Nature Inspired Solutions for Polymers: Will Cutinase Enzymes Make Polyesters and Polyamides Greener?
Abstract
:1. Introduction
2. The Cutinase Family
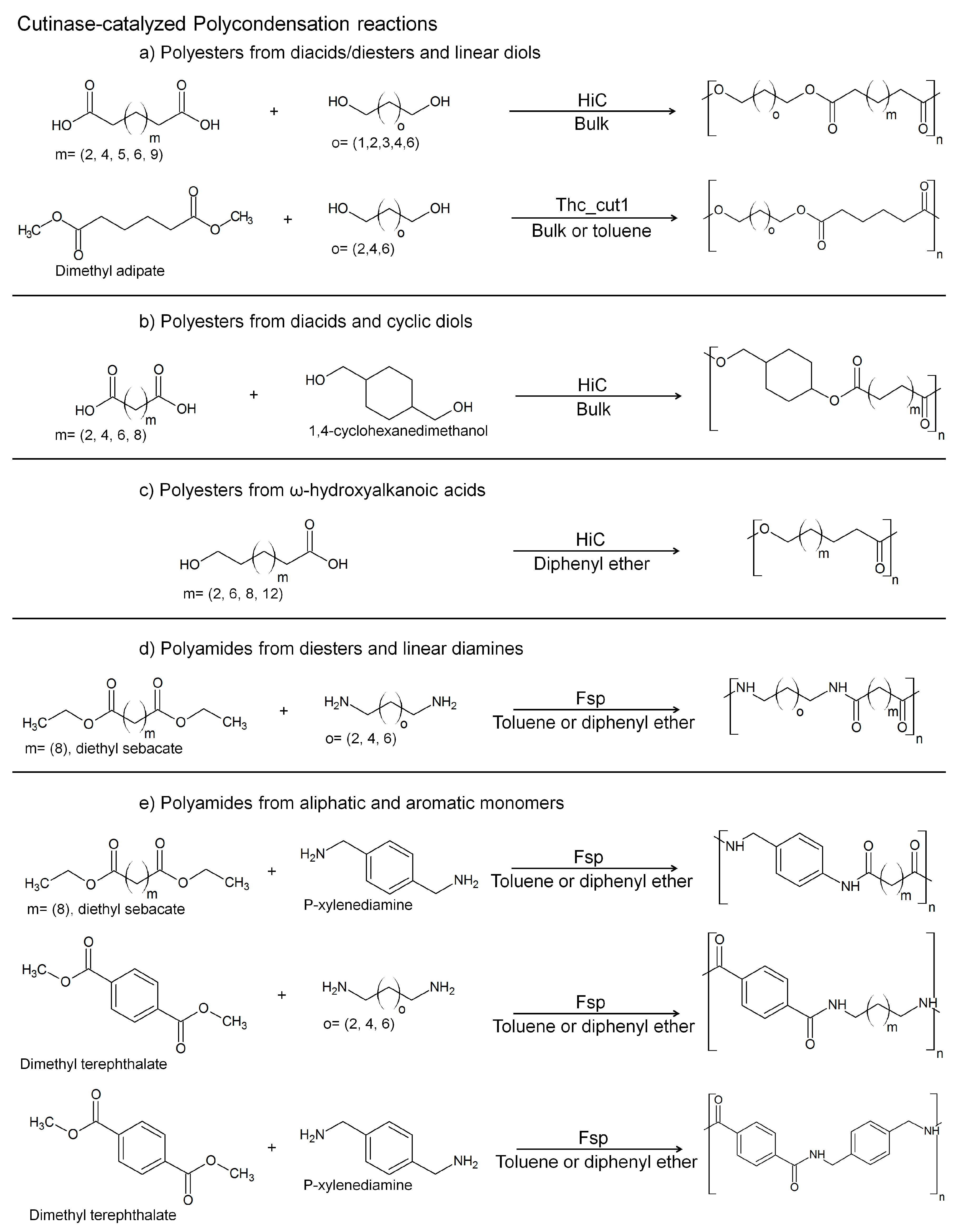
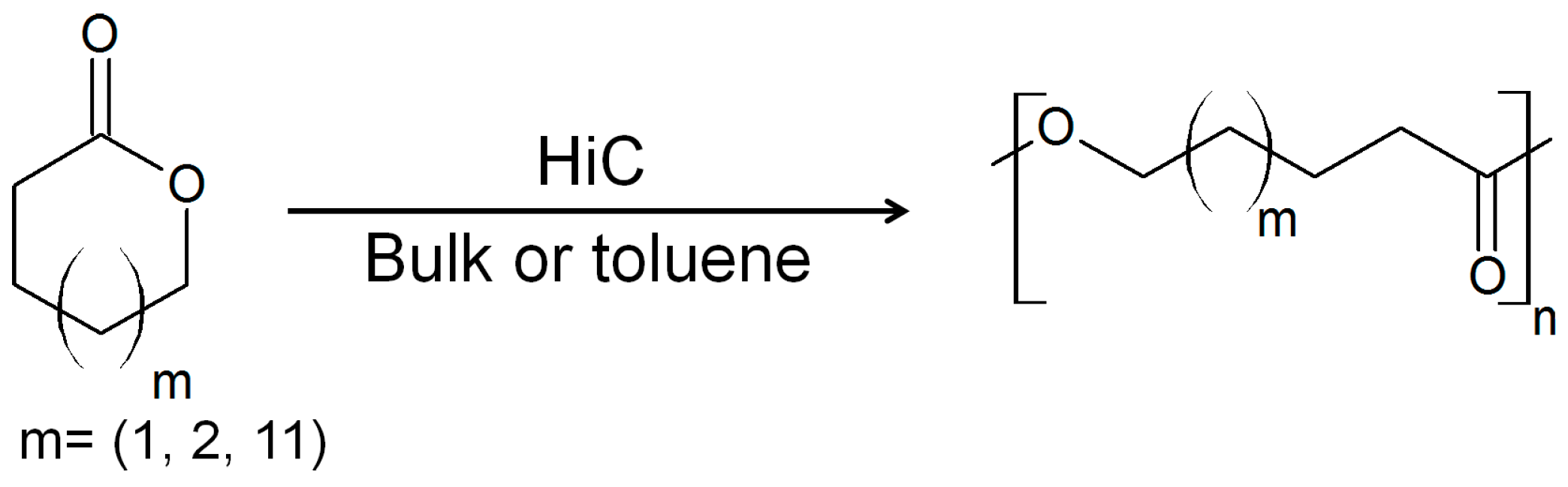
3. Cutinases as Biocatalysts for Polymerization Reactions
4. Cutinases as Biocatalysts for Polymers Hydrolysis
5. Conclusions and Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Guebitz, G.M.; Cavaco-Paulo, A. Enzymes go big: Surface hydrolysis and functionalisation of synthetic polymers. Trends Biotechnol. 2008, 46, 32–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellis, A.; Herrero Acero, E.; Ferrario, V.; Ribitsch, D.; Guebitz, G.M.; Gardossi, L. The closure of the cycle: Enzymatic synthesis and functionalization of bio-based polyesters. Trends Biotechnol. 2016, 34, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Pellis, A.; Herrero Acero, E.; Gardossi, L.; Ferrario, V.; Guebitz, G.M. Renewable building blocks for sustainable polyesters: New biotechnological routes for greener plastics. Polym. Int. 2016, 65, 861–871. [Google Scholar] [CrossRef]
- Gross, R.A.; Ganesh, M.; Lu, W. Enzyme-catalysis breathes new life into polyester condensation polymerizations. Trends Biotechnol. 2010, 28, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Nyyssola, A. Which properties of cutinases are important for applications? Appl. Microbiol. Biotechnol. 2015, 99, 4931–4942. [Google Scholar] [CrossRef] [PubMed]
- Dutta, K.; Sen, S.; Veeranki, V.K. Production, characterization and applications of microbial cutinases. Proc. Biochem. 2009, 44, 127–134. [Google Scholar] [CrossRef]
- Briggs, D.E.G. Molecular taphonomy of animal and plant cuticles: Selective preservation and diagenesis. Philos. Trans. R. Soc. B 1999, 354, 7–17. [Google Scholar] [CrossRef]
- Kolattukudy, P.E. Polyesters in higher plants. Adv. Biochem. Eng. Biotechnol. 2001, 71, 1–4. [Google Scholar] [PubMed]
- Gross, R.A. Overview: Polyester synthesis catalyzed by Candida antarctica lipase B and the cutinase from Humicola insolens. Polym. Prepr. 2006, 47, 263–264. [Google Scholar]
- Pellis, A.; Ferrario, V.; Zartl, B.; Brandauer, M.; Gamerith, C.; Herrero Acero, E.; Ebert, C.; Gardossi, L.; Guebitz, G.M. Enlarging the tools for efficient enzymatic polycondensation: Structural and catalytic features of cutinase 1 from Thermobifida cellulosilytica. Catal. Sci. Technol. 2016, 6, 3430–3442. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, C.M.L.; Aires-Barros, M.R.; Cabral, J.M.S. Cutinase structure, function and biocatalytic applications. Electron. J. Biotechnol. 1998, 1, 160–173. [Google Scholar] [CrossRef]
- Carvalho, C.M.L.; Aires-Barros, M.R.; Cabral, J.M.S. Cutinase: From molecular level to bioprocess development. Biotechnol. Bioeng. 1999, 66, 17–34. [Google Scholar] [CrossRef]
- Poulose, A.; Boston, M. Enzyme Assisted Degradation of Surface Membranes of Harvested Fruits and Vegetables. U.S. Patent 5,298,265, 29 March 1994. [Google Scholar]
- Andersen, K.E.; Borch, K.; Krebs, L.N.E.; Steffen, E.; Landvik, S.; Schnorr, K.M. Plant Extraction Process. WO Patent 2,006,111,163, 26 October 2006. [Google Scholar]
- Regado, M.A.; Cristóvão, B.M.; Moutinho, C.G.; Balcão, V.M.; Aires-Barros, R.; Ferreira, J.P.M.; Xavier Malcata, F. Flavour development via lipolysis of milkfats: Changes in free fatty acid pool. Int. J. Food Sci. Technol. 2007, 42, 961–968. [Google Scholar] [CrossRef]
- De Barros, D.P.C.; Fonseca, L.P.; Cabral, J.M.S.; Weiss, C.K.; Landfester, K. Synthesis of alkyl esters by cutinase in miniemulsion and organic solvent media. Biotechnol. J. 2009, 4, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Badenes, S.M.; Lemos, F.; Cabral, J.M.S. Transesterification of oil mixtures catalyzed by microencapsulated cutinase in reversed micelles. Biotechnol. Lett. 2010, 32, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Badenes, S.M.; Lemos, F.; Cabral, J.M.S. Kinetics and mechanism of the cutinase-catalyzed transesterification of oils in AOT reversed micellar system. Bioprocess. Biosyst. Eng. 2011, 34, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Flipsen, J.A.C.; Appel, A.C.M.; van der Hijden, H.T.W.M.; Verrips, C.T. Mechanism of removal of immobilized triacylglycerol by lipolytic enzymes in a sequential laundry wash process. Enzym. Microb. Technol. 1998, 23, 274–280. [Google Scholar] [CrossRef]
- Ahn, J.Y.; Kim, Y.H.; Min, J.; Lee, J. Accelerated degradation of dipentyl phthalate by Fusarium oxysporum f. sp. pisi cutinase and toxicity evaluation of its degradation products using bioluminescent bacteria. Curr. Microbiol. 2006, 52, 340–344. [Google Scholar] [PubMed]
- Kim, Y.H.; Lee, J.; Ahn, J.Y.; Gu, M.B.; Moon, S.H. Enhanced degradation of an endocrine-disrupting chemical, butyl benzyl phthalate, by Fusarium oxysporum f. sp. pisi cutinase. Appl. Environ. Microbiol. 2002, 68, 4684–4688. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Lee, J.; Moon, S.H. Degradation of an endocrine disrupting chemical, DEHP [di-(2-ethylhexyl)-phthalate], by Fusarium oxysporum f. sp. pisi cutinase. Appl. Microbiol. Biotechnol. 2003, 63, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Su, L.; Chen, J.; Wu, J. Cutinase: Characteristics, preparation, and application. Biotechnol. Adv. 2013, 31, 1754–1767. [Google Scholar] [CrossRef] [PubMed]
- Soliday, C.L.; Kolattukudy, P.E. Primary structure of the active site region of fungal cutinase, an enzyme involved in phytopathogenesis. Biochem. Biophys. Res. Commun. 1983, 114, 1017–1022. [Google Scholar] [CrossRef]
- Purdy, R.E.; Kolattukudy, P.E. Hydrolysis of plant cuticle by plant pathogens. Properties of cutinase I, cutinase II, and a nonspecific esterase isolated from Fusarium solani pisi. Biochemistry 1975, 14, 2832–2840. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Köller, W. Diversity of cutinases from plant pathogenic fungi: Different cutinases are expressed during saprophytic and pathogenic stages of Alternaria brassicicola. Mol. Plant Microbe Interact. 1995, 8, 122–130. [Google Scholar] [CrossRef]
- Castro-Ochoa, D.; Peña-Montes, C.; González-Canto, A.; Alva-Gasca, A.; Esquivel-Bautista, R.; Navarro-Ocaña, A.; Farrés, A. An extracellular cutinase from Aspergillus nidulans induced by olive oil. Appl. Biochem. Biotechnol. 2012, 166, 1275–1290. [Google Scholar] [CrossRef] [PubMed]
- Nyyssölä, A.; Pihlajaniemi, V.; Järvinen, R.; Mikander, S.; Kontkanen, H.; Kruus, K.; Kallio, H.; Buchert, J. Screening of microbes for novel acidic cutinases and cloning and expression of an acidic cutinase from Aspergillus niger CBS 513.88. Enzym. Microb. Technol. 2013, 52, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Yamagata, Y.; Abe, K.; Hasegawa, F.; Machida, M.; Ishioka, R.; Gomi, K.; Nakajima, T. Purification and characterization of a biodegradable plastic-degrading enzyme from Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2005, 67, 778–788. [Google Scholar] [CrossRef] [PubMed]
- Shishiyama, J.; Araki, F.; Akai, S. Studies on cutin-esterase II. Characteristics of cutin-esterase from Botrytis cinerea and its activity on tomato-cutin. Plant Cell Physiol. 1970, 11, 937–945. [Google Scholar]
- Ettinger William, F.; Thukral Sushi, K.; Kolattukudy Pappachan, E. Structure of cutinase gene, cDNA, and the derived amino acid sequence from phytopathogenic Fungi. Biochemistry 1987, 26, 7883–7892. [Google Scholar] [CrossRef]
- Chen, Z.; Franco, C.F.; Baptista, R.P.; Cabral, J.M.S.; Coelho, A.V.; Rodrigues, C.J.; Melo, E.P. Purification and identification of cutinases from Colletotrichum kahawae and Colletotrichum gloeosporioides. Appl. Microbiol. Biotechnol. 2007, 73, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Merz, J.; Schembecker, G.; Riemer, S.; Nimtz, M.; Zorn, H. Purification and identification of a novel cutinase from Coprinopsis. cinerea by adsorptive bubble separation. Sep. Purif. Technol. 2009, 69, 57–62. [Google Scholar] [CrossRef]
- Kodama, Y.; Masaki, K.; Kondo, H.; Suzuki, M.; Tsuda, S.; Nagura, T.; Shimba, N.; Suzuki, E.; Iefuji, H. Crystal structure and enhanced activity of a cutinase-like enzyme from Cryptococcus sp. strain S-2. Proteins 2009, 77, 710–717. [Google Scholar] [CrossRef] [PubMed]
- Dimarogona, M.; Nikolaivits, E.; Kanelli, M.; Christakopoulos, P.; Sandgren, M.; Topakas, E. Structural and functional studies of a Fusarium oxysporum cutinase with polyethylene terephthalate modification potential. Biochim. Biophys. Acta 2015, 1850, 2308–2317. [Google Scholar] [CrossRef] [PubMed]
- Soliday, C.L.; Kolattukudy, P.E. Isolation and characterization of a cutinase from Fusarium roseum culmorum and its immunological comparison with cutinases from F. solani pisi. Arch. Biochem. Biophys. 1976, 176, 334–343. [Google Scholar] [CrossRef]
- Longhi, S.; Czjzek, M.; Lamzin, V.; Nicolas, A.; Cambillau, C. Atomic resolution (1.0 A) crystal structure of Fusarium solani cutinase: Stereochemical analysis. J. Mol. Biol. 1997, 268, 779–799. [Google Scholar] [CrossRef] [PubMed]
- Nyon, M.P.; Rice, D.W.; Berrisford, J.M.; Hounslow, A.M.; Moir, A.J.G.; Huang, H.; Nathan, S.; Mahadi, N.M.; Bakar, F.D.A.; Craven, C.J. Catalysis by Glomerella cingulata cutinase requires conformational cycling between the active and inactive states of its catalytic triad. J. Mol. Biol. 2009, 385, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.S.; Kolattukudy, P.E. Structural studies on cutinase, a glycoprotein containing novel amino acids and glucuronic acid amide at the N terminus. Eur. J. Biochem. 1980, 106, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Kold, D.; Dauter, Z.; Laustsen, A.K.; Brzozowski, A.M.; Turkenburg, J.P.; Nielsen, A.D.; Kolds, H.; Petersen, E.; Schitt, B.; de Maria, L.; et al. Thermodynamic and structural investigation of the specific SDS binding of Humicola insolens cutinase. Protein Sci. 2014, 23, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Sweigard, J.A.; Chumley, F.G.; Valent, B. Disruption of a Magnaporthe grisea cutinase gene. Mol. Gen. Genet. 1992, 232, 183–190. [Google Scholar] [PubMed]
- Wang, G.Y.; Michailides, T.J.; Hammock, B.D.; Lee, Y.M.; Bostock, R.M. Affinity purification and characterization of a cutinase from the fungal plant pathogen Monilinia fructicola (Wint.) honey. Arch. Biochem. Biophys. 2000, 382, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Liebminger, S.; Eberl, A.; Sousa, F.; Heumann, S.; Fischer-Colbrie, G.; Cavaco-Paulo, A.; Guebitz, G.M. Hydrolysis of PET and bis-(benzoyloxyethyl) terephthalate with a new polyesterase from Penicillium citrinum. Biocatal. Biotransform. 2007, 25, 171–177. [Google Scholar] [CrossRef]
- Sebastian, J.; Kolattukudy, P.E. Purification and characterization of cutinase from a fluorescent Pseudomonas putida bacterial strain isolated from phyllosphere. Arch. Biochem. Biophys. 1988, 263, 77–85. [Google Scholar] [CrossRef]
- Davies, K.A.; Lorono, I.; Foster, S.J.; Li, D.; Johnstone, K.; Ashby, A.M. Evidence for a role of cutinase in pathogenicity of Pyrenopeziza brassicae on brassicas. Physiol. Mol. Plant Pathol. 2000, 57, 63–75. [Google Scholar] [CrossRef]
- Parker, D.M.; Köller, W. Cutinase and other lipolytic esterases protect bean leaves from infection by Rhizoctonia solani. Mol. Plant Microbe Interact. 1998, 11, 514–522. [Google Scholar] [CrossRef]
- Miyakawa, T.; Mizushima, H.; Ohtsuka, J.; Oda, M.; Kawai, F.; Tanokura, M. Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotechnol. 2015, 99, 4297–4307. [Google Scholar] [CrossRef] [PubMed]
- Kitadokoro, K.; Thumarat, U.; Nakamura, R.; Nishimura, K.; Karatani, H.; Suzuki, H.; Kawai, F. Crystal structure of cutinase Est119 from Thermobifida alba AHK119 that can degrade modified polyethylene terephthalate at 1.76 Å resolution. Polym. Degrad. Stab. 2012, 97, 771–775. [Google Scholar] [CrossRef]
- Roth, C.; Wei, R.; Oeser, T.; Then, J.; Foellner, C.; Zimmermann, W.; Straeter, N. Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca. Appl. Microbiol. Biotechnol. 2014, 98, 7815–7823. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yan, Q.; Duan, X.; Yang, S.; Jiang, Z. Characterization of an acidic cold-adapted cutinase from Thielavia terrestris and its application in flavor ester synthesis. Food Chem. 2015, 188, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Roussel, A.; Amara, S.; Nyyssola, A.; Mateos-Diaz, E.; Blangy, S.; Kontkanen, H.; Westerholm-Parvinen, A.; Carriere, F.; Cambillau, C. A Cutinase from Trichoderma reesei with a lid-covered active site and kinetic properties of true lipases. J. Mol. Biol. 2014, 426, 3757–3772. [Google Scholar] [CrossRef] [PubMed]
- Maiti, I.B.; Kolattukudy, P.E.; Shaykh, M. Purification and characterization of a novel cutinase from nasturtium (Tropaeolum majus) pollen. Arch. Biochem. Biophys. 1979, 196, 412–423. [Google Scholar] [CrossRef]
- Köller, W.; Parker, D.M. Purification and characterization of cutinase from Venturia inaequalis. Phytopathology 1989, 79, 278–283. [Google Scholar] [CrossRef]
- Ferrario, V.; Ebert, C.; Knapic, L.; Fattor, D.; Basso, A.; Spizzo, P.; Gardossi, L. Conformational changes of lipases in aqueous media: A comparative computational study and experimental implications. Adv. Synth. Catal. 2011, 353, 2466–2480. [Google Scholar] [CrossRef]
- Ferrario, V.; Siragusa, L.; Ebert, C.; Baroni, M.; Foscato, M.; Cruciani, G.; Gardossi, L. BioGPS descriptors for rational engineering of enzyme promiscuity and structure based bioinformatic analysis. PLoS ONE 2014, 9, e109354. [Google Scholar] [CrossRef] [PubMed]
- Pellis, A.; Ferrario, V.; Cespugli, M.; Corici, L.; Guarnieri, A.; Zartl, B.; Herrero-Acero, E.; Ebert, C.; Guebitz, G.M.; Gardossi, L. Fully renewable polyesters via polycondensation catalyzed by Thermobifida cellulosilytica cutinase 1: An integrated approach. Green Chem. 2017. [Google Scholar] [CrossRef]
- Farmer, T.J.; Castle, R.L.; Clark, J.H.; Macquarrie, D.J. Synthesis of unsaturated polyester resins from various bio-derived platform molecules. Int. J. Mol. Sci. 2015, 16, 14912–14932. [Google Scholar] [CrossRef] [PubMed]
- Bassanini, I.; Hult, K.; Riva, S. Dicarboxylic esters: Useful tools for the biocatalyzed synthesis of hybrid compounds and polymers. Beilstein J. Org. Chem. 2015, 11, 1583–1595. [Google Scholar] [CrossRef] [PubMed]
- Pellis, A.; Corici, L.; Sinigoi, L.; D’amelio, N.; Fattor, D.; Ferrario, V.; Ebert, C.; Gardossi, L. Towards feasible and scalable solvent-free enzymatic polycondensations: Integrating robust biocatalysts with thin film reactions. Green Chem. 2015, 17, 1756–1766. [Google Scholar] [CrossRef] [Green Version]
- Hunsen, M.; Azim, A.; Mang, H.; Wallner, S.R.; Ronkvist, A.; Xie, W.; Gross, R.A. A cutinase with polyester synthesis activity. Macromolecules 2007, 40, 148–150. [Google Scholar] [CrossRef]
- Feder, D.; Gross, R.A. Exploring chain length selectivity in HIC-catalyzed polycondensation reactions. Biomacromolecules 2010, 11, 690–697. [Google Scholar] [CrossRef] [PubMed]
- Stavila, E.; Arsyi, R.Z.; Petrovic, D.M.; Loos, K. Fusarium solani pisi cutinase-catalyzed synthesis of polyamides. Eur. Polym. J. 2013, 49, 834–842. [Google Scholar] [CrossRef]
- Stavila, E.; Alberda van Ekenstein, G.O.R.; Loos, K. Enzyme-catalyzed synthesis of aliphatic-aromatic oligoamides. Biomacromolecules 2013, 14, 1600–1606. [Google Scholar] [CrossRef] [PubMed]
- Binns, F.; Harffey, P.; Roberts, S.M.; Taylor, A. Studies leading to the large scale synthesis of polyesters using enzymes. J. Chem. Soc. Perkin Trans. 1999, 1, 2671–2676. [Google Scholar] [CrossRef]
- Cantone, S.; Ferrario, V.; Corici, L.; Ebert, C.; Fattor, D.; Spizzo, P.; Gardossi, L. Efficient immobilisation of industrial biocatalysts: Criteria and constraints for the selection of organic polymeric carriers and immobilisation methods. Chem. Soc. Rev. 2013, 42, 6262–6276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corici, L.; Ferrario, V.; Pellis, A.; Ebert, C.; Lotteria, S.; Cantone, S.; Voinovich, D.; Gardossi, L. Large scale applications of immobilized enzymes call for sustainable and inexpensive solutions: Rice husks as renewable alternatives to fossil-based organic resins. RSC Adv. 2016, 6, 63256–63270. [Google Scholar] [CrossRef]
- Barth, M.; Oeser, T.; Wei, R.; Then, J.; Schmidt, J.; Zimmermann, W. Effect of hydrolysis products on the enzymatic degradation of polyethylene terephthalate nanoparticles by a polyester hydrolase from Thermobifida fusca. Biochem. Eng. J. 2015, 93, 222–228. [Google Scholar] [CrossRef]
- Alisch-Mark, M.; Herrmann, A.; Zimmermann, W. Increase of the hydrophilicity of polyethylene terephthalate fibres by hydrolases from Thermomonospora fusca and Fusarium solani f. sp. pisi. Biotechnol. Lett. 2006, 28, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Donelli, I.; Taddei, P.; Smet, P.F.; Poelman, D.; Nierstrasz, V.A.; Freddi, G. Enzymatic surface modification and functionalization of PET: A water contact angle, FTIR, and fluorescence spectroscopy study. Biotechnol. Bioeng. 2009, 103, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Pellis, A.; Herrero Acero, E.; Weber, H.; Obersriebnig, M.; Breinbauer, R.; Srebotnik, E.; Guebitz, G.M. Biocatalyzed approach for the surface functionalization of poly(L-lactic acid) films using hydrolytic enzymes. Biotechnol. J. 2015, 10, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Mueller, R.J. Biological degradation of synthetic polyesters—ENZYMES as potential catalysts for polyester recycling. Proc. Biochem. 2006, 41, 2124–2128. [Google Scholar] [CrossRef]
- Herrero Acero, E.; Ribitsch, D.; Steinkellner, G.; Gruber, K.; Greimel, K.; Eiteljoerg, I.; Trotscha, E.; Wei, R.; Zimmermann, W.; Zinn, M.; et al. Enzymatic surface hydrolysis of PET: Effect of structural diversity on kinetic properties of cutinases from Thermobifida. Macromolecules 2011, 44, 4632–4640. [Google Scholar] [CrossRef] [Green Version]
- Pellis, A.; Gamerith, C.; Ghazaryan, G.; Ortner, A.; Herrero Acero, E.; Guebitz, G.M. Ultrasound-enhanced enzymatic hydrolysis of poly (ethylene terephthalate). Biores. Technol. 2016, 218, 1298–1302. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Xu, H.; Yan, Q.; Liu, Y.; Zhou, P.; Jiang, Z. A low molecular mass cutinase of Thielavia. terrestris efficiently hydrolyzes poly(esters). J. Ind. Microbiol. Biotechnol. 2013, 40, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Ronkvist, A.M.; Xie, W.; Lu, W.; Gross, R.A. Cutinase-catalyzed hydrolysis of poly(ethylene terephtalate). Maceomolecules 2009, 42, 5128–5138. [Google Scholar] [CrossRef]
- Pellis, A.; Silvestrini, L.; Scaini, D.; Coburn, J.; Gardossi, L.; Kaplan, D.; Herrero Acero, E.; Guebitz, G.M. Enzyme-catalyzed functionalization of poly (l-lactic acid) for drug delivery applications. Process Biochem. 2016. [Google Scholar] [CrossRef]
- Ribitsch, D.; Herrero Acero, E.; Greimel, K.; Dellacher, A.; Zitzenbacher, S.; Marold, A.; Diaz Rodriguez, R.; Steinkellner, G.; Gruber, K.; Schwab, H.; et al. A new esterase from Thermobifida halotolerans hydrolyses polyethylene terephthalate (PET) and polylactic acid (PLA). Polymers 2012, 4, 617–629. [Google Scholar] [CrossRef]
- Pellis, A.; Haernvall, K.; Pichler, C.M.; Ghazaryan, G.; Breinbauer, R.; Guebitz, G.M. Enzymatic hydrolysis of poly (ethylene furanoate). J. Biotechnol. 2016, 235, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Perz, V.; Bleymaier, K.; Sinkel, C.; Kueper, U.; Bonnekessel, M.; Ribitsch, D.; Guebitz, G.M. Substrate specificities of cutinases on aliphatic–aromatic polyesters and on their model substrates. New Biotechnol. 2016, 33, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.M.; Carnerio, F.; O’Neill, A.; Fonseca, L.P.; Cabral, J.S.M.; Guebitz, G.M.; Cavaco-Paulo, A. Cutinase—A new tool for biomodification of synthetic fibers. J. Polym. Sci. Part A Polym. Chem. 2005, 43, 2448–2450. [Google Scholar] [CrossRef] [Green Version]
- Ribitsch, D.; Oracal Yebra, A.; Zitzenbacher, S.; Wu, J.; Nowitsch, S.; Steinkellner, G.; Greimel, K.; Doliska, A.; Oberdorfer, G.; Gruber, C.C.; et al. Fusion of binding domains to Thermobifida cellulosilytica cutinase to tune sorption characteristics and enhancing PET hydrolysis. Biomacromolecules 2013, 14, 1769–1776. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.; Da, S.; Silva, N.; Matamá, T.; Araújo, R.; Martins, M.; Chen, S.; Chen, J.; Wu, J.; Casal, M.; et al. Engineered Thermobifida fusca cutinase with increased activity on polyester substrates. Biotechnol. J. 2011, 6, 1230–1239. [Google Scholar] [CrossRef] [PubMed]
- Araújo, R.; Silva, C.; O’Neill, A.; Micaelo, N.; Guebitz, G.M.; Soares, C.M.; Casal, M.; Cavaco-Paulo, A. Tailoring cutinase activity towards polyethylene terephthalate and polyamide 6,6 fibers. J. Biotechnol. 2007, 128, 849–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilal, M.H.; Prehm, M.; Njau, A.E.; Samiullah, M.H.; Meister, A.; Kressler, J. Enzymatic synthesis and characterization of hydrophilic sugar based polyesters and their modification with stearic acid. Polymers 2016, 8, 80. [Google Scholar] [CrossRef]
- Clark, J.H.; Farmer, T.J.; Herrero-Davila, L.; Sherwood, J. Circular economy design considerations for research and process development in the chemical sciences. Green Chem. 2016, 18, 3914–3934. [Google Scholar] [CrossRef]

| Source | Molar Masses (kDa) | Ref. |
|---|---|---|
| Alternaria brassicicola | 26 | [26] |
| Aspergillus nidulans | 29 | [27] |
| Aspergillus niger | 22.8 | [28] |
| Aspergillus oryzae | 19.6 | [29] |
| Botrytis cinerea | 18 | [30] |
| Colletotrichum capsici | 23.7 | [31] |
| Colletotrichum gloeosporioides | 40 | [32] |
| Colletotrichum kahawae | 21 | [32] |
| Coprinopsis cinerea | 29.6 | [33] |
| Cryptococcus sp. | 21.2 | [34] |
| Fusarium oxysporum | 23.4 | [35] |
| Fusarium roseum culmorum | 24.3 | [36] |
| Fusarium solani | 20.8 | [37] |
| Glomerella cingulata | 21.1 | [38] |
| Helminthosporium sativum | 25 | [39] |
| Humicola insolens | 20.3 | [40] |
| Magnaporthe grisea | 24.3 | [41] |
| Monilinia fructicola | 18.6 | [42] |
| Pennicillium citrinum | 14.1 | [43] |
| Pseudomonas putida | 30 | [44] |
| Pyrenopeziza brassicae | 21 | [45] |
| Rhizoctonia solani | 19.8 | [46] |
| Saccharomonospora virdis | 30.3 | [47] |
| Thermobifida alba | 33.5 | [48] |
| Thermobifida cellulosilytica | 30.8 | [10] |
| Thermobifida fusca | 30.8 | [49] |
| Thielavia terrestris | 27 | [50] |
| Trichoderma reesei | 27.3 | [51] |
| Tropaeolum majus | 40 | [52] |
| Venturia inaequalis | 21.7 | [53] |
| Polymer | Enzyme | Ref. |
|---|---|---|
| Poly(ethylene terephthalate) (PET) | Thermobifida cellulosilytica | [72,73] |
| Thermobifida fusca | [67] | |
| Thermobifida alba | ||
| Thielavia terrestris | [74] | |
| Humicola insolens | [75] | |
| Pseudomonas mendocina | ||
| Fusarium solani | ||
| Penicillum mendocin | [69,74] | |
| Penicillum citrinum | ||
| Poly(l-lactic acid) (PLA) | Humicola insolens | [70,76] |
| Thermobifida halotolerans | [77] | |
| Poly(ethylene furanoate) (PEF) | Thermobifida cellulosilytica | [78] |
| Poly(butylene adipate-co-terephthalate) (PBAT) | Humicola insolens | [79] |
| Thermobifida cellulosilytica | ||
| Poly(caprolactone) (PCL) | Thielavia terrestris | [79] |
| Poly(butylene succinate) (PBS) | Thielavia terrestris | [79] |
| Aspergillus oryzae | ||
| Fusarium solani | ||
| Humicola insolens | ||
| Alternaria brassicicola | ||
| Polyamide 6,6 | Fusarium solani | [80] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrario, V.; Pellis, A.; Cespugli, M.; Guebitz, G.M.; Gardossi, L. Nature Inspired Solutions for Polymers: Will Cutinase Enzymes Make Polyesters and Polyamides Greener? Catalysts 2016, 6, 205. https://doi.org/10.3390/catal6120205
Ferrario V, Pellis A, Cespugli M, Guebitz GM, Gardossi L. Nature Inspired Solutions for Polymers: Will Cutinase Enzymes Make Polyesters and Polyamides Greener? Catalysts. 2016; 6(12):205. https://doi.org/10.3390/catal6120205
Chicago/Turabian StyleFerrario, Valerio, Alessandro Pellis, Marco Cespugli, Georg M. Guebitz, and Lucia Gardossi. 2016. "Nature Inspired Solutions for Polymers: Will Cutinase Enzymes Make Polyesters and Polyamides Greener?" Catalysts 6, no. 12: 205. https://doi.org/10.3390/catal6120205
APA StyleFerrario, V., Pellis, A., Cespugli, M., Guebitz, G. M., & Gardossi, L. (2016). Nature Inspired Solutions for Polymers: Will Cutinase Enzymes Make Polyesters and Polyamides Greener? Catalysts, 6(12), 205. https://doi.org/10.3390/catal6120205