Improved Stability and Hydrolysates of Hyperthermophilic GH57 Type II Pullulanase from the Deep-Sea Archaeon Thermococcus siculi HJ21 by Truncation
Abstract
1. Introduction
2. Results and Discussion
2.1. Pullulanase Bioinformatics Analysis
2.2. Expression of Truncated Pullulanase
2.3. Characteristics of Truncated Pullulanase
2.4. Hydrolysates of Truncated Pullulanase
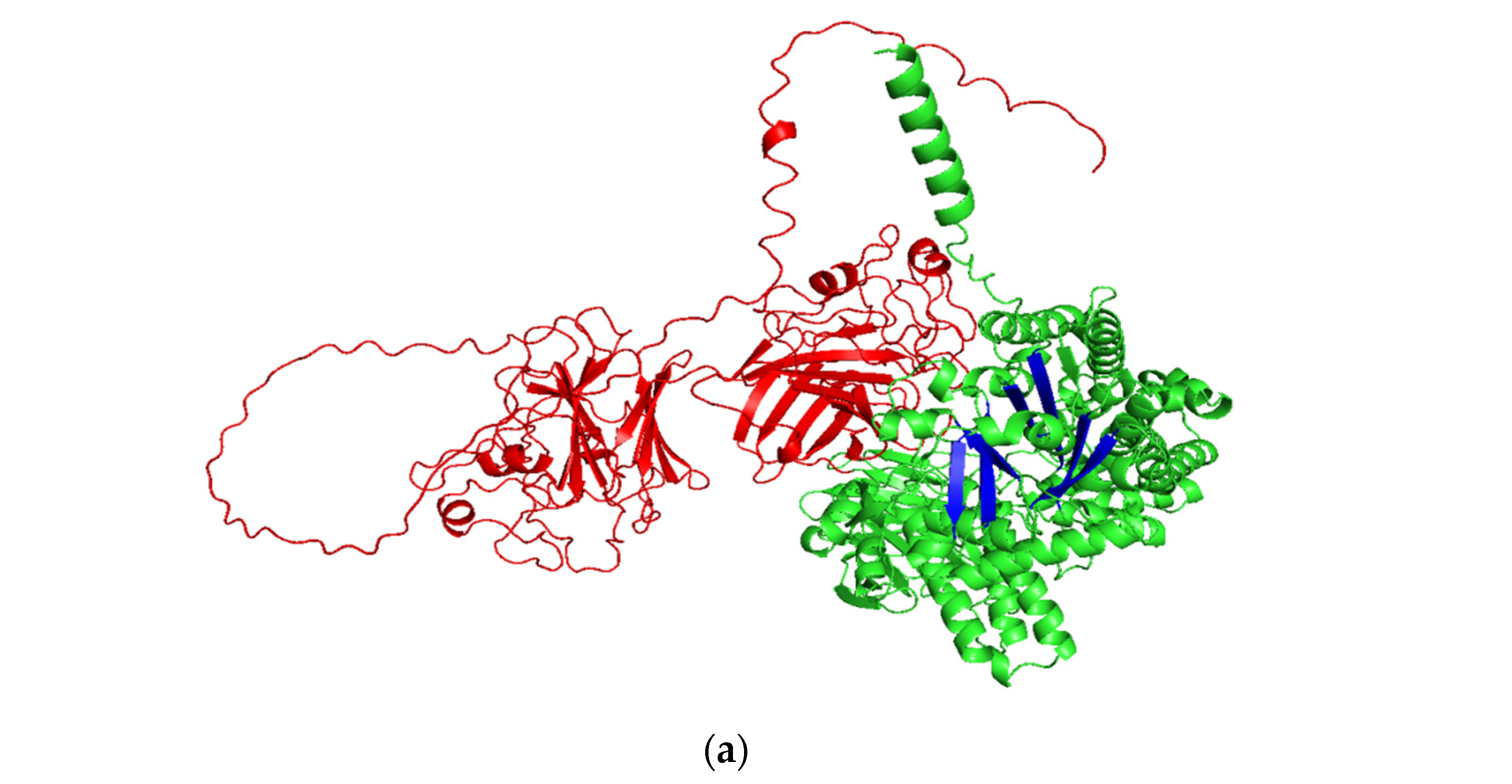
2.5. Molecular Mechanism of Truncated Pullulanase
3. Materials and Methods
3.1. Materials and Reagents
3.1.1. Strain and Plasmid
3.1.2. Reagents
3.2. Methods
3.2.1. Pullulanase Gene Acquisition
3.2.2. Bioinformatics Analysis of Pullulanase
3.2.3. Truncated Expression of the Pullulanase Gene
3.2.4. Detection of Pullulanase Activity
3.2.5. Analysis of Enzymatic Properties
3.2.6. Analysis of Enzymatic Hydrolysates
3.2.7. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Apriyanto, A.; Compart, J.; Fettke, J. A review of starch, a unique biopolymer–Structure, metabolism and in planta modifications. Plant Sci. 2022, 318, 111223. [Google Scholar] [CrossRef] [PubMed]
- Kahar, U.M.; Latif, N.A.; Amran, S.I.; Liew, K.J.; Goh, K.M. A Bibliometric Analysis and Review of Pullulan-Degrading Enzymes—Past and Current Trends. Catalysts 2022, 12, 143. [Google Scholar] [CrossRef]
- Bangar, S.P.; Ashogbon, A.O.; Singh, A.; Chaudhary, V.; Whiteside, W.S. Whiteside, Enzymatic modification of starch: A green approach for starch applications. Carbohyd. Polym. 2022, 287, 119265. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhang, S.Y.; Luo, Z.G.; Zong, M.H.; Li, X.X.; Lou, W.Y. Biotechnology and bioengineering of pullulanase: State of the art and perspectives. World J. Microbiol. Biotechnol. 2021, 37, 43. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Zhang, K.; Su, L.; Wu, J. Microbial starch debranching enzymes: Developments and applications. Biotechnol. Adv. 2021, 50, 107786. [Google Scholar] [CrossRef]
- Wang, M.; Hu, H.; Zhang, B.; Zheng, Y.; Wu, P.; Lu, Z.; Zhang, G. Discovery of a New Microbial Origin Cold-Active Neopullulanase Capable for Effective Conversion of Pullulan to Panose. Int. J. Mol. Sci. 2022, 23, 6928. [Google Scholar] [CrossRef]
- Wang, X.; Nie, Y.; Xu, Y. Industrially produced pullulanases with thermostability: Discovery, engineering, and heterologous expression. Bioresour. Technol. 2019, 278, 360–371. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Moller, M.S.; Henriksen, A.; Svensson, B. Structure and function of alpha-glucan debranching enzymes. Cell. Mol. Life Sci. 2016, 73, 2619–2641. [Google Scholar] [CrossRef]
- Møller, M.S.; Goh, Y.J.; Rasmussen, K.B.; Cypryk, W.; Celebioglu, H.U.; Klaenhammer, T.R.; Abou Hachem, M. An Extracellular Cell-Attached Pullulanase Confers Branched alpha-Glucan Utilization in Human Gut Lactobacillus acidophilus. Appl. Environ. Microbiol. 2017, 83, e00402–e00417. [Google Scholar] [CrossRef]
- Kuriki, T.; Imanaka, T. The concept of the alpha-amylase family: Structural similarity and common catalytic mechanism. J. Biosci. Bioeng. 1999, 87, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, Y.; Kusunoki, M.; Harada, W.; Kakudo, M. Structure and possible catalytic residues of Taka-amylase A. J. Biochem. 1984, 95, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Janecek, S.; Svensson, B.; MacGregor, E.A. Alpha-Amylase: An enzyme specificity found in various families of glycoside hydrolases. Cell. Mol. Life Sci. 2014, 71, 1149–1170. [Google Scholar] [CrossRef] [PubMed]
- Stam, M.R.; Danchin, E.G.; Rancurel, C.; Coutinho, P.M.; Henrissat, B. Dividing the large glycoside hydrolase family 13 into subfamilies: Towards improved functional annotations of alpha-amylase-related proteins. Protein Eng. Des. Sel. 2006, 19, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Xu, J.; Fu, X.; Tan, M.; Liu, F.; Zheng, H.; Song, H. Effects of different carbohydrate-binding modules on the enzymatic properties of pullulanase. Int. J. Biol. Macromol. 2019, 137, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Sunako, M.; Ono, H.; Murooka, Y.; Fukusaki, E.; Yamashita, M. Characterization of gene encoding amylopullulanase from plant-originated lactic acid bacterium, Lactobacillus plantarum L137. J. Biosci. Bioeng. 2008, 106, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Chuang, H.H.; Lin, F.P. Biochemical characterization of engineered amylopullulanase from Thermoanaerobacter ethanolicus 39E-implicating the non-necessity of its 100 C-terminal amino acid residues. Extremophiles 2008, 12, 641–650. [Google Scholar] [CrossRef]
- Pang, B.; Zhou, L.; Cui, W.; Liu, Z.; Zhou, S.; Xu, J.; Zhou, Z. A Hyperthermostable Type II Pullulanase from a Deep-Sea Microorganism Pyrococcus yayanosii CH1. J. Agric. Food Chem. 2019, 67, 9611–9617. [Google Scholar] [CrossRef]
- Zona, R.; Chang-Pi-Hin, F.; O’Donohue, M.J.; Janecek, S. Bioinformatics of the glycoside hydrolase family 57 and identification of catalytic residues in amylopullulanase from Thermococcus hydrothermalis. Eur. J. Biochem. 2004, 271, 2863–2872. [Google Scholar] [CrossRef]
- Li, X.; Li, D. Preparation of linear maltodextrins using a hyperthermophilic amylopullulanase with cyclodextrin- and starch-hydrolysing activities. Carbohydr. Polym. 2015, 119, 134–141. [Google Scholar] [CrossRef]
- Dong, G.; Vieille, C.; Zeikus, J.G. Cloning, sequencing, and expression of the gene encoding amylopullulanase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl. Environ. Microbiol. 1997, 63, 3577–3584. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.H.; Cha, J. Membrane-bound amylopullulanase is essential for starch metabolism of Sulfolobus acidocaldarius DSM639. Extremophiles 2015, 19, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, D.; Park, K.H. An extremely thermostable amylopullulanase from Staphylothermus marinus displays both pullulan- and cyclodextrin-degrading activities. Appl. Microbiol. Biotechnol. 2013, 97, 5359–5369. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.L.; Wang, S.J.; Lv, M.S.; Xu, J.L.; Fang, Y.W.; Liu, S. A GH57 family amylopullulanase from deep-sea Thermococcus siculi: Expression of the gene and characterization of the recombinant enzyme. Curr. Microbiol. 2011, 62, 222–228. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Akassou, M.; Groleau, D. Advances and challenges in the production of extracellular thermoduric pullulanases by wild-type and recombinant microorganisms: A review. Crit. Rev. Biotechnol. 2019, 39, 337–350. [Google Scholar] [CrossRef]
- Bi, J.; Chen, S.; Zhao, X.; Nie, Y.; Xu, Y. Computation-aided engineering of starch-debranching pullulanase from Bacillus thermoleovorans for enhanced thermostability. Appl. Microbiol. Biotechnol. 2020, 104, 7551–7562. [Google Scholar] [CrossRef]
- Lingmeng, L.; Fengying, D.; Lin, L.; Dannong, H.; Jingwen, C.; Wei, W.; Dongzhi, W. Biochemical Characterization of a Novel Thermostable Type I Pullulanase Produced Recombinantly in Bacillus subtilis. Starch–Stärke 2018, 70, 1700179. [Google Scholar]
- Tanimoto, T.; Omatsu, M.; Ikuta, A.; Nishi, Y.; Murakami, H.; Nakano, H.; Kitahata, S. Synthesis of novel heterobranched beta-cyclodextrins having beta-D-N-acetylglucosaminyl-maltotriose on the side chain. Biosci. Biotechnol. Biochem. 2005, 69, 732–739. [Google Scholar] [CrossRef]
- Shaw, J.F.; Sheu, J.R. Production of High-maltose Syrup and High-protein Flour from Rice by an Enzymatic Method. Biosci. Biotechnol. Biochem. 1992, 56, 1071–1073. [Google Scholar] [CrossRef]
- Bertoldo, C.; Antranikian, G. Starch-hydrolyzing enzymes from thermophilic archaea and bacteria. Curr. Opin. Chem. Biol. 2002, 6, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Fujita, K.; Kuwahara, N.; Tanimoto, T.; Hashimoto, H.; Koizumi, K.; Kitahata, S. Galactosylation of cyclodextrins and branched cyclodextrins by alpha-galactosidases. Biosci. Biotechnol. Biochem. 1994, 58, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Schallmey, M.; Singh, A.; Ward, O.P. Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 2004, 50, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Furiga, A.; Dols-Lafargue, M.; Heyraud, A.; Chambat, G.; Lonvaud-Funel, A.; Badet, C. Effect of antiplaque compounds and mouthrinses on the activity of glucosyltransferases from Streptococcus sobrinus and insoluble glucan production. Oral Microbiol. Immunol. 2008, 23, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Zillig, W.; Holz, I.; Janekovic, D.; Schafer, W.; Reiter, W.D. The Archaebacterium Thermococcus celer Represents, a Novel Genus within the Thermophilic Branch of the Archaebacteria. Syst. Appl. Microbiol. 1983, 4, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Palomo, M.; Pijning, T.; Booiman, T.; Dobruchowska, J.M.; van der Vlist, J.; Kralj, S.; Leemhuis, H. Thermus thermophilus glycoside hydrolase family 57 branching enzyme: Crystal structure, mechanism of action, and products formed. J. Biol. Chem. 2011, 286, 3520–3530. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.R.; Tonoli, C.C.; Trindade, D.M.; Betzel, C.; Takata, H.; Kuriki, T.; Murakami, M.T. Structural basis for branching-enzyme activity of glycoside hydrolase family 57: Structure and stability studies of a novel branching enzyme from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. Proteins 2011, 79, 547–557. [Google Scholar] [CrossRef]
- Na, S.; Park, M.; Jo, I.; Cha, J.; Ha, N.C. Structural basis for the transglycosylase activity of a GH57-type glycogen branching enzyme from Pyrococcus horikoshii. Biochem. Biophys. Res. Commun. 2017, 484, 850–856. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, H.; Kim, Y.J.; Jung, D.H.; Seo, D.H.; Jung, J.H.; Park, C.S. Enzymatic analysis of truncation mutants of a type II pullulanase from Bifidobacterium adolescentis P2P3, a resistant starch-degrading gut bacterium. Int. J. Biol. Macromol. 2021, 193, 1340–1349. [Google Scholar] [CrossRef]
- Imamura, H.; Fushinobu, S.; Yamamoto, M.; Kumasaka, T.; Jeon, B.S.; Wakagi, T.; Matsuzawa, H. Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor. J. Biol. Chem. 2003, 278, 19378–19386. [Google Scholar] [CrossRef]
- Henrissat, B. Glycosidase families. Biochem. Soc. Trans. 1998, 26, 153–156. [Google Scholar] [CrossRef] [PubMed]
- Erra-Pujada, M.; Debeire, P.; Duchiron, F.; O’Donohue, M.J. The type II pullulanase of Thermococcus hydrothermalis: Molecular characterization of the gene and expression of the catalytic domain. J. Bacteriol. 1999, 181, 3284–3287. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Vieille, C.; Zeikus, J.G. Identification of Pyrococcus furiosus amylopullulanase catalytic residues. Appl. Microbiol. Biotechnol. 2005, 66, 408–413. [Google Scholar] [CrossRef]
- Dao, V.L.; Chan, S.; Zhang, J.; Ngo, R.; Poh, C.L. Single 3′-exonuclease-based multifragment DNA assembly method (SENAX). Sci. Rep. 2022, 12, 4004. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Hassabis, D. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Santos-Martins, D.; Solis-Vasquez, L.; Tillack, A.F.; Sanner, M.F.; Koch, A.; Forli, S. Accelerating AutoDock4 with GPUs and Gradient-Based Local Search. J. Chem. Theory Comput. 2021, 17, 1060–1073. [Google Scholar] [CrossRef]
- Lu, T.; Tan, H.; Lee, D.; Chen, G.; Jia, Z. New insights into the activation of Escherichia coli tyrosine kinase revealed by molecular dynamics simulation and biochemical analysis. Biochemistry 2009, 48, 7986–7995. [Google Scholar] [CrossRef]










| Characteristics | Pul-HJΔ782 | Pul-HJ21 |
|---|---|---|
| Number of amino acids | 782 | 1372 |
| Molecular weight (Da) | 90,046.50 | 153,712.72 |
| Theoretical pI | 4.65 | 4.58 |
| Asp +Glu | 120 | 198 |
| Arg + Lys | 69 | 109 |
| Formula | C411H6159N1021O1218S21 | C6972H10528N1758O2107S32 |
| Instability index | 25.79 | 28.83 |
| Ext. coefficient/(mol·cm)−1 | 210,970 | 311,240 |
| Aliphatic index | 82.26 | 81.78 |
| Grand average of hydropathicity (GRAVY) | −0.410 | −0.37 |
| Mutant and Temperature | kd a | T1/2 b |
|---|---|---|
| Pul-HJ21-100 °C | 0.176 ± 0.010 | 3.95 ± 0.3 |
| Pul-HJ21-90 °C | 0.051 ± 0.003 | 13.64 ± 1.1 |
| Pul-HJ21-80 °C | 0.019 ± 0.003 | 37.41 ± 8.4 |
| Pul-HJΔ782-100 °C | 0.132 ± 0.001 | 5.25 ± 0.1 |
| Pul-HJΔ782-90 °C | 0.048 ± 0.012 | 15.40 ± 5.4 |
| Pul-HJΔ782-80 °C | 0.011 ± 0.001 | 63.54 ± 8.2 |
| Enzyme and Time | Oligosaccharides (mg/mL) | ||||
|---|---|---|---|---|---|
| G7 | G5 | G3 | G2 | G1 | |
| Pul-HJ21-15 min | 11.21 ± 0.48 | 0.78 ± 0.05 | 0.11 ± 0.03 | 0.12 ± 0.03 | 0.41 ± 0.02 |
| Pul-HJ21-30 min | 12.06 ± 0.38 | 0.76 ± 0.01 | 0.13 ± 0.02 | 0.19 ± 0.01 | 0.70 ± 0.20 |
| Pul-HJ21-1 h | 12.37 ± 0.30 | 0.92 ± 0.07 | 0.34 ± 0.05 | 0.38 ± 0.03 | 0.46 ± 0.04 |
| Pul-HJ21-3 h | 13.64 ± 0.17 | 1.09 ± 0.07 | 0.58 ± 0.14 | 1.06 ± 0.10 | 0.87 ± 0.02 |
| Pul-HJ21-5 h | 11.94 ± 0.72 | 0.88 ± 0.15 | 0.55 ± 0.17 | 0.85 ± 0.40 | 1.32 ± 0.04 |
| Pul-HJΔ782-15 min | 11.27 ± 0.39 | 0.82 ± 0.03 | 0.18 ± 0.10 | 0.28 ± 0.11 | 0.68 ± 0.05 |
| Pul-HJΔ782-30 min | 10.71 ± 0.61 | 0.76 ± 0.20 | 0.11 ± 0.03 | 0.35 ± 0.03 | 0.63 ± 0.10 |
| Pul-HJΔ782-1 h | 11.84 ± 0.53 | 0.96 ± 0.10 | 0.12 ± 0.01 | 0.28 ± 0.03 | 1.00 ± 0.05 |
| Pul-HJΔ782-3 h | 11.14 ± 0.33 | 0.64 ± 0.40 | 0.19 ± 0.03 | 0.36 ± 0.03 | 1.92 ± 0.39 |
| Pul-HJΔ782-5 h | 13.39 ± 0.56 | 1.17 ± 0.03 | 0.35 ± 0.11 | 0.60 ± 0.05 | 3.00 ± 0.18 |
| Enzyme and Time | Oligosaccharides (mg/mL) | ||||
|---|---|---|---|---|---|
| G7 | G5 | G3 | G2 | G1 | |
| Pul-HJ21-15 min | 13.12 ± 0.56 | 0.95 ± 0.01 | 0.37 ± 0.14 | / | 0.42 ± 0.01 |
| Pul-HJ21-30 min | 14.62 ± 0.09 | 0.98 ± 0.02 | 0.50 ± 0.03 | / | 0.47 ± 0.03 |
| Pul-HJ21-1 h | 16.61 ± 0.72 | 0.88 ± 0.01 | 0.81 ± 0.09 | / | 0.48 ± 0.01 |
| Pul-HJ21-3 h | 14.88 ± 0.30 | 0.87 ± 0.03 | 2.03 ± 0.17 | / | 0.43 ± 0.03 |
| Pul-HJ21-5 h | 19.60 ± 0.78 | 1.06 ± 0.02 | 5.27 ± 0.28 | / | 0.52 ± 0.03 |
| Pul-HJΔ782-15 min | 13.86 ± 0.68 | 0.84 ± 0.02 | 0.14 ± 0.01 | / | 0.14 ± 0.01 |
| Pul-HJΔ782-30 min | 15.54 ± 0.74 | 0.81 ± 0.02 | 0.18 ± 0.03 | / | 0.11 ± 0.03 |
| Pul-HJΔ782-1 h | 17.84 ± 1.95 | 0.32 ± 0.73 | 0.36 ± 0.01 | / | 0.11 ± 0.02 |
| Pul-HJΔ782-3 h | 19.52 ± 0.91 | 0.75 ± 0.03 | 1.22 ± 0.13 | / | 0.14 ± 0.02 |
| Pul-HJΔ782-5 h | 18.90 ± 0.47 | 0.69 ± 0.69 | 1.99 ± 0.02 | / | 0.15 ± 0.02 |
| Primer | (5′ to 3′) | Primer |
|---|---|---|
| His-F | atgcaccaccaccaccaccacAGGCGGGTGGTTGCC | His-F |
| T-His-F | taagaaggagatatacatatgATGCACCACCACCACCACCACAGG | T-His-F |
| 477-R | acggagctcgaattcggatccCATCATCTTGGGGGTGAGCTT | 477-R |
| 526-R | acggagctcgaattcggatccGCCTATCCAGGTGGAGAGCG | 526-R |
| 576-R | acggagctcgaattcggatccCCCATACCACCAGAACCAGTCG | 576-R |
| 636-R | acggagctcgaattcggatccATTCTTCATCTCCCCCTCCTTG | 636-R |
| 782-R | acggagctcgaattcggatccCTTCAGCTCGACGGGGGT | 782-R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Dou, B.; Wang, B.; Liu, M.; Shao, R.; Lu, J.; Lyu, M.; Wang, S. Improved Stability and Hydrolysates of Hyperthermophilic GH57 Type II Pullulanase from the Deep-Sea Archaeon Thermococcus siculi HJ21 by Truncation. Catalysts 2023, 13, 453. https://doi.org/10.3390/catal13030453
Wu X, Dou B, Wang B, Liu M, Shao R, Lu J, Lyu M, Wang S. Improved Stability and Hydrolysates of Hyperthermophilic GH57 Type II Pullulanase from the Deep-Sea Archaeon Thermococcus siculi HJ21 by Truncation. Catalysts. 2023; 13(3):453. https://doi.org/10.3390/catal13030453
Chicago/Turabian StyleWu, Xudong, Baojie Dou, Boyan Wang, Mingwang Liu, Ruxue Shao, Jing Lu, Mingsheng Lyu, and Shujun Wang. 2023. "Improved Stability and Hydrolysates of Hyperthermophilic GH57 Type II Pullulanase from the Deep-Sea Archaeon Thermococcus siculi HJ21 by Truncation" Catalysts 13, no. 3: 453. https://doi.org/10.3390/catal13030453
APA StyleWu, X., Dou, B., Wang, B., Liu, M., Shao, R., Lu, J., Lyu, M., & Wang, S. (2023). Improved Stability and Hydrolysates of Hyperthermophilic GH57 Type II Pullulanase from the Deep-Sea Archaeon Thermococcus siculi HJ21 by Truncation. Catalysts, 13(3), 453. https://doi.org/10.3390/catal13030453







