Tuning Almond Lipase Features by Using Different Immobilization Supports
Abstract
:1. Introduction
2. Results
2.1. Immobilization of Lipase from Almond on Different Supports
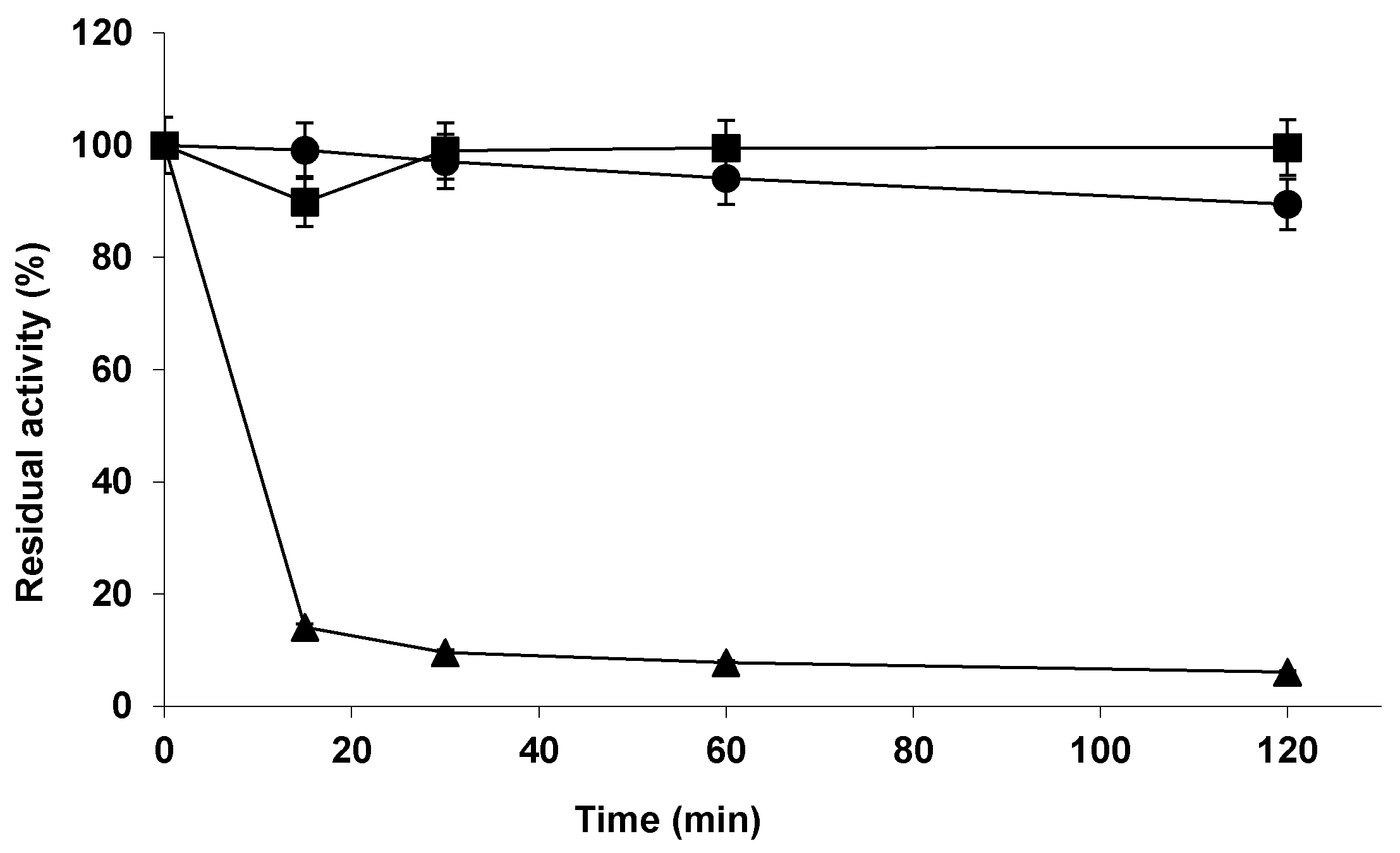
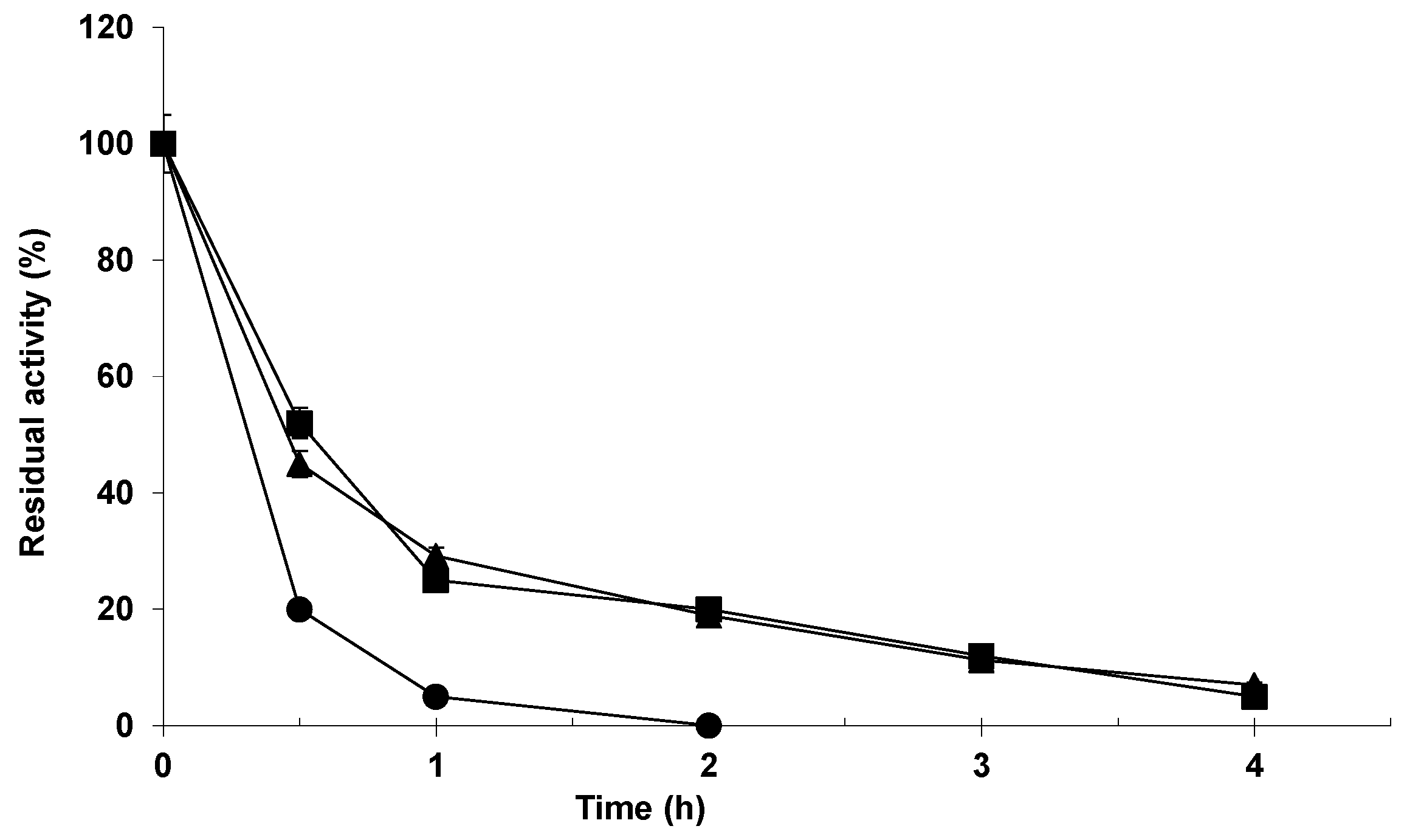
2.2. Stability of the Different Biocatalysts of Lipase from Almonds
2.3. Activity of the Different Almond Lipase Biocatalysts versus Different Substrates under Different Experimental Conditions
3. Material and Methods
3.1. Materials
3.2. Methods
3.2.1. Preparation of Lipase Extract
3.2.2. Determination of the Enzyme Activity Using Different Substrates
Hydrolysis of p-NPB
Hydrolysis of Triacetin
R- or S-Methyl Mandelate Hydrolysis
3.2.3. Immobilization of Lipase
3.2.4. Thermal Inactivations of the Biocatalysts
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, A.; Verma, V.; Dubey, V.K.; Srivastava, A.; Garg, S.K.; Singh, V.P.; Arora, P.K. Industrial applications of fungal lipases: A review. Front. Microbiol. 2023, 14, 1142536. [Google Scholar] [CrossRef]
- Ismail, A.R.; Kashtoh, H.; Baek, K.H. Temperature-resistant and solvent-tolerant lipases as industrial biocatalysts: Biotechnological approaches and applications. Int. J. Biol. Macromol. 2021, 187, 127–142. [Google Scholar] [CrossRef]
- Ali, S.; Khan, S.A.; Hamayun, M.; Lee, I.J. The Recent Advances in the Utility of Microbial Lipases: A Review. Microorganisms 2023, 11, 510. [Google Scholar] [CrossRef]
- dos Santos, L.N.; Perna, R.F.; Vieira, A.C.; de Almeida, A.F.; Ferreira, N.R. Trends in the Use of Lipases: A Systematic Review and Bibliometric Analysis. Foods 2023, 12, 3058. [Google Scholar] [CrossRef]
- Akram, F.; Mir, A.S.; ul Haq, I.; Roohi, A. An Appraisal on Prominent Industrial and Biotechnological Applications of Bacterial Lipases. Mol. Biotechnol. 2023, 65, 521–543. [Google Scholar] [CrossRef]
- Mahfoudhi, A.; Benmabrouk, S.; Fendri, A.; Sayari, A. Fungal lipases as biocatalysts: A promising platform in several industrial biotechnology applications. Biotechnol. Bioeng. 2022, 119, 3370–3392. [Google Scholar] [CrossRef]
- Goswami, D.; Basu, J.K.; De, S. Lipase applications in oil hydrolysis with a case study on castor oil: A review. Crit. Rev. Biotechnol. 2013, 33, 81–96. [Google Scholar] [CrossRef] [PubMed]
- Murty, V.R.; Bhat, J.; Muniswaran, P.K.A. Hydrolysis of oils by using immobilized lipase enzyme: A review. Biotechnol. Bioprocess Eng. 2002, 7, 57–66. [Google Scholar] [CrossRef]
- Kumar, A.; Dhar, K.; Kanwar, S.S.; Arora, P.K. Lipase catalysis in organic solvents: Advantages and applications. Biol. Proced. Online 2016, 18, 2. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Qian, J. Lipase catalysis in ionic liquids/supercritical carbon dioxide and its applications. J. Mol. Catal. B Enzym. 2010, 66, 1–7. [Google Scholar] [CrossRef]
- Dias, A.L.B.; dos Santos, P.; Martínez, J. Supercritical CO2 technology applied to the production of flavor ester compounds through lipase-catalyzed reaction: A review. J. CO2 Util. 2018, 23, 159–178. [Google Scholar] [CrossRef]
- Šibalić, D.; Šalić, A.; Zelić, B.; Tran, N.N.; Hessel, V.; Nigam, K.D.P.; Tišma, M. Synergism of ionic liquids and lipases for lignocellulosic biomass valorization. Chem. Eng. J. 2023, 461, 142011. [Google Scholar] [CrossRef]
- Alvarez, E.; Rodriguez, J.; Villa, R.; Gomez, C.; Nieto, S.; Donaire, A.; Lozano, P. Clean enzymatic production of flavor esters in spongelike ionic liquids. ACS Sustain. Chem. Eng. 2019, 7, 13307–13314. [Google Scholar] [CrossRef]
- Lozano, P.; Villa, R.; Nieto, S.; Donaire, A.; García-Verdugo, E. Clean biocatalysis in sponge-like ionic liquids. In Biocatalysis in Green Solvents; Academic Press: Cambridge, MA, USA, 2022; pp. 155–182. [Google Scholar] [CrossRef]
- Nieto, S.; Villa, R.; Donaire, A.; Lozano, P. Ultrasound-assisted enzymatic synthesis of xylitol fatty acid esters in solvent-free conditions. Ultrason. Sonochem. 2021, 75, 105606. [Google Scholar] [CrossRef] [PubMed]
- Elgharbawy, A.A.; Riyadi, F.A.; Alam, M.Z.; Moniruzzaman, M. Ionic liquids as a potential solvent for lipase-catalysed reactions: A review. J. Mol. Liq. 2018, 251, 150–166. [Google Scholar] [CrossRef]
- Tan, J.-N.; Dou, Y. Deep eutectic solvents for biocatalytic transformations: Focused lipase-catalyzed organic reactions. Appl. Microbiol. Biotechnol. 2020, 104, 1481–1496. [Google Scholar] [CrossRef] [PubMed]
- Durand, E.; Lecomte, J.; Villeneuve, P. Deep eutectic solvents: Synthesis, application, and focus on lipase-catalyzed reactions. Eur. J. Lipid Sci. Technol. 2013, 115, 379–385. [Google Scholar] [CrossRef]
- Sousa, R.R.; Silva, A.S.A.; Fernandez-Lafuente, R.; Ferreira-Leitão, V.S. Solvent-free esterifications mediated by immobilized lipases: A review from thermodynamic and kinetic perspectives. Catal. Sci. Technol. 2021, 11, 5696–5711. [Google Scholar] [CrossRef]
- Liu, Y.Q.; WeiZhuo, X.; Wei, X. A review on lipase-catalyzed synthesis of geranyl esters as flavor additives for food, pharmaceutical and cosmetic applications. Food Chem. Adv. 2022, 1, 100052. [Google Scholar] [CrossRef]
- Nimkande, V.D.; Bafana, A. A review on the utility of microbial lipases in wastewater treatment. J. Water Process Eng. 2022, 46, 102591. [Google Scholar] [CrossRef]
- Salgado, C.A.; dos Santos, C.I.A.; Vanetti, M.C.D. Microbial lipases: Propitious biocatalysts for the food industry. Food Biosci. 2022, 45, 101509. [Google Scholar] [CrossRef]
- Cheng, W.; Nian, B. Computer-Aided Lipase Engineering for Improving Their Stability and Activity in the Food Industry: State of the Art. Molecules 2023, 28, 5848. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.d.S.; de Souza, A.H.; Fraga, J.L.; Villeneuve, P.; Torres, A.G.; Amaral, P.F.F. Lipases as Effective Green Biocatalysts for Phytosterol Esters’ Production: A Review. Catalysts 2022, 12, 88. [Google Scholar] [CrossRef]
- Moya Joëlle Carole, A.; Konan Edmond, K.; Abollé, A.; Esaie Kouadio Appiah, K.; Kouassi Benjamin, Y. Transesterification of vegetable oils into biodiesel by an immobilized lipase: A review. Biofuels 2023, 14, 1087–1101. [Google Scholar] [CrossRef]
- Monteiro, R.R.C.; Arana-Peña, S.; da Rocha, T.N.; Miranda, L.P.; Berenguer-Murcia, Á.; Tardioli, P.W.; dos Santos, J.C.S.; Fernandez-Lafuente, R. Liquid lipase preparations designed for industrial production of biodiesel. Is it really an optimal solution? Renew. Energy 2021, 164, 1566–1587. [Google Scholar] [CrossRef]
- Alvarez, E.; Villa, R.; Nieto, S.; Donaire, A.; García-Verdugo, E.; Luis, S.V.; Lozano, P. The Suitability of Lipases for the Synthesis of Bioactive Compounds with Cosmeceutical Applications. Mini-Rev. Org. Chem. 2020, 18, 515–528. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Q.; Yu, X.; Liang, J.; Zhang, Y.; Jiang, Y.; Su, W. Application of Lipase B from Candida antarctica in the Pharmaceutical Industry. Ind. Eng. Chem. Res. 2023, 62, 15733–15751. [Google Scholar] [CrossRef]
- Pinto, G.B.; Mendes, F.M.L.; Antunes, A.M.d.S. Technological Profile of Lipases in the Pharmaceutical Industry. Mini-Rev. Org. Chem. 2019, 17, 701–716. [Google Scholar] [CrossRef]
- Verma, S.; Choudhary, R.N.; Kanadje, A.P.; Banerjee, U.C. Diversifying arena of drug synthesis: In the realm of lipase mediated waves of biocatalysis. Catalysts 2021, 11, 1328. [Google Scholar] [CrossRef]
- Patti, A.; Sanfilippo, C. Stereoselective Promiscuous Reactions Catalyzed by Lipases. Int. J. Mol. Sci. 2022, 23, 2675. [Google Scholar] [CrossRef]
- Budhiraja, M.; Ali, A.; Tyagi, V. Recent Advances in Lipase Catalyzed Multicomponent Reactions to Synthesize N-Heterocycles. Asian J. Org. Chem. 2023, 12, e202300427. [Google Scholar] [CrossRef]
- Monteiro, R.R.C.; Berenguer-Murcia, Á.; Rocha-Martin, J.; Vieira, R.S.; Fernandez-Lafuente, R. Biocatalytic production of biolubricants: Strategies, problems and future trends. Biotechnol. Adv. 2023, 68, 108215. [Google Scholar] [CrossRef]
- Bolina, I.C.A.; Gomes, R.A.B.; Mendes, A.A. Biolubricant Production from Several Oleaginous Feedstocks Using Lipases as Catalysts: Current Scenario and Future Perspectives. Bioenergy Res. 2021, 14, 1039–1057. [Google Scholar] [CrossRef]
- Vilas Bôas, R.N.; Castro, H.F. A review of synthesis of esters with aromatic, emulsifying, and lubricant properties by biotransformation using lipases. Biotechnol. Bioeng. 2022, 119, 725–742. [Google Scholar] [CrossRef] [PubMed]
- Serra, S.; De Simeis, D. Stereoselective Synthesis of Terpenoids through Lipase-Mediated Resolution Approaches. Catalysts 2020, 10, 504. [Google Scholar] [CrossRef]
- Zou, X.; Su, H.; Zhang, F.; Zhang, H.; Yeerbolati, Y.; Xu, X.; Chao, Z.; Zheng, L.; Jiang, B. Bioimprinted lipase-catalyzed synthesis of medium- and long-chain structured lipids rich in docosahexaenoic acid for infant formula. Food Chem. 2023, 424, 136450. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, T.; Kuroiwa, T.; Wakahara, K.; Ozono, S. Accelerated acidolysis for triglyceride modification using commercial lipases activated by a hydration–aggregation pretreatment. Process Biochem. 2023, 132, 130–139. [Google Scholar] [CrossRef]
- Utama, Q.D.; Sitanggang, A.B.; Adawiyah, D.R.; Hariyadi, P. Lipase-catalyzed interesterification for the synthesis of medium-long-medium (MLM) structured lipids. Food Technol. Biotechnol. 2019, 57, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Soumanou, M.M.; Pérignon, M.; Villeneuve, P. Lipase-catalyzed interesterification reactions for human milk fat substitutes production: A review. Eur. J. Lipid Sci. Technol. 2013, 115, 270–285. [Google Scholar] [CrossRef]
- Stergiou, P.Y.; Foukis, A.; Filippou, M.; Koukouritaki, M.; Parapouli, M.; Theodorou, L.G.; Hatziloukas, E.; Afendra, A.; Pandey, A.; Papamichael, E.M. Advances in lipase-catalyzed esterification reactions. Biotechnol. Adv. 2013, 31, 1846–1859. [Google Scholar] [CrossRef]
- Cha, H.J.; Park, J.B.; Park, S. Esterification of secondary alcohols and multi-hydroxyl compounds by Candida antarctica lipase B and subtilisin. Biotechnol. Bioprocess Eng. 2019, 24, 41–47. [Google Scholar] [CrossRef]
- Toldrá-Reig, F.; Mora, L.; Toldrá, F. Developments in the use of lipase transesterification for biodiesel production from animal fat waste. Appl. Sci. 2020, 10, 5085. [Google Scholar] [CrossRef]
- Gotor, V. Non-conventional hydrolase chemistry: Amide and carbamate bond formation catalyzed by lipases. Bioorg. Med. Chem. 1999, 7, 2189–2197. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Nie, K.; Wang, F.; Deng, L. Optimization of the Lipase-Catalyzed Selective Amidation of Phenylglycinol. Front. Bioeng. Biotechnol. 2020, 7, 486. [Google Scholar] [CrossRef] [PubMed]
- Nian, B.; Liao, G.; Song, Y.; Su, Y.; Cao, C.; Liu, Y. Ionic hydrogen-bonding interaction controlled electrophilicity and nucleophilicity: Mechanistic insights into the synergistic catalytic effect of lipase and natural deep eutectic solvents in amidation reaction. J. Catal. 2020, 384, 159–168. [Google Scholar] [CrossRef]
- Kapoor, M.; Gupta, M.N. Lipase promiscuity and its biochemical applications. Process Biochem. 2012, 47, 555–569. [Google Scholar] [CrossRef]
- Dwivedee, B.P.; Soni, S.; Sharma, M.; Bhaumik, J.; Laha, J.K.; Banerjee, U.C. Promiscuity of lipase-catalyzed reactions for organic synthesis: A recent update. ChemistrySelect 2018, 3, 2441–2466. [Google Scholar] [CrossRef]
- Vivek, K.; Sandhia, G.S.; Subramaniyan, S. Extremophilic lipases for industrial applications: A general review. Biotechnol. Adv. 2022, 60, 108002. [Google Scholar] [CrossRef]
- Mhetras, N.; Mapare, V.; Gokhale, D. Cold active lipases: Biocatalytic tools for greener technology. Appl. Biochem. Biotechnol. 2021, 193, 2245–2266. [Google Scholar] [CrossRef]
- Schmid, R.D.; Verger, R. Lipases: Interfacial enzymes with attractive applications. Angew. Chem. 1998, 37, 1608–1633. [Google Scholar] [CrossRef]
- Verger, R. ‘Interfacial activation’ of lipases: Facts and artifacts. Trends Biotechnol. 1997, 15, 32–38. [Google Scholar] [CrossRef]
- Martinelle, M.; Holmquist, M.; Hult, K. On the interfacial activation of Candida antarctica lipase A and B as compared with Humicola lanuginosa lipase. Biochim. Biophys. Acta Lipids Lipid Metab. 1995, 1258, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Grochulski, P.; Li, Y.; Schrag, J.D.; Bouthillier, F.; Smith, P.; Harrison, D.; Rubin, B.; Cygler, M. Insights into interfacial activation from an open structure of Candida rugosa lipase. J. Biol. Chem. 1993, 268, 12843–12847. [Google Scholar] [CrossRef] [PubMed]
- Brzozowski, A.M.; Derewenda, U.; Derewenda, Z.S.; Dodson, G.G.; Lawson, D.M.; Turkenburg, J.P.; Bjorkling, F.; Huge-Jensen, B.; Patkar, S.A.; Thim, L. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature 1991, 351, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Jiang, T.; Wu, Y.; Cui, L.; Qin, S.; He, B. Elucidation of lid open and orientation of lipase activated in interfacial activation by amphiphilic environment. Int. J. Biol. Macromol. 2018, 119, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Angkawidjaja, C.; Matsumura, H.; Koga, Y.; Takano, K.; Kanaya, S. X-ray crystallographic and MD simulation studies on the mechanism of interfacial activation of a family I.3 lipase with two lids. J. Mol. Biol. 2010, 400, 82–95. [Google Scholar] [CrossRef]
- Wang, S.; Xu, Y.; Yu, X.W. A phenylalanine dynamic switch controls the interfacial activation of Rhizopus chinensis lipase. Int. J. Biol. Macromol. 2021, 173, 1–12. [Google Scholar] [CrossRef]
- Palomo, J.M.; Fuentes, M.; Fernández-Lorente, G.; Mateo, C.; Guisan, J.M.; Fernández-Lafuente, R. General trend of lipase to self-assemble giving bimolecular aggregates greatly modifies the enzyme functionality. Biomacromolecules 2003, 4, 1–6. [Google Scholar] [CrossRef]
- Fernández-Lorente, G.; Palomo, J.M.; Fuentes, M.; Mateo, C.; Guisán, J.M.; Fernández-Lafuente, R. Self-assembly of Pseudomonas fluorescens lipase into bimolecular aggregates dramatically affects functional properties. Biotechnol. Bioeng. 2003, 82, 232–237. [Google Scholar] [CrossRef]
- Palomo, J.M.; Peñas, M.M.; Fernández-Lorente, G.; Mateo, C.; Pisabarro, A.G.; Fernández-Lafuente, R.; Ramírez, L.; Guisán, J.M. Solid-phase handling of hydrophobins: Immobilized hydrophobins as a new tool to study lipases. Biomacromolecules 2003, 4, 204–210. [Google Scholar] [CrossRef]
- Uppenberg, J.; Hansen, M.T.; Patkar, S.; Jones, T.A. The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica. Structure 1994, 2, 293–308. [Google Scholar] [CrossRef]
- Stauch, B.; Fisher, S.J.; Cianci, M. Open and closed states of Candida antarctica lipase B: Protonation and the mechanism of interfacial activation. J. Lipid Res. 2015, 56, 2348–2358. [Google Scholar] [CrossRef]
- Zisis, T.; Freddolino, P.L.; Turunen, P.; Van Teeseling, M.C.F.; Rowan, A.E.; Blank, K.G. Interfacial Activation of Candida antarctica Lipase B: Combined Evidence from Experiment and Simulation. Biochemistry 2015, 54, 5969–5979. [Google Scholar] [CrossRef] [PubMed]
- Foresti, M.L.; Ferreira, M.L. Computational Approach to Solvent-Free Synthesis of Ethyl Oleate Using Candida rugosa Candida antarctica B lipases. I. Interfacial Activation and Substrate (Ethanol, Oleic Acid) Adsorption. Biomacromolecules 2004, 5, 2366–2375. [Google Scholar] [CrossRef] [PubMed]
- Kublicki, M.; Koszelewski, D.; Brodzka, A.; Ostaszewski, R. Wheat germ lipase: Isolation, purification and applications. Crit. Rev. Biotechnol. 2022, 42, 184–200. [Google Scholar] [CrossRef]
- Kumar, R.R.; Bhargava, D.V.; Pandit, K.; Goswami, S.; Mukesh Shankar, S.; Singh, S.P.; Rai, G.K.; Satyavathi, C.T.; Praveen, S. Lipase—The fascinating dynamics of enzyme in seed storage and germination—A real challenge to pearl millet. Food Chem. 2021, 361, 130031. [Google Scholar] [CrossRef] [PubMed]
- Barros, M.; Fleuri, L.F.; MacEdo, G.A. Seed lipases: Sources, applications and properties—A review. Braz. J. Chem. Eng. 2010, 27, 15–29. [Google Scholar] [CrossRef]
- Jung, H.; Moon, S.J. Purification, distribution, and characterization activity of lipase from oat seeds (Avena sativa L.). J. Korean Soc. Appl. Biol. Chem. 2013, 56, 639–645. [Google Scholar] [CrossRef]
- Polizelli, P.P.; Facchini, F.D.A.; Cabral, H.; Bonilla-Rodriguez, G.O. A new lipase isolated from oleaginous seeds from Pachira aquatica (Bombacaceae). Appl. Biochem. Biotechnol. 2008, 150, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Nanssou Kouteu, P.A.; Baréa, B.; Barouh, N.; Blin, J.; Villeneuve, P. Lipase Activity of Tropical Oilseed Plants for Ethyl Biodiesel Synthesis and Their Typo- and Regioselectivity. J. Agric. Food Chem. 2016, 64, 8838–8847. [Google Scholar] [CrossRef]
- Moussavou, M.R.W.; Brunschwig, C.; Baréa, B.; Villeneuve, P.; Blin, J. Assessing the Enzyme Activity of Different Plant Extracts of Biomasses from Sub-Saharan Africa for Ethyl Biodiesel Production. Energy Fuels 2016, 30, 2356–2364. [Google Scholar] [CrossRef]
- Seth, S.; Chakravorty, D.; Dubey, V.K.; Patra, S. An insight into plant lipase research—Challenges encountered. Protein Expr. Purif. 2014, 95, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Jorge, N.; Luzia, D.M.M. Characterization of seed oil Pachira aquatica Aublet for food utilization. Acta Amaz. 2012, 42, 149–156. [Google Scholar] [CrossRef]
- Yeşiloǧlu, Y.; Başkurt, L. Partial Purification and Characterization of Almond Seed Lipase. Prep. Biochem. Biotechnol. 2008, 38, 397–410. [Google Scholar] [CrossRef]
- Sheldon, R.A.; van Pelt, S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef]
- Di Cosimo, R.; Mc Auliffe, J.; Poulose, A.J.; Bohlmann, G. Industrial use of immobilized enzymes. Chem. Soc. Rev. 2013, 42, 6437–6474. [Google Scholar] [CrossRef]
- Liese, A.; Hilterhaus, L. Evaluation of immobilized enzymes for industrial applications. Chem. Soc. Rev. 2013, 42, 6236–6249. [Google Scholar] [CrossRef] [PubMed]
- Wancura, J.H.C.; Tres, M.V.; Jahn, S.L.; Oliveira, J.V. Lipases in liquid formulation for biodiesel production: Current status and challenges. Biotechnol. Appl. Biochem. 2019, 67, 648–667. [Google Scholar] [CrossRef]
- Kim, B.H.; Hwang, J.; Akoh, C.C. Liquid microbial lipase—Recent applications and expanded use through immobilization. Curr. Opin. Food Sci. 2023, 50, 100987. [Google Scholar] [CrossRef]
- Bhatt, C.; Nielsen, P.M.; Rancke-Madsen, A.; Woodley, J.M. Combining technology with liquid-formulated lipases for in-spec biodiesel production. Biotechnol. Appl. Biochem. 2022, 69, 7–19. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Guzik, U.; Hupert-Kocurek, K.; Wojcieszyńska, D. Immobilization as a strategy for improving enzyme properties—Application to oxidoreductases. Molecules 2014, 19, 8995–9018. [Google Scholar] [CrossRef]
- Boudrant, J.; Woodley, J.M.; Fernandez-Lafuente, R. Parameters necessary to define an immobilized enzyme preparation. Process Biochem. 2020, 90, 66–80. [Google Scholar] [CrossRef]
- Klibanov, A.M. Enzyme stabilization by immobilization. Anal. Biochem. 1979, 93, 1–25. [Google Scholar] [CrossRef]
- Iyer, P.V.; Ananthanarayan, L. Enzyme stability and stabilization—Aqueous and non-aqueous environment. Process Biochem. 2008, 43, 1019–1032. [Google Scholar] [CrossRef]
- Hwang, E.T.; Gu, M.B. Enzyme stabilization by nano/microsized hybrid materials. Eng. Life Sci. 2013, 13, 49–61. [Google Scholar] [CrossRef]
- Fernandez-Lafuente, R. Stabilization of multimeric enzymes: Strategies to prevent subunit dissociation. Enzym. Microb. Technol. 2009, 45, 405–418. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Berenguer-Murcia, Á.; Carballares, D.; Morellon-Sterling, R.; Fernandez-Lafuente, R. Stabilization of enzymes via immobilization: Multipoint covalent attachment and other stabilization strategies. Biotechnol. Adv. 2021, 52, 107821. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Strategies for the one-step immobilization–purification of enzymes as industrial biocatalysts. Biotechnol. Adv. 2015, 33, 435–456. [Google Scholar] [CrossRef]
- Chen, N.; Zhang, C.; Liu, Y.; Dong, X.; Sun, Y. Cysteine-modified poly(glycidyl methacrylate) grafted onto silica nanoparticles: New supports for significantly enhanced performance of immobilized lipase. Biochem. Eng. J. 2019, 145, 137–144. [Google Scholar] [CrossRef]
- de Morais Júnior, W.G.; Moura Maia, A.; Alves Martins, P.; Fernández-Lorente, G.; Guisán, J.M.; Pessela, B.C. Influence of different immobilization techniques to improve the enantioselectivity of lipase from Geotrichum candidum applied on the resolution of mandelic acid. Mol. Catal. 2018, 458, 89–96. [Google Scholar] [CrossRef]
- Moure, V.R.; Fabrício, C.; Frensch, G.; Marques, F.A.; Mitchell, D.A.; Krieger, N. Enhancing the enantioselectivity of the lipase from Burkholderia cepacia LTEB11 towards the resolution of secondary allylic alcohols. Biocatal. Agric. Biotechnol. 2014, 3, 146–153. [Google Scholar] [CrossRef]
- Ghattas, N.; Filice, M.; Abidi, F.; Guisan, J.M.; Ben Salah, A. Purification and improvement of the functional properties of Rhizopus oryzae lipase using immobilization techniques. J. Mol. Catal. B Enzym. 2014, 110, 111–116. [Google Scholar] [CrossRef]
- Naya, M.; Imai, M. Regulation of the hydrolysis reactivity of immobilized Candida rugosa lipase with the aid of a hydrophobic porous carrier. Asia-Pac. J. Chem. Eng. 2012, 7 (Suppl. S1), S157–S165. [Google Scholar] [CrossRef]
- Uyanik, A.; Sen, N.; Yilmaz, M. Improvement of catalytic activity of lipase from Candida rugosa via sol-gel encapsulation in the presence of calix(aza)crown. Bioresour. Technol. 2011, 102, 4313–4318. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Zhao, B.; Li, C.; Wang, P.; Wang, Z.; Tang, J.; Wang, L. Improvement of the enantioselectivity and activity of lipase from Pseudomonas sp. via adsorption on a hydrophobic support: Kinetic resolution of 2-octanol. Biocatal. Biotransform. 2009, 27, 340–347. [Google Scholar] [CrossRef]
- Volpato, G.; Filice, M.; Rodrigues, R.C.; Heck, J.X.; Guisan, J.M.; Mateo, C.; Ayub, M.A.Z. Modulation of a lipase from Staphylococcus warneri EX17 using immobilization techniques. J. Mol. Catal. B Enzym. 2009, 60, 125–132. [Google Scholar] [CrossRef]
- Yang, G.; Wu, J.; Xu, G.; Yang, L. Enhancement of the activity and enantioselectivity of lipase in organic systems by immobilization onto low-cost support. J. Mol. Catal. B Enzym. 2009, 57, 96–103. [Google Scholar] [CrossRef]
- Wang, P.Y.; Tsai, S.W.; Chen, T.L. Improvement of enantioselectivity and stability of Klebsiella oxytoca hydrolase immobilized on Eupergit C 250L. J. Chem. Technol. Biotechnol. 2008, 83, 1518–1525. [Google Scholar] [CrossRef]
- Takaç, S.; Bakkal, M. Impressive effect of immobilization conditions on the catalytic activity and enantioselectivity of Candida rugosa lipase toward S-Naproxen production. Process Biochem. 2007, 42, 1021–1027. [Google Scholar] [CrossRef]
- Chaubey, A.; Parshad, R.; Koul, S.; Taneja, S.C.; Qazi, G.N. Enantioselectivity modulation through immobilization of Arthrobacter sp. lipase: Kinetic resolution of fluoxetine intermediate. J. Mol. Catal. B Enzym. 2006, 42, 39–44. [Google Scholar] [CrossRef]
- Sabbani, S.; Hedenström, E.; Nordin, O. The enantioselectivity of Candida rugosa lipase is influenced by the particle size of the immobilising support material Accurel. J. Mol. Catal. B Enzym. 2006, 42, 1–9. [Google Scholar] [CrossRef]
- Yu, H.; Wu, J.; Chi, B.C. Enhanced activity and enantioselectivity of Candida rugosa lipase immobilized on macroporous adsorptive resins for ibuprofen resolution. Biotechnol. Lett. 2004, 26, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Bolivar, J.M.; Woodley, J.M.; Fernandez-Lafuente, R. Is enzyme immobilization a mature discipline? Some critical considerations to capitalize on the benefits of immobilization. Chem. Soc. Rev. 2022, 51, 6251–6290. [Google Scholar] [CrossRef] [PubMed]
- Jesionowski, T.; Zdarta, J.; Krajewska, B. Enzyme immobilization by adsorption: A review. Adsorption 2014, 20, 801–821. [Google Scholar] [CrossRef]
- Pessela, B.C.C.; Munilla, R.; Betancor, L.; Fuentes, M.; Carrascosa, A.V.; Vian, A.; Fernández-Lafuente, R.; Guisán, J.M. Ion exchange using poorly activated supports, an easy way for purification of large proteins. J. Chromatogr. A 2004, 1034, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Manoel, E.A.; dos Santos, J.C.S.; Freire, D.M.G.; Rueda, N.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports involves the open form of the enzyme. Enzym. Microb. Technol. 2015, 71, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.C.; Virgen-Ortíz, J.J.; dos Santos, J.C.S.; Berenguer-Murcia, Á.; Alcantara, A.R.; Barbosa, O.; Ortiz, C.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports: Immobilization mechanism, advantages, problems, and solutions. Biotechnol. Adv. 2019, 37, 746–770. [Google Scholar] [CrossRef]
- Kornecki, J.F.; Carballares, D.; Morellon-Sterling, R.; Siar, E.H.; Kashefi, S.; Chafiaa, M.; Arana-Peña, S.; Rios, N.S.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Influence of phosphate anions on the stability of immobilized enzymes. Effect of enzyme nature, immobilization protocol and inactivation conditions. Process Biochem. 2020, 95, 288–296. [Google Scholar] [CrossRef]
- Abellanas-Perez, P.; Carballares, D.; Fernandez-Lafuente, R.; Rocha-Martin, J. Glutaraldehyde modification of lipases immobilized on octyl agarose beads: Roles of the support enzyme loading and chemical amination of the enzyme on the final enzyme features. Int. J. Biol. Macromol. 2023, 248, 125853. [Google Scholar] [CrossRef]
- Fernandez-Lopez, L.; Bartolome-Cabrero, R.; Rodriguez, M.D.; Dos Santos, C.S.; Rueda, N.; Fernandez-Lafuente, R. Stabilizing effects of cations on lipases depend on the immobilization protocol. RSC Adv. 2015, 5, 83868–83875. [Google Scholar] [CrossRef]
- Arana-Peña, S.; Rios, N.S.; Carballares, D.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Immobilization of lipases via interfacial activation on hydrophobic supports: Production of biocatalysts libraries by altering the immobilization conditions. Catal. Today 2021, 362, 130–140. [Google Scholar] [CrossRef]
- Tacias-Pascacio, V.G.; Peirce, S.; Torrestiana-Sanchez, B.; Yates, M.; Rosales-Quintero, A.; Virgen-Ortíz, J.J.; Fernandez-Lafuente, R. Evaluation of different commercial hydrophobic supports for the immobilization of lipases: Tuning their stability, activity and specificity. RSC Adv. 2016, 6, 100281–100294. [Google Scholar] [CrossRef]
- Cygler, M.; Schrag, J.D. Structure and conformational flexibility of Candida rugosa lipase. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1999, 1441, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, K.-E.; Ransac, S.; Koch, H.B.; Ferrato, F.; Dijkstra, B.W. Topological characterization and modeling of the 3D structure of lipase from Pseudomonas aeruginosa. FEBS Lett. 1993, 332, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.K.; Song, H.K.; Shin, D.H.; Hwang, K.Y.; Suh, S.W. The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor. Structure 1997, 5, 173–185. [Google Scholar] [CrossRef]
- Palomo, J.M.; Fernandez-Lorente, G.; Mateo, C.; Ortiz, C.; Fernandez-Lafuente, R.; Guisan, J.M. Modulation of the enantioselectivity of lipases via controlled immobilization and medium engineering: Hydrolytic resolution of mandelic acid esters. Enzym. Microb. Technol. 2002, 31, 775–783. [Google Scholar] [CrossRef]
- Guisán, J.M. Aldehyde-agarose gels as activated supports for immobilization-stabilization of enzymes. Enzym. Microb. Technol. 1988, 10, 375–382. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fuentes, M.; Betancor, L.; Grazu, V.; López-Gallego, F.; Pessela, B.C.C.; Hidalgo, A.; Fernández-Lorente, G.; Fernández-Lafuente, R.; et al. Glyoxyl agarose: A fully inert and hydrophilic support for immobilization and high stabilization of proteins. Enzym. Microb. Technol. 2006, 39, 274–280. [Google Scholar] [CrossRef]
- Fernandez-Lafuente, R.; Rosell, C.M.; Rodriguez, V.; Santana, C.; Soler, G.; Bastida, A.; Guisán, J.M. Preparation of activated supports containing low pK amino groups. A new tool for protein immobilization via the carboxyl coupling method. Enzym. Microb. Technol. 1993, 15, 546–550. [Google Scholar] [CrossRef]
- Tacias-Pascacio, V.G.; Virgen-Ortíz, J.J.; Jiménez-Pérez, M.; Yates, M.; Torrestiana-Sanchez, B.; Rosales-Quintero, A.; Fernandez-Lafuente, R. Evaluation of different lipase biocatalysts in the production of biodiesel from used cooking oil: Critical role of the immobilization support. Fuel 2017, 200, 1–10. [Google Scholar] [CrossRef]
- Bahri, S. Lipolytic activity and chilling requirement for germination of some almond cultivars. Afr. J. Biotechnol. 2012, 11, 14096–14101. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.N.P.; Fernandez-Lafuente, R.; Cowan, D.A. Purification and partial characterization of a novel thermophilic carboxylesterase with high mesophilic specific activity. Enzym. Microb. Technol. 1995, 17, 816–825. [Google Scholar] [CrossRef]
- Hernandez, K.; Garcia-Verdugo, E.; Porcar, R.; Fernandez-Lafuente, R. Hydrolysis of triacetin catalyzed by immobilized lipases: Effect of the immobilization protocol and experimental conditions on diacetin yield. Enzym. Microb. Technol. 2011, 48, 510–517. [Google Scholar] [CrossRef]

| Phosphate Buffer, pH 7 | |||
|---|---|---|---|
| Supports | Substrates | ||
| R-methyl mandelate | S-methyl mandelate | Triacetin | |
| Purolite | 4.75 ± 0.30 | 4.62 ± 0.34 | 10.81 ± 0.97 |
| MANAE | 0.16 ± 0.02 | 0.27 ± 0.02 | 0.95 ± 0.03 |
| Bicarbonate buffer, pH 9 | |||
| Purolite | 0 | 16.43 ± 0.89 | 2.09 ± 0.37 |
| MANAE | 0 | 0 | 6.33 ± 0.23 |
| Phosphate buffer, pH 9 | |||
| Purolite | 13.57 ± 0. 93 | 11.7 ± 0.34 | 31.39 ± 1.29 |
| MANAE | 0.23 ± 0.02 | 0.24 ± 0.01 | 11.7 ± 0.74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cherni, O.; Carballares, D.; Siar, E.H.; Abellanas-Perez, P.; de Andrades, D.; Rocha-Martin, J.; Bahri, S.; Fernandez-Lafuente, R. Tuning Almond Lipase Features by Using Different Immobilization Supports. Catalysts 2024, 14, 115. https://doi.org/10.3390/catal14020115
Cherni O, Carballares D, Siar EH, Abellanas-Perez P, de Andrades D, Rocha-Martin J, Bahri S, Fernandez-Lafuente R. Tuning Almond Lipase Features by Using Different Immobilization Supports. Catalysts. 2024; 14(2):115. https://doi.org/10.3390/catal14020115
Chicago/Turabian StyleCherni, Oumaima, Diego Carballares, El Hocine Siar, Pedro Abellanas-Perez, Diandra de Andrades, Javier Rocha-Martin, Sellema Bahri, and Roberto Fernandez-Lafuente. 2024. "Tuning Almond Lipase Features by Using Different Immobilization Supports" Catalysts 14, no. 2: 115. https://doi.org/10.3390/catal14020115









