Site-Directed Mutagenesis of Two-Domain Laccase ScaSL for Obtaining a Biocatalyst with Improved Characteristics
Abstract
:1. Introduction
2. Results
2.1. Alignment of Laccase Sequences and Modeling Their Three-Dimensional Structure
2.2. Production and Physic-Chemical Properties of Mutant Proteins
2.3. Redox Potential and Kinetic Characteristics of Mutant Laccases
2.4. Bioelectrocatalysis
2.5. Dye Decolorization
2.6. Crystal Structure of H286T Mutant
3. Discussion
4. Materials and Methods
4.1. Alignment and Modeling of Three-Dimensional Structure of Laccase
4.2. Construction of the ScaSL Mutants and Proteins Purification
4.3. Characterization of ScaSL Mutants
4.4. Redox Potential Determination by Redox Titration
4.5. Bioelectrocatalysis
4.5.1. MWCNTox Characteristics
4.5.2. Preparation of Modified Electrodes
4.5.3. Electrochemical Measurements
4.5.4. Redox Potential Determination by Chronoamperometric Measurements
4.6. Dye Decolorization
4.7. Crystallization and Crystallography
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Palmer, A.E.; Lee, S.K.; Solomon, E.I. Decay of the Peroxide Intermediate in Laccase: Reductive Cleavage of the O−O Bond. J. Am. Chem. Soc. 2001, 123, 6591–6599. [Google Scholar] [CrossRef] [PubMed]
- Mot, A.C.; Silaghi-Dumitrescu, R. Laccases: Complex architectures for one-electron oxidations. Biochemistry 2012, 77, 1395–1407. [Google Scholar] [CrossRef] [PubMed]
- Janusz, G.; Pawlik, A.; Swiderska-Burek, U.; Polak, J.; Sulej, J.; Jarosz-Wilkolazka, A.; Paszczynski, A. Laccase Properties, Physiological Functions, and Evolution. Int. J. Mol. Sci. 2020, 21, 966. [Google Scholar] [CrossRef]
- Chauhan, P.S.; Goradia, B.; Saxena, A. Bacterial laccase: Recent update on production, properties and industrial applications. 3 Biotech 2017, 7, 323. [Google Scholar] [CrossRef]
- Arregui, L.; Ayala, M.; Gómez-Gil, X.; Gutiérrez-Soto, G.; Hernández-Luna, C.E.; Herrera de los Santos, M.; Levin, L.; Rojo-Domínguez, A.; Romero-Martínez, D.; Saparrat, M.C.N.; et al. Laccases: Structure, function, and potential application in water bioremediation. Microb. Cell Factories 2019, 18, 200. [Google Scholar] [CrossRef]
- Upadhyay, P.; Shrivastava, R.; Agrawal, P.K. Bioprospecting and biotechnological applications of fungal laccase. 3 Biotech 2016, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Shleev, S.; Tkac, J.; Christenson, A.; Ruzgas, T.; Yaropolov, A.I.; Whittaker, J.W.; Gorton, L. Direct electron transfer between copper-containing proteins and electrodes. Biosens. Bioelectron. 2005, 20, 2517–2554. [Google Scholar] [CrossRef]
- Berezin, I.V.; Bogdanovskaya, V.A.; Varfolomeev, S.D.; Tarasevich, M.R.; Yaropolov, A.I. Bioelectrocatalysis—Equilibrium oxygen potential in the presence of laccase. Dokl. Akad. Nauk SSSR 1978, 240, 615–618. [Google Scholar]
- Le Goff, A.; Holzinger, M.; Cosnier, S. Recent progress in oxygen-reducing laccase biocathodes for enzymatic biofuel cells. Cell. Mol. Life Sci. 2015, 72, 941–952. [Google Scholar] [CrossRef]
- Haque, S.u.; Duteanu, N.; Ciocan, S.; Nasar, A.; Inamuddin. A review: Evolution of enzymatic biofuel cells. J. Environ. Manag. 2021, 298, 113483. [Google Scholar] [CrossRef]
- Zhang, Y.; Lv, Z.Y.; Zhou, J.; Xin, F.X.; Ma, J.F.; Wu, H.; Fang, Y.; Jiang, M.; Dong, W.L. Application of eukaryotic and prokaryotic laccases in biosensor and biofuel cells: Recent advances and electrochemical aspects. Appl. Microbiol. Biotechnol. 2018, 102, 10409–10423. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Delgado, M.M.; Alemán-Nava, G.S.; Rodríguez-Delgado, J.M.; Dieck-Assad, G.; Martínez-Chapa, S.O.; Barceló, D.; Parra, R. Laccase-based biosensors for detection of phenolic compounds. TrAC Trends Anal. Chem. 2015, 74, 21–45. [Google Scholar] [CrossRef]
- Gunne, M.; Höppner, A.; Hagedoorn, P.L.; Urlacher, V.B. Structural and redox properties of the small laccase Ssl1 from Streptomyces sviceus. FEBS J. 2014, 281, 4307–4318. [Google Scholar] [CrossRef] [PubMed]
- Trubitsina, L.I.; Tishchenko, S.V.; Gabdulkhakov, A.G.; Lisov, A.V.; Zakharova, M.V.; Leontievsky, A.A. Structural and functional characterization of two-domain laccase from Streptomyces viridochromogenes. Biochimie 2015, 112, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Blánquez, A.; Rodríguez, J.; Brissos, V.; Mendes, S.; Martins, L.O.; Ball, A.S.; Arias, M.E.; Hernández, M. Decolorization and detoxification of textile dyes using a versatile Streptomyces laccase-natural mediator system. Saudi J. Biol. Sci. 2019, 26, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-X.; Vilbert, A.C.; Cui, C.; Mirts, E.N.; Williams, L.H.; Kim, W.; Jessie Zhang, Y.; Lu, Y. Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations. Angew. Chem. Int. Ed. 2023, 62, e202314019. [Google Scholar] [CrossRef]
- Kaur, R.; Salwan, R.; Sharma, V. Structural properties, genomic distribution of laccases from Streptomyces and their potential applications. Process Biochem. 2022, 118, 133–144. [Google Scholar] [CrossRef]
- Dey, B.; Dutta, T. Laccases: Thriving the domain of bio-electrocatalysis. Bioelectrochemistry 2022, 146, 108144. [Google Scholar] [CrossRef] [PubMed]
- Prins, A.; Kleinsmidt, L.; Khan, N.; Kirby, B.; Kudanga, T.; Vollmer, J.; Pleiss, J.; Burton, S.; Le Roes-Hill, M. The effect of mutations near the T1 copper site on the biochemical characteristics of the small laccase from Streptomyces coelicolor A3 (2). Enzym. Microb. Technol. 2015, 68, 23–32. [Google Scholar] [CrossRef]
- Olbrich, A.C.; Schild, J.N.; Urlacher, V.B. Correlation between the T1 copper reduction potential and catalytic activity of a small laccase. J. Inorg. Biochem. 2019, 201, 110843. [Google Scholar] [CrossRef]
- Toscano, M.D.; De Maria, L.; Lobedanz, S.; Ostergaard, L.H. Optimization of a small laccase by active-site redesign. Chembiochem 2013, 14, 1209–1211. [Google Scholar] [CrossRef] [PubMed]
- Trubitsina, L.I.; Trubitsin, I.V.; Lisov, A.V.; Gabdulkhakov, A.G.; Zavarzina, A.G.; Belova, O.V.; Larionova, A.P.; Tishchenko, S.V.; Leontievsky, A.A. A Novel Two-Domain Laccase with Middle Redox Potential: Physicochemical and Structural Properties. Biochemistry 2023, 88, 1658–1667. [Google Scholar] [CrossRef] [PubMed]
- Trubitsina, L.I.; Abdullatypov, A.V.; Larionova, A.P.; Trubitsin, I.V.; Alferov, S.V.; Ponamoreva, O.N.; Leontievsky, A.A. Expression of thermophilic two-domain laccase from Catenuloplanes japonicus in Escherichia coli and its activity against triarylmethane and azo dyes. PeerJ 2021, 9, e11646. [Google Scholar] [CrossRef] [PubMed]
- Lisov, A.V.; Trubitsina, L.I.; Lisova, Z.A.; Trubitsin, I.V.; Zavarzina, A.G.; Leontievsky, A.A. Transformation of humic acids by two-domain laccase from Streptomyces anulatus. Process Biochem. 2019, 76, 128–135. [Google Scholar] [CrossRef]
- Nathani, A.; Vishnu, N.; Sharma, C.S. Review-Pencil Graphite Electrodes as Platform for Enzyme and Enzyme-Like Protein Immobilization for Electrochemical Detection. J. Electrochem. Soc. 2020, 167, 11. [Google Scholar] [CrossRef]
- Abdullatypov, A.; Oskin, P.; Fedina, V.; Trubitsina, L.; Yakimovich, S.; Shuvalova, E.; Verma, P.; Dyachkova, T.; Ponamoreva, O.; Alferov, S. Functionalization of MWCNTs for Bioelectrocatalysis by Bacterial Two-Domain Laccase from Catenuloplanes japonicus. Nanomaterials 2023, 13, 3019. [Google Scholar] [CrossRef]
- Ben Tahar, A.; Żelechowska, K.; Biernat, J.F.; Paluszkiewicz, E.; Cinquin, P.; Martin, D.; Zebda, A. High catalytic performance of laccase wired to naphthylated multiwall carbon nanotubes. Biosens. Bioelectron. 2020, 151, 111961. [Google Scholar] [CrossRef]
- Che, A.-F.; Germain, V.; Cretin, M.; Cornu, D.; Innocent, C.; Tingry, S. Fabrication of free-standing electrospun carbon nanofibers as efficient electrode materials for bioelectrocatalysis. New J. Chem. 2011, 35, 2848–2853. [Google Scholar] [CrossRef]
- Jones, S.M.; Solomon, E.I. Electron transfer and reaction mechanism of laccases. Cell. Mol. Life Sci. 2015, 72, 869–883. [Google Scholar] [CrossRef]
- Gabdulkhakov, A.; Kolyadenko, I.; Kostareva, O.; Mikhaylina, A.; Oliveira, P.; Tamagnini, P.; Lisov, A.; Tishchenko, S. Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase from Streptomyces griseoflavus Ac-993. Int. J. Mol. Sci. 2019, 20, 3184. [Google Scholar] [CrossRef]
- Liu, J.; Xie, Y.; Peng, C.; Yu, G.; Zhou, J. Molecular Understanding of Laccase Adsorption on Charged Self-Assembled Monolayers. J. Phys. Chem. B 2017, 121, 10610–10617. [Google Scholar] [CrossRef] [PubMed]
- Hoshikawa, Y.; Castro-Muñiz, A.; Tawata, H.; Nozaki, K.; Yamane, S.; Itoh, T.; Kyotani, T. Orientation control of Trametes laccases on a carbon electrode surface to understand the orientation effect on the electrocatalytic activity. Bioconjugate Chem. 2018, 29, 2927–2935. [Google Scholar] [CrossRef] [PubMed]
- Gentil, S.; Rousselot-Pailley, P.; Sancho, F.; Robert, V.; Mekmouche, Y.; Guallar, V.; Tron, T.; Le Goff, A. Efficiency of Site-Specific Clicked Laccase-Carbon Nanotubes Biocathodes towards O2 Reduction. Chem.—Eur. J. 2020, 26, 4798–4804. [Google Scholar] [CrossRef]
- Milton, R.D.; Minteer, S.D. Direct enzymatic bioelectrocatalysis: Differentiating between myth and reality. J. R. Soc. Interface 2017, 14, 20170253. [Google Scholar] [CrossRef]
- Aleksejeva, O.; Mateljak, I.; Ludwig, R.; Alcalde, M.; Shleev, S. Electrochemistry of a high redox potential laccase obtained by computer-guided mutagenesis combined with directed evolution. Electrochem. Commun. 2019, 106, 106511. [Google Scholar] [CrossRef]
- Kolyadenko, I.; Scherbakova, A.; Kovalev, K.; Gabdulkhakov, A.; Tishchenko, S. Engineering the catalytic properties of two-domain laccase from Streptomyces griseoflavus Ac-993. Int. J. Mol. Sci. 2022, 23, 65. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.; Eramian, D.; Shen, M.Y.; Pieper, U.; Sali, A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinform. 2006, 54, 5–6. [Google Scholar] [CrossRef]
- Skálová, T.; Dohnálek, J.; Østergaard, L.H.; Østergaard, P.R.; Kolenko, P.; Dušková, J.; Štěpánková, A.; Hašek, J. The Structure of the Small Laccase from Streptomyces coelicolor Reveals a Link between Laccases and Nitrite Reductases. J. Mol. Biol. 2009, 385, 1165–1178. [Google Scholar] [CrossRef]
- Land, H.; Humble, M.S. YASARA: A Tool to Obtain Structural Guidance in Biocatalytic Investigations. In Protein Engineering: Methods and Protocols; Bornscheuer, U.T., Höhne, M., Eds.; Springer: New York, NY, USA, 2018; pp. 43–67. [Google Scholar]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Wariishi, H.; Valli, K.; Gold, M.H. Manganese (II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. Kinetic mechanism and role of chelators. J. Biol. Chem. 1992, 267, 23688–23695. [Google Scholar] [CrossRef] [PubMed]
- Heinfling, A.; Martínez, M.a.J.; Martínez, A.T.; Bergbauer, M.; Szewzyk, U. Purification and characterization of peroxidases from the dye-decolorizing fungus Bjerkandera adusta. FEMS Microbiol. Lett. 1998, 165, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Britton, H.T.S.; Robinson, R.A. CXCVIII.—Universal buffer solutions and the dissociation constant of veronal. J. Chem. Soc. (Resumed) 1931, 1456–1462. [Google Scholar] [CrossRef]
- Xu, F.; Shin, W.; Brown, S.H.; Wahleithner, J.A.; Sundaram, U.M.; Solomon, E.I. A study of a series of recombinant fungal laccases and bilirubin oxidase that exhibit significant differences in redox potential, substrate specificity, and stability. Biochim. Biophys. Acta (BBA)—Protein Struct. Mol. Enzymol. 1996, 1292, 303–311. [Google Scholar] [CrossRef]
- Xu, F. Effects of Redox Potential and Hydroxide Inhibition on the pH Activity Profile of Fungal Laccases. J. Biol. Chem. 1997, 272, 924–928. [Google Scholar] [CrossRef] [PubMed]
- Rigaku XtaLAB Synergy-S. Available online: https://www.rigaku.com/products/crystallography/synergys (accessed on 20 October 2023).
- Rigaku Oxford Diffraction. CrysAlisPro; Rigaku Oxford Diffraction: Yarnton, UK, 2019. [Google Scholar]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallographica. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallographica. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallographica. Sect. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System. Available online: http://www.pymol.org/ (accessed on 20 October 2023).




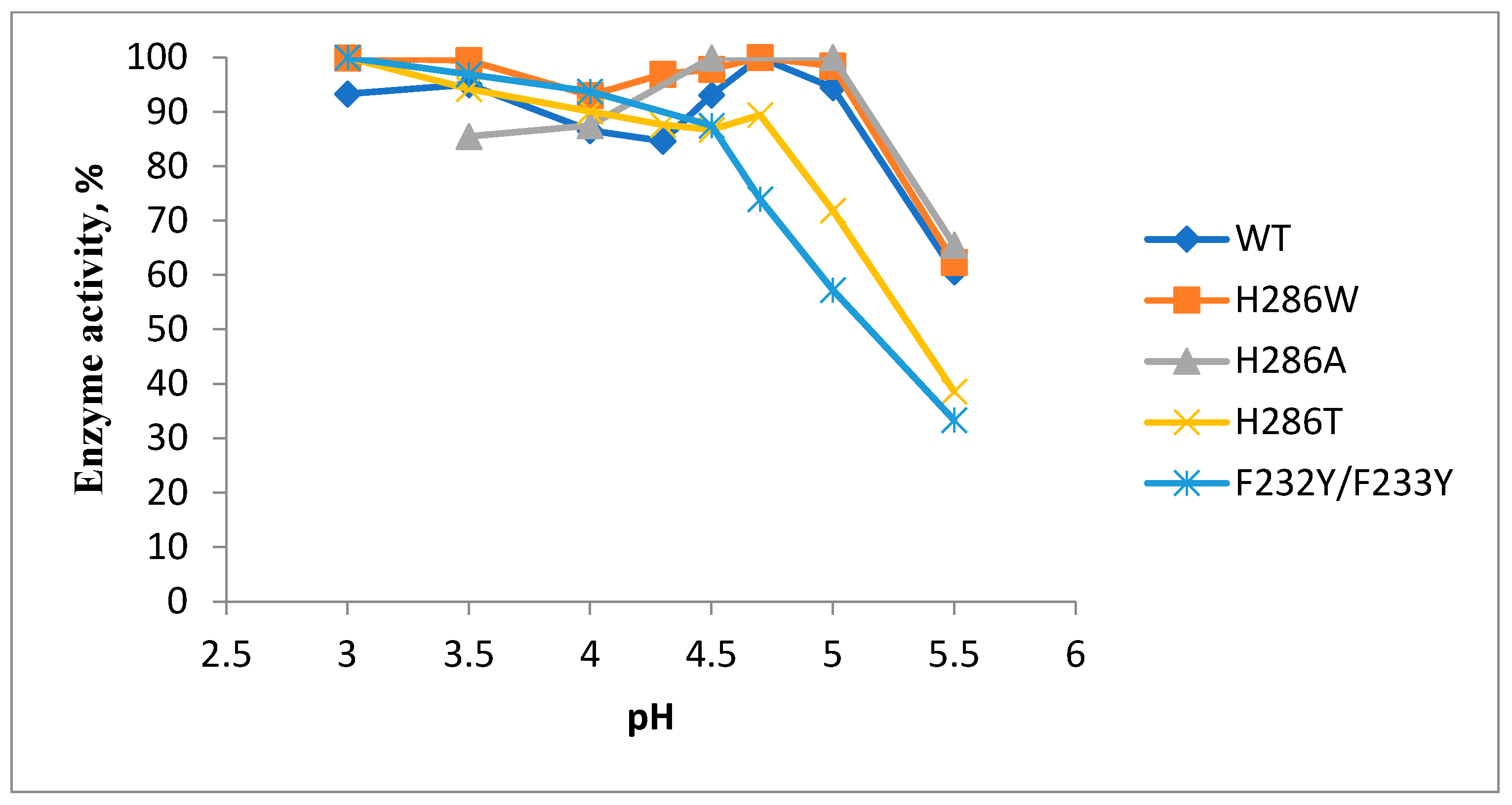

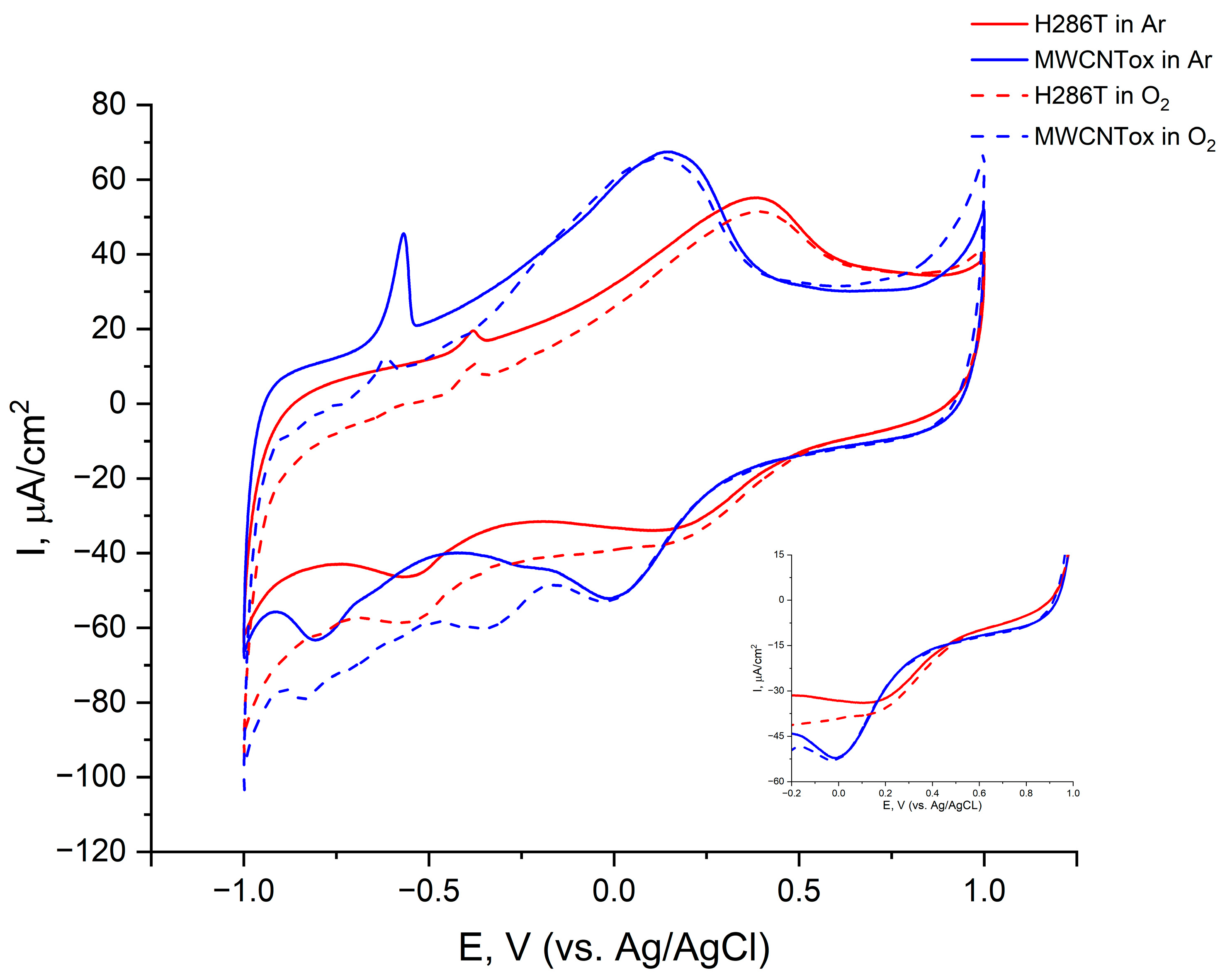


| Laccase | pH Optimum | Residual Activity after 60 h Incubation, % | Thermostability after Incubation at 70, 80, and 90 °C, Respectively, % | ||
|---|---|---|---|---|---|
| ABTS | 2,6-DMP | at pH 9.0 | at pH 11.0 | ||
| ScaSL (WT) | 4.5–5.0 | 7.5 | 70 | 66 | 62; 40; 33 |
| H286A | 4.5–5.0 | 7.5 | 95 | 82 | 67; 43; 35 |
| H286T | 3.0–4.0 | 7.5 | 77 | 57 | 58; 36; 21 |
| H286W | 4.5–5.0 | 7.5 | 72 | 61 | 62; 40; 28 |
| F232Y/F233Y | 3.0–4.0 | 7.0 | 54 | 49 | 69; 29; 12 |
| Laccase | Kinetic Constants for ABTS | Kinetic Constants for 2,6-DMP | Redox Potential, V 1 | ||||
|---|---|---|---|---|---|---|---|
| Km, mM | Vmax | kcat, s−1 | Km, mM | Vmax | kcat, s−1 | ||
| ScaSL (WT) | 0.100 0.006 | 40 ± 1 | 19.7 ± 0.7 | 0.840 ± 0.002 | 1.99 ± 0.01 | 0.36 ± 0.01 | 0.472 ± 0.007/0.47 |
| H286A | 0.170 ± 0.004 | 65 ± 2 | 74 ± 3 | 0.62 ± 0.04 | 2.03 ± 0.01 | 1.16 ± 0.01 | 0.455 ± 0.002/0.45 |
| H286T | 0.180 ± 0.008 | 76 ± 3 | 67 ± 2 | 0.84 ± 0.11 | 2.52 ± 0.15 | 1.11 ± 0.06 | >0.47/0.50 |
| H286W | 0.150 ± 0.007 | 72 ± 3 | 18 ± 3 | 0.89 ± 0.05 | 2.35 ± 0.04 | 1.37 ± 0.01 | >0.47/0.48 |
| F232Y/F233Y | 0.025 ± 0.001 | 46 ± 1 | 36.0 ± 0.2 | 0.600 ± 0.003 | 1.24 ± 0.01 | 0.98 ± 0.01 | >0.47/Undetermined |
| Laccase | ScaSL | H286T | H286A | F232Y/F233Y |
|---|---|---|---|---|
| Enzyme quantity, E | 8.0 ± 0.1 | 8.6 ± 1.1 | 8.2 ± 0.2 | 5.0 ± 0.6 |
| Direct electron transport current, nA | 108 ± 6 | 173 ± 24 | 95 ± 34 | 70 ± 19 |
| Specific current of direct electron transfer, mA·E−1 | 1.3 ± 0.1 | 1.9 ± 0.4 | 1.2 ± 0.5 | 1.5 ± 0.5 |
| Decolorization of Dye, % | ||||||
|---|---|---|---|---|---|---|
| ScaSL (WT) | H286A | H286T | H286W | F232Y/ F233Y | ||
| Brilliant green | E | 95.3 ± 0.2 | 95.4 ± 0.1 | 92.4 ± 0.4 | 93.5 ± 0.3 | 0 |
| E+ABTS | 91.5 ± 0.2 | 91.4 ± 0.6 | 92.0 ± 0.3 | 92.1 ± 0.6 | 81.9 ± 0.5 | |
| E+SA | 96.0 ± 0.1 | 96.6 ± 0.2 | 96.0± 0.4 | 95.2 ± 0.3 | 17.0 ± 1.1 | |
| Malachite green | E | 87.7 ± 2.9 | 89.8 ± 0.9 | 77.7 ± 0.9 | 77.6 ± 2 | 0.6 ± 2 |
| E+ABTS | 88.8 ± 0.4 | 89.5 ± 0.3 | 89.3 ± 1 | 89.0 ± 0.4 | 90.2 ± 0.3 | |
| E+SA | 92.8 ± 0.4 | 88.0 ± 1.5 | 92.5 ± 0.8 | 88.1 ± 0.9 | 2.8 ± 0.4 | |
| Methyl orange, pH 4/4.5 | E | 6.6 ± 3.4 | 8.4 ± 1.5 | 5.9 ± 1.2 | 4.9 ± 0.8 | 0.1± 4 |
| E+ABTS | 49.8 ± 1.9 | 52.3 ± 0.7 | 50.6 ± 0.1 | 52.2 ± 1.3 | 60.6 ± 1 | |
| E+SA | 31.3 ± 0.8 | 28.2 ± 0.9 | 29.2 ± 1.7 | 25.7 ± 0.9 | 25.6 ± 2 | |
| Methyl orange, pH 7/7.5 | E | 0 | 0 | 0 | 0 | 2.5±0.7 |
| E+ABTS | 13.2 ± 0.7 | 12.2 ± 1.7 | 11.8 ± 1.8 | 10.7 ± 2.1 | 41.4 ± 7 | |
| E+SA | 1.7 ± 0.7 | 2.2 ± 0.7 | 2.0 ± 0.8 | 1.6 ± 1.1 | 10.0 ± 2 | |
| Congo red, pH 7/7.5 | E | 0 | 0 | 0 | 0 | 0.2 ± 0.5 |
| E+ABTS | 46.9 ± 0.6 | 46.8 ± 2.1 | 45.6 ± 0.9 | 42.8 ± 3.1 | 60.7 ± 1 | |
| E+SA | 12.9 ± 0.9 | 13.8 ± 0.7 | 13.6 ± 0.8 | 13.5 ± 1.4 | 20.6 ± 1 | |
| Data Collection | |
| Wavelength (Å) | 1.54179 |
| Resolution range (Å) | 30.00–2.00 (2.07–2.00) a |
| Space group | I23 |
| Cell parameters | |
| a = b = c (Å) | 196.54 |
| α = β = γ (◦) | 90.0 |
| Collection temperature (K) | 120 |
| Total reflections | 1,789,093 (182,322) |
| Unique reflections | 84,698 (8403) |
| Rint (%) | 26.2 (74.2) |
| Multiplicity | 21.1 (21.7) |
| Completeness (%) | 99.9 (99.9) |
| Mean I/sigma(I) | 8.17 (1.51) |
| Wilson B-factor (Å2) | 21.1 |
| CC1/2 | 0.99 (0.88) |
| Refinement | |
| Resolution range | 25.81–2.05 (2.08–2.05) |
| Reflections used in refinement | 74,749 (2645) |
| Reflections used for R-free | 3793 (145) |
| R-work, % | 20.91 (28.18) |
| R-free, % | 25.83 (36.57) |
| RMSD bond lengths (Å) | 0.007 |
| RMSD bond angles (◦) | 0.843 |
| Ramachandran favored (%) | 98.08 |
| Ramachandran allowed (%) | 1.92 |
| Ramachandran outliers (%) | 0.00 |
| Average B-factor (Å2) | 22.80 |
| Macromolecules | 22.72 |
| Ligands | 27.75 |
| Solvent | 23.62 |
| PDB ID | 9IX9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trubitsina, L.; Trubitsin, I.; Gabdulkhakov, A.; Abdullatypov, A.; Larionova, A.; Petrakova, M.; Trubitsyn, V.; Egorov, K.; Tishchenko, S.; Leontievsky, A.; et al. Site-Directed Mutagenesis of Two-Domain Laccase ScaSL for Obtaining a Biocatalyst with Improved Characteristics. Catalysts 2024, 14, 694. https://doi.org/10.3390/catal14100694
Trubitsina L, Trubitsin I, Gabdulkhakov A, Abdullatypov A, Larionova A, Petrakova M, Trubitsyn V, Egorov K, Tishchenko S, Leontievsky A, et al. Site-Directed Mutagenesis of Two-Domain Laccase ScaSL for Obtaining a Biocatalyst with Improved Characteristics. Catalysts. 2024; 14(10):694. https://doi.org/10.3390/catal14100694
Chicago/Turabian StyleTrubitsina, Liubov, Ivan Trubitsin, Azat Gabdulkhakov, Azat Abdullatypov, Anna Larionova, Marina Petrakova, Vladimir Trubitsyn, Konstantin Egorov, Svetlana Tishchenko, Alexey Leontievsky, and et al. 2024. "Site-Directed Mutagenesis of Two-Domain Laccase ScaSL for Obtaining a Biocatalyst with Improved Characteristics" Catalysts 14, no. 10: 694. https://doi.org/10.3390/catal14100694
APA StyleTrubitsina, L., Trubitsin, I., Gabdulkhakov, A., Abdullatypov, A., Larionova, A., Petrakova, M., Trubitsyn, V., Egorov, K., Tishchenko, S., Leontievsky, A., Alferov, S., & Ponamoreva, O. (2024). Site-Directed Mutagenesis of Two-Domain Laccase ScaSL for Obtaining a Biocatalyst with Improved Characteristics. Catalysts, 14(10), 694. https://doi.org/10.3390/catal14100694








