, 73, 2661–2679. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Meng, X.; Dijkhuizen, L.; Liu, W. Structures, physico-chemical properties, production and (potential) applications of sucrose-derived α-d-glucans synthesized by glucansucrases. Carbohydr. Polym. 2020, 249, 116818. [Google Scholar] [CrossRef]
- Côté, G.L.; Skory, C.D. Effects of mutations at threonine-654 on the insoluble glucan synthesized by Leuconostoc mesenteroides NRRL B-1118 glucansucrase. Appl. Microbiol. Biotechnol. 2014, 98, 6651–6658. [Google Scholar] [CrossRef]
- Wangpaiboon, K.; Waiyaseesang, N.; Panpetch, P.; Charoenwongpaiboon, T.; Nepogodiev, S.A.; Ekgasit, S.; Field, R.A.; Pichayangkura, R. Characterisation of insoluble α-1,3-/α-1,6 mixed linkage glucan produced in addition to soluble α-1,6-linked dextran by glucansucrase (DEX-N) from Leuconostoc citreum ABK-1. Int. J. Biol. Macromol. 2020, 152, 473–482. [Google Scholar] [CrossRef]
- Ogawa, K.; Yui, T.; Okamura, K.; Misaki, A. Crystalline features of streptococcal (1→3)-α-D-glucans of human saliva. Biosci. Biotech. Biochem. 1994, 58, 1326–1327. [Google Scholar] [CrossRef]
- Yui, T.; Goto, K.; Kawano, Y.; Ogawa, K. Molecular modeling study of highly branching (1→3)-α-D-glucan, a model polysaccharide for cariogenic glucan, using the N-H mapping method. Biosci. Biotechnol. Biochem. 2000, 64, 52–60. [Google Scholar] [CrossRef]
- Côté, G.L. Alternan. In Polysaccharides, I. Polysaccharides from Prokaryotes, 5th ed.; Vandamme, E.J., DeBaets, S., Steinbüchel, A., Eds.; Wiley: Weinheim, Germany, 2002; Volume 5, pp. 323–350. [Google Scholar]
- Ito, K.; Ito, S.; Shimamura, T.; Weyand, S.; Kawarasaki, Y.; Misaka, T.; Abe, K.; Ko-bayashi, T.; Cameron, A.D.; Iwata, S. Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans. J. Mol. Biol. 2011, 408, 177–186. [Google Scholar] [CrossRef]
- Vujičić-Žagar, A.; Pijning, T.; Kralj, S.; López, C.A.; Eeuwema, W.; Dijkhuizen, L.; Dijkstra, B.W. Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes. Proc. Natl. Acad. Sci. USA 2010, 107, 21406–21411. [Google Scholar] [CrossRef]
- Pijning, T.; Vujičić-Žagar, A.; Kralj, S.; Dijkhuizen, L.; Dijkstra, B.W. Flexibility of truncated and full-length glucansucrase GTF180 enzymes from Lactobacillus reuteri 180. FEBS J. 2014, 281, 2159–2171. [Google Scholar] [CrossRef]
- Molina, M.; Cioci, G.; Moulis, C.; Séverac, E.; Remaud-Siméon, M. Bacterial α-Glucan and Branching Sucrases from GH70 Family: Discovery, Structure-Function Relationship Studies and Engineering. Microorganisms 2021, 9, 1607. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, H.B.; Li, M.Q.; Hu, X.Q.; Li, Y. Functional analysis of truncated and site-directed mutagenesis dextransucrases to produce different type dextrans. Enzyme Microb. Technol. 2017, 102, 26–34. [Google Scholar] [CrossRef]
- Swistowska, A.M.; Gronert, S.; Wittrock, S.; Collisi, W.; Hecht, H.-J.; Hofer, B. Identification of structural determinants for sub-strate binding and turnover by glucosyltransferase R supports the permutation hypothesis. FEBS Lett. 2007, 581, 4036–4042. [Google Scholar] [CrossRef]
- Tsumori, H.; Minami, T.; Kuramitsu, H.K. Identification of essential amino acids in the Streptococcus mutans glucosyltransfer-ases. J. Bacteriol. 1997, 179, 3391–3396. [Google Scholar] [CrossRef]
- Claverie, M.; Cioci, G.; Guionnet, M.; Schörghuber, J.; Lichtenecker, R.; Moulis, C.; Remaud-Siméon, M.; Lippens, G. Futile Encounter Engineering of the DSR-M Dextransucrase Modifies the Resulting Polymer Length. Biochemistry 2019, 58, 2853–2859. [Google Scholar] [CrossRef]
- Meng, X.; Pijning, T.; Tietema, M.; Dobruchowska, J.M.; Yin, H.; Gerwig, G.J.; Kralj, S.; Dijkhuizen, L. Characterization of the glucansucrase GTF180 W1065 mutant enzymes producing polysaccharides and oligosaccharides with altered linkage composition. Food Chem. 2017, 217, 81–90. [Google Scholar] [CrossRef]
- Hellmuth, H.; Wittrock, S.; Kralj, S.; Dijkhuizen, L.; Hofer, B.; Seibel, J. Engineering the glucansucrase GTFR enzyme reaction and glycosidic bond specificity: Toward tailor-made polymer and oligosaccharide products. Biochemistry 2008, 47, 6678–6684. [Google Scholar] [CrossRef]
- Moulis, C.; Joucla, G.; Harrison, D.; Fabre, E.; Potocki-Veronese, G.; Monsan, P.; Remaud-Simeon, M. Understanding the polymerization mechanism of glycoside-hydrolase family 70 glucansucrases. J. Biol. Chem. 2006, 281, 31254–31267. [Google Scholar] [CrossRef]
- Bozonnet, S.; Dols-Laffargue, M.; Fabre, E.; Pizzut, S.; Remaud-Simeon, M.; Monsan, P.; Willemot, R.M. Molecular characterization of DSR-E, an alpha-1,2 linkage-synthesizing dextransucrase with two catalytic domains. J. Bacteriol. 2002, 184, 5753–5761. [Google Scholar] [CrossRef]
- Brison, Y.; Fabre, E.; Moulis, C.; Portais, J.C.; Monsan, P.; Remaud-Siméon, M. Synthesis of dextrans with controlled amounts of alpha-1,2 linkages using the transglucosidase GBD-CD2. Appl. Microbiol. Biotechnol. 2010, 86, 545–554. [Google Scholar] [CrossRef]
- Meng, X.; Gangoiti, J.; Wang, X.; Grijpstra, P.; van Leeuwen, S.S.; Pijning, T.; Dijkhuizen, L. Biochemical characterization of a GH70 protein from Lactobacillus kunkeei DSM 12361 with two catalytic domains involving branching sucrase activity. Appl. Microbiol. Biotechnol. 2018, 102, 7935–7950. [Google Scholar] [CrossRef]
- Bivolarski, V.; Vasileva, T.; Valerie, G.; Iliev, I. Synthesis of glucooligosaccharides with prebiotic potential by glucansucrase URE 13-300 acceptor reactions with maltose, raffinose and lactose. Eng. Life Sci. 2018, 18, 904–913. [Google Scholar] [CrossRef]
- Iliev, I.; Vasileva, T.; Bivolarski, V.; Yovcheva, T.; Marudova, M.; Viraneva, A.; Bodurov, I. Obtaining of Water-Insoluble Glucan Through Transferase Enzyme Reaction. BG Patent 67404 B1, 21 November 2023. [Google Scholar]
- Meng, X.; Dobruchowska, J.M.; Pijning, T.; López, C.A.; Kamerling, J.P.; Dijkhuizen, L. Residue Leu940 has a crucial role in the linkage and reaction specificity of the glucansucrase GTF180 of the probiotic bacterium Lactobacillus reuteri 180. J. Biol. Chem. 2014, 289, 32773–32782. [Google Scholar] [CrossRef]
- Funane, K.; Ishii, T.; Ono, H.; Kobayashi, M. Changes in linkage pattern of glucan products induced by substitution of Lys residues in the dextransucrase. FEBS Lett. 2005, 579, 4739–4745. [Google Scholar] [CrossRef]
- Molina, M.; Moulis, C.; Monties, N.; Pizzut-Serin, S.; Guieysse, D.; Morel, S.; Cioci, G.; Remaud-Simeon, M. Deciphering an Undecided Enzyme: Investigations of the Structural Determinants Involved in the Linkage Specificity of Alternansucrase. ACS Catal. 2019, 9, 2222–2237. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Miao, M.; Ma, Y.; Jiang, B.; Cui, S.W.; Jin, Z.; Zhang, T. Characterisations of Lactobacillus reuteri SK24. 003 glucansucrase implications for a-gluco-poly-and oligosaccharides biosynthesis. Food Chem. 2017, 222, 105–112. [Google Scholar] [CrossRef]
- van Leeuwen, S.S.; Kralj, S.; Eeuwema, W.; Gerwig, G.; Dijkhuizen, L.; Kamerling, J.P. Structural characterization of bioengineered alpha-D-glucans produced by mutant glucansucrase GTF180 enzymes of Lactobacillus reuteri strain 180. Biomacromolecules 2009, 10, 580–588. [Google Scholar] [CrossRef]
- van Leeuwen, S.S.; Kralj, S.; Gerwig, G.J.; Dijkhuizen, L.; Kamerling, J.P. Structural Analysis of Bioengineered α-d-Glucan Produced by a Triple Mutant of the Glucansucrase GTF180 Enzyme from Lactobacillus reuteri Strain 180: Generation of (α1→4) Linkages in a Native (1→3) (1→6)-α-d-Glucan. Biomacromolecules 2008, 9, 2251–2258. [Google Scholar] [CrossRef][Green Version]
- Vuillemin, M.; Grimaud, F.; Claverie, M.; Rolland-Sabaté, A.; Garnier, C.; Lucas, P.; Monsan, P.; Dols-Lafargue, M.; Remaud-Siméon, M.; Moulis, C. A dextran with unique rheological properties produced by the dextransucrase from Oenococcus kitaharae DSM 17330. Carbohydr Polym. 2018, 179, 10–18. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Miller, A.W.; Robyt, J.F. Detection of dextransucrase and levansucrase on polyacrylamide gels by the periodic acid-Schiff stain: Staining artifacts and their prevention. Anal. Biochem. 1986, 156, 357–363. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ahmed, Z.; Wang, Y.; Anjum, N.; Ahmad, A.; Khan, S.T. Characterization of exopolysaccharide produced by Lactobacillus kefiranofaciens ZW3 isolated from Tibet kefir—Part II. Food Hydrocoll. 2013, 30, 343–350. [Google Scholar] [CrossRef]
- Schormann, N.; Patel, M.; Thannickal, L.; Purushotham, S.; Wu, R.; Mieher, J.L.; Wu, H.; Deivanayagam, C. The catalytic domains of Streptococcus mutans glucosyltransferases: A structural analysis. Acta Cryst. 2023, F79, 119–127. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Meng, X.; Pijning, T.; Dobruchowska, J.M.; Gerwig, G.J.; Dijkhuizen, L. Characterization of the Functional Roles of Amino Acid Residues in Acceptor-binding Subsite +1 in the Active Site of the Glucansucrase GTF180 from Lactobacillus reuteri 180. J. Biol. Chem. 2015, 290, 30131–30141. [Google Scholar] [CrossRef]
- Kang, H.K.; Oh, J.S.; Kim, D. Molecular characterization and expression analysis of the glucansucrase DSRWC from Weissella cibaria synthesizing α-(1→6) glucan. FEMS Microbiol. Lett. 2009, 292, 33–41. [Google Scholar] [CrossRef]
- İspirli, H.; Yüzer, M.O.; Skory, C.; Colquhoun, I.J.; Sağdıç, O.; Dertli, E. Characterization of a glucansucrase from Lactobacillus reuteri E81 and production of malto-oligosaccharides. Biocatal. Biotransformation 2019, 37, 421–430. [Google Scholar] [CrossRef]
- Yu, L.; Qian, Z.; Ge, J.; Du, R. Glucansucrase Produced by Lactic Acid Bacteria: Structure, Properties, and Applications. Fermentation 2022, 8, 629. [Google Scholar] [CrossRef]
- Li, M.Q.; Zhang, H.B.; Li, Y.; Hu, X.-Q.; Yang, J.W. The thermoduric effects of site-directed mutagenesis of proline and lysine on dextransucrase from Leuconostoc mesenteroides 0326. Int. J. Biol. Macromol. 2018, 107, 1641–1649. [Google Scholar] [CrossRef]
- Robyt, J.F.; Walseth, T.F. Production, purification, and properties of dextransucrase from Leuconostoc mesenteroides NRRL B-512F. Carbohydr. Res. 1979, 68, 95–111. [Google Scholar] [CrossRef]
- Yi, A.-R.; Lee, S.-R.; Jang, M.-U.; Park, J.-M.; Eom, H.-J.; Han, N.S.; Kim, T.-J. Cloning of dextransucrase gene from Leuconostoc citreum HJ-P4 and its high-level expression in E. coli by low temperature induction. J. Microbiol. Biotechnol. 2009, 19, 829–835. [Google Scholar]
- van Leeuwen, S.S.; Kralj, S.; van Geel-Schutten, I.H.; Gerwig, G.J.; Dijkhuizen, L.; Kamerling, J.P. Structural analysis of the α-D-glucan (EPS180) produced by the Lactobacillus reuteri strain 180 glucansucrase GTF180 enzyme. Carbohydr. Res. 2008, 343, 1237–1250. [Google Scholar] [CrossRef]
- Patel, S.; Majumder, A.; Goyal, A. Potentials of exopolysaccharides from lactic acid bacteria. Indian J. Microbiol. 2012, 52, 3–12. [Google Scholar] [CrossRef]
- Naessens, M.; Cerdobbel, A.; Soetaert, W.; Vandamme, E.J. Leuconostoc dextransucrase and dextran: Production, properties and applications. J. Chem. Technol. Biotechnol. 2005, 80, 8445–8860. [Google Scholar] [CrossRef]
- Saravanan, C.; Kumar, H.; Shetty, P.K.H. Isolation and characterization of exopolysaccharide from Leuconostoc lactis KC117496 isolated from idli batter. Int. J. Biol. Macromol. 2016, 90, 100–106. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).