Enhanced Catalytic Synthesis of Flavonoid by UV-B Radiation in Artemisia argyi
Abstract
:1. Introduction
2. Results and Discussion
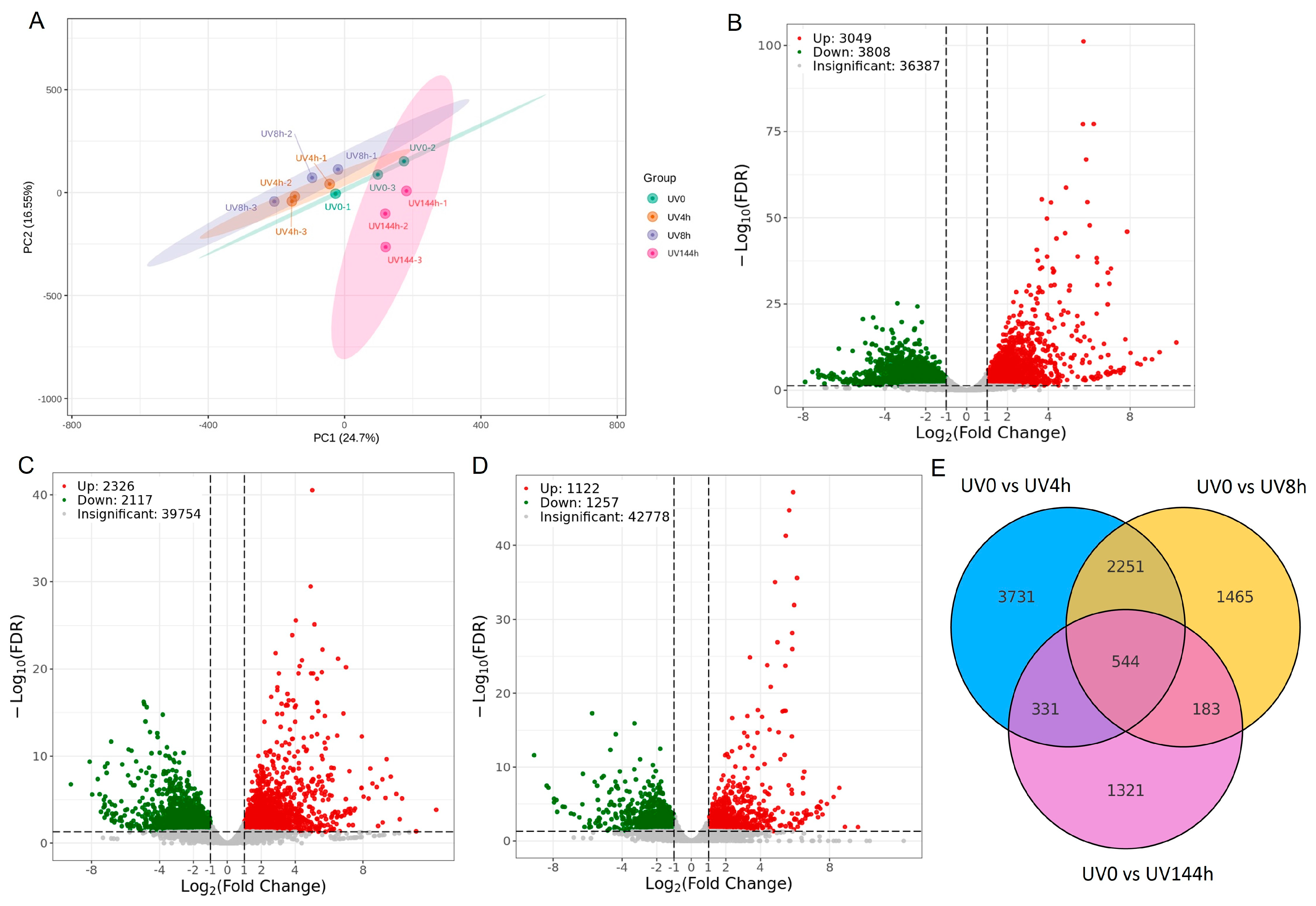
2.1. Illumina Sequencing and DEG Analysis
2.2. Metabolic Pathway Enrichment Analysis for DEGs
2.3. Metabolomic Analysis
2.4. Candidate DAMs with Potential Roles in UV-B Resistance
2.5. Integrated Analysis of Genes and Metabolites Involved in Flavonoid Biosynthesis
2.6. The Potential Regulation of Flavonoids by Salicylic Acid and Jasmonic Acid
2.7. Verification of Enhanced Flavonoid Synthesis by UV Radiation
3. Materials and Methods
3.1. Plant Material and Cultivation Method
3.2. RNA Isolation and Transcriptome Sequencing
3.3. Metabolite Extraction and Widely Targeted Metabolome Analysis
3.4. Quantitative Real-Time PCR
3.5. Quantitation of Total Flavonoids in A. argyi Leaves
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bora, K.S.; Sharma, A. The genus Artemisia: A comprehensive review. Pharm. Biol. 2011, 49, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wen, X.; He, J.; Zhao, H.; Li, S.; Wang, M. Phytochemical components and biological activities of Artemisia argyi. J. Funct. Foods 2019, 52, 648–662. [Google Scholar] [CrossRef]
- Qiao, Y.; Wu, L.; Yang, S.; Wang, Q.; Gu, H.; Wei, L.; Liu, G.; Zhou, S.; Wang, P.; Song, M. Metabolomic and transcriptomic analyses provide insights into variations in flavonoids contents between two Artemisia cultivars. BMC Plant Biol. 2023, 23, 288. [Google Scholar] [CrossRef]
- Rashid, M.U.; Alamzeb, M.; Ali, A.; Ullah, Z.; Shah, Z.A.; Naz, I.; Khan, M.R. The chemistry and pharmacology of alkaloids and allied nitrogen compounds from Artemisia species: A review. Phytother. Res. 2019, 33, 2661–2684. [Google Scholar] [CrossRef] [PubMed]
- Panahirad, S.; Morshedloo, M.R.; Ali, S.; Hano, C.; Kulak, M. Secondary metabolites and their potential roles in plant tolerance against abiotic and biotic stress. Plant Stress 2023, 10, 100292. [Google Scholar] [CrossRef]
- Nigam, M.; Atanassova, M.; Mishra, A.P.; Pezzani, R.; Devkota, H.P.; Plygun, S.; Salehi, B.; Setzer, W.N.; Sharifi-Rad, J. Bioactive compounds and health benefits of Artemisia species. Nat. Prod. Commun. 2019, 14, 1934578X19850354. [Google Scholar]
- Nabavi, S.M.; Samec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef] [PubMed]
- Tuenter, E.; Creylman, J.; Verheyen, G.; Pieters, L.; Van Miert, S. Development of a classification model for the antigenotoxic activity of flavonoids. Bioorg. Chem. 2020, 98, 103705. [Google Scholar] [CrossRef] [PubMed]
- Marone, D.; Mastrangelo, A.M.; Borrelli, G.M.; Mores, A.; Laido, G.; Russo, M.A.; Ficco, D.B.M. Specialized metabolites: Physiological and biochemical role in stress resistance, strategies to improve their accumulation, and new applications in crop breeding and management. Plant Physiol. Biochem. 2022, 172, 48–55. [Google Scholar] [CrossRef]
- DiBiase, C.; Godtfredsen, E.; Dahl, J.; Shapiro, A.; Brown, K.; Martin, A.; Wermuth, A.; Heschel, M.S. Maternal flower color, ultraviolet protection, and germination in Ipomopsis aggregate (Polemoniaceae). Popul. Ecol. 2022, 64, 176–187. [Google Scholar] [CrossRef]
- Wang, F.; Xu, Z.; Fan, X.; Zhou, Q.; Cao, J.; Ji, G.; Jing, S.; Feng, B.; Wang, T. Transcriptome analysis reveals complex molecular mechanisms underlying UV tolerance of wheat (Triticum aestivum, L.). J. Agric. Food Chem. 2019, 67, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Gómez, J.D.; Vital, C.E.; Oliveira, M.G.A.; Ramos, H.J.O. Broad range flavonoid profiling by LC/MS of soybean genotypes contrasting for resistance to Anticarsia gemmatalis (Lepidoptera: Noctuidae). PLoS ONE 2018, 13, e0205010. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chen, Q.; Bu, Y.; Luo, R.; Hao, S.; Zhang, J.; Tian, J.; Yao, Y. Flavonoid accumulation plays an important role in the rust resistance of Malus plant leaves. Front. Plant Sci. 2017, 8, 1286. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Xu, X.; Li, Y.; Wang, Y.; Li, M.; Wang, Y.; Ding, X.; Chu, Z. Rutin-mediated priming of plant resistance to three bacterial pathogens initiating the early SA signal pathway. PLoS ONE 2016, 11, e0146910. [Google Scholar] [CrossRef] [PubMed]
- Marhava, P. Recent developments in the understanding of PIN polarity. New Phytologist. 2022, 233, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Kacprzyk, J.; Burke, R.; Schwarze, J.; McCabe, P.F. Plant programmed cell death meets auxin signaling. FEBS J. 2022, 289, 1731–1745. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, J.; Zhang, G.; Ding, W.; Duan, L.; Yang, J.; Kui, L.; Cheng, X.; Ruan, J.; Fan, W.; et al. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat. Commun. 2018, 9, 448. [Google Scholar] [CrossRef]
- Waki, T.; Takahashi, S.; Nakayama, T. Managing enzyme promiscuity in plant specialized metabolism: A lesson from flavonoid biosynthesis. BioEssays 2021, 43, 2000164. [Google Scholar] [CrossRef]
- Zhang, F.; Ma, Z.; Qiao, Y.; Wang, Z.; Chen, W.; Zheng, S.; Yu, C.; Song, L.; Lou, H.; Wu, J. Transcriptome sequencing and metabolomics analyses provide insights into the flavonoid biosynthesis in Torreya grandis kernels. Food Chem. 2022, 16, 131558. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, F.; Lian, Y.; Xiao, H.; Zheng, J. Biosynthesis of citrus flavonoids and their health effects. Crit. Rev. Food Sci. Nutr. 2020, 60, 566–583. [Google Scholar] [CrossRef]
- Gu, N.; Liu, S.M.; Qiu, C.; Zhao, L.G.; Pei, J.J. Biosynthesis of 3′-O-methylisoorientin from luteolin by selecting O-methylation/C-glycosylation motif. Enzyme Microb. Technol. 2021, 150, 109862. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.B.; Liu, S.K.; Gao, S.; Zhou, J.W. Identification and characterization of three flavonoid 3-O-glycosyltransferases from Epimedium koreanum Nakai. Biochem. Eng. J. 2020, 163, 107759. [Google Scholar] [CrossRef]
- Jan, R.; Asaf, S.; Paudel, S.; Lubna; Lee, S.; Kim, K.M. Discovery and validation of a novel step catalyzed by OsF3H in the flavonoid biosynthesis pathway. Biology 2021, 10, 32. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.X.; Gao, Y.; Han, M.L.; Liu, P.Y.; Yang, C.; Shen, T.; Li, H.H. In vitro anthocyanin induction and metabolite analysis in Malus spectabilis leaves under low nitrogen conditions. Hortic. Plant J. 2020, 6, 284–292. [Google Scholar] [CrossRef]
- William, R.; Chezem, A.M.; Li, F.S.; Weng, J.K.; Nicole, K.C. SG2-Type R2R3-MYB transcription factor MYB15 controls defense-induced lignification and basal immunity in Arabidopsis. Plant Cell. 2017, 17, 1907–1926. [Google Scholar]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Neugart, S.; Krumbein, A.; Zrenner, R. Influence of light and temperature on gene expression leading to accumulation of specific flavonol glycosides and hydroxycinnamic acid derivatives in kale (Brassica oleracea var. sabellica). Front. Plant Sci. 2016, 7, 326. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.L.; Yang, L.; Yang, X.; Zhang, T.; Lan, Y.M.; Zhao, Y.; Han, M.; Yang, L.M. Drought stress induces biosynthesis of flavonoids in leaves and saikosaponins in roots of Bupleurum chinense DC. Phytochemistry 2020, 177, 112434. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.L.; Li, Y.; Tian, W.; Sun, Y.; Chen, F.; Zhang, Y.; Zhai, Y.; Zhang, J.; Su, H.; Wang, L. A novel dark septate fungal endophyte positively affected blueberry growth and changed the expression of plant genes involved in phytohormone and flavonoid biosynthesis. Tree Physiol. 2020, 40, 1080–1094. [Google Scholar] [CrossRef]
- Chen, H.; Guo, M.; Dong, S.; Wu, X.; Zhang, G.; He, L.; Jiao, Y.; Chen, S.; Li, L.; Luo, H. A chromosome-scale genome assembly of Artemisia argyi reveals unbiased subgenome evolution and key contributions of gene duplication to volatile terpenoid diversity. Plant Commun. 2023, 4, 100516. [Google Scholar] [CrossRef]
- Saradhi, P.P.; Alia, A.S.; Prasad, K.V. Proline accumulates in plants exposed to UV radiation and protects them against UV induced peroxidation. Biochem. Biophys. Res. Commun. 1995, 209, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Huang, J.; Tian, X.; Zheng, J.; He, X. Transcriptome analysis reveals dynamic changes in the salt stress response in Salix. J. For. Res. 2020, 31, 12. [Google Scholar] [CrossRef]
- Ferreyra, M.L.F.; Serra, P.; Casati, P. Recent advances on the roles of flavonoids as plant protective molecules after UV and high light exposure. Physiol. Plant. 2021, 173, 736–749. [Google Scholar] [CrossRef]
- Yadav, V.; Wang, Z.; Wei, C.; Amo, A.; Ahmed, B.; Yang, X.; Zhang, X. Phenylpropanoid pathway engineering: An emerging approach towards plant defense. Pathogens 2020, 9, 312. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Dong, A.; Deng, L.; Wang, F.; Liu, J. Deciphering the change pattern of lipid metabolism in Saccharomyces cerevisiae responding to low temperature. Biochem. Eng. J. 2023, 194, 108884. [Google Scholar] [CrossRef]
- Feng, S.; Yao, Y.T.; Wang, B.B.; Li, Y.M.; Li, L.; Bao, A.K. Flavonoids are involved in salt tolerance through ROS scavenging in the halophyte Atriplex canescens. Plant Cell Rep. 2024, 43, 5. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wei, L.; Huang, S.; Yang, C.; Wang, Y.; Yuan, H.; Xu, Q.; Zhang, W.; Wang, M.; Zeng, X.; et al. Drought resistance in Qingke involves a reprogramming of the phenylpropanoid pathway and UDP-glucosyltransferase regulation of abiotic stress tolerance targeting flavonoid biosynthesis. J. Agric. Food Chem. 2021, 69, 3992–4005. [Google Scholar] [CrossRef]
- Li, P.; Ruan, Z.; Fei, Z.; Yan, J.; Tang, G. Integrated transcriptome and metabolome analysis revealed that flavonoid biosynthesis may dominate the resistance of Zanthoxylum bungeanum against stem canker. J. Agric. Food Chem. 2021, 69, 6360–6378. [Google Scholar] [CrossRef]
- Górniak, I.; Bartoszewski, R.; Króliczewski, J. Comprehensive review of antimicrobial activities of plant flavonoids. Phytochem. Rev. 2018, 18, 241–272. [Google Scholar] [CrossRef]
- Olsson, L.C.; Veit, M.; Weissenböck, G.; Bornman, J.F. Differential flavonoid response to enhanced UV-B radiation in Brassica napus. Phytochemistry 1998, 49, 1021–1028. [Google Scholar] [CrossRef]
- Schnitzler, J.P.; Jungblut, T.P.; Heller, W.; Koferlein, M.; Hutzler, P.; Heinzmann, U.; Schmelzer, E.; Ernst, D.; Langebartels, C.; Sandermann, H. Tissue localization of u.v.-B-screening pigments and of chalcone synthase mRNA in needles of Scots pine seedlings. New Phytol. 1996, 132, 247–258. [Google Scholar] [CrossRef]
- Fini, A.; Brunetti, C.; Di Ferdinando, M.; Ferrini, F.; Tattini, M. Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant Signal. Behav. 2011, 6, 709–711. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Wang, X.; Gao, S.; Li, D.; Zhou, J. Production of natural pigments using microorganisms. J. Agric. Food Chem. 2023, 71, 9243–9254. [Google Scholar] [CrossRef] [PubMed]
- Lorenc-Kukuła, K.; Wróbel-Kwiatkowska, M.; Starzycki, M.; Szopa, J. Engineering flax with increased flavonoid content and thus Fusarium resistance. Physiol. Mol. Plant Pathol. 2007, 70, 38–48. [Google Scholar] [CrossRef]
- Torrens-Spence, M.P.; Bobokalonova, A.; Carballo, V.; Glinkerman, C.M.; Weng, J.K. Pbs3 and eps1 complete salicylic acid biosynthesis from isochorismate in Arabidopsis. Mol. Plant. 2019, 12, 10. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhu, W.T.; Zhao, Q. Salicylic acid biosynthesis is not from phenylalanine in Arabidopsis. J. Integr. Plant Biol. 2023, 65, 881–887. [Google Scholar] [CrossRef]
- Hu, Z.; Zhong, X.; Zhang, H.; Luo, X.; Wang, Y.; Wang, Y.; Liu, T.; Zhang, Y.; Wang, X.; An, H.; et al. GhMYB18 confers Aphis gossypii glover resistance through regulating the synthesis of salicylic acid and flavonoids in cotton plants. Plant Cell Rep. 2023, 42, 355–369. [Google Scholar] [CrossRef]
- Zhao, M.; Li, J.; Zhu, L.; Chang, P.; Li, L.; Zhang, L. Identification and characterization of MYB-bHLH-WD40 regulatory complex members controlling anthocyanidin biosynthesis in blueberry fruits development. Genes 2019, 10, 496. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X.; Wang, J.; Zou, G.; Wang, L.; Li, X. Two responses to MeJA induction of R2R3-MYB transcription factors regulate flavonoid accumulation in Glycyrrhiza uralensis Fisch. PLoS ONE 2020, 15, e0236565. [Google Scholar] [CrossRef]
- Li, W.; Wen, L.; Chen, Z.; Zhang, Z.; Pang, X.; Deng, Z.; Liu, T.; Guo, Y.F. Study on metabolic variation in whole grains of four proso millet varieties reveals metabolites important for antioxidant properties and quality traits. Food Chem. 2021, 357, 129791. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yu, D.; Wu, Z.; Wang, C.; Yu, L.; Wei, A.; Wang, D. Comparative transcriptome analysis and expression of genes reveal the biosynthesis and accumulation patterns of key flavonoids in different varieties of Zanthoxylum bungeanum leaves. J. Agric. Food Chem. 2019, 67, 13258–13268. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, Y.; Chen, S.; Min, Y.; Tang, Y.; Ma, X.; Li, H.; Li, J.; Liu, Z. Spraying chitosan on cassava roots reduces postharvest deterioration by promoting wound healing and inducing disease resistance. Carbohydr. Polym. 2023, 318, 121133. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, H.; Liu, S.; Li, G.; Hou, L.; Shen, T.; Song, M.; Liu, J. Enhanced Catalytic Synthesis of Flavonoid by UV-B Radiation in Artemisia argyi. Catalysts 2024, 14, 504. https://doi.org/10.3390/catal14080504
Gu H, Liu S, Li G, Hou L, Shen T, Song M, Liu J. Enhanced Catalytic Synthesis of Flavonoid by UV-B Radiation in Artemisia argyi. Catalysts. 2024; 14(8):504. https://doi.org/10.3390/catal14080504
Chicago/Turabian StyleGu, Haike, Shuang Liu, Guoyu Li, Li Hou, Tengyuan Shen, Meifang Song, and Junfeng Liu. 2024. "Enhanced Catalytic Synthesis of Flavonoid by UV-B Radiation in Artemisia argyi" Catalysts 14, no. 8: 504. https://doi.org/10.3390/catal14080504




