Bacterial Biodegradation of 4-Monohalogenated Diphenyl Ethers in One-Substrate and Co-Metabolic Systems
Abstract
:1. Introduction
2. Results
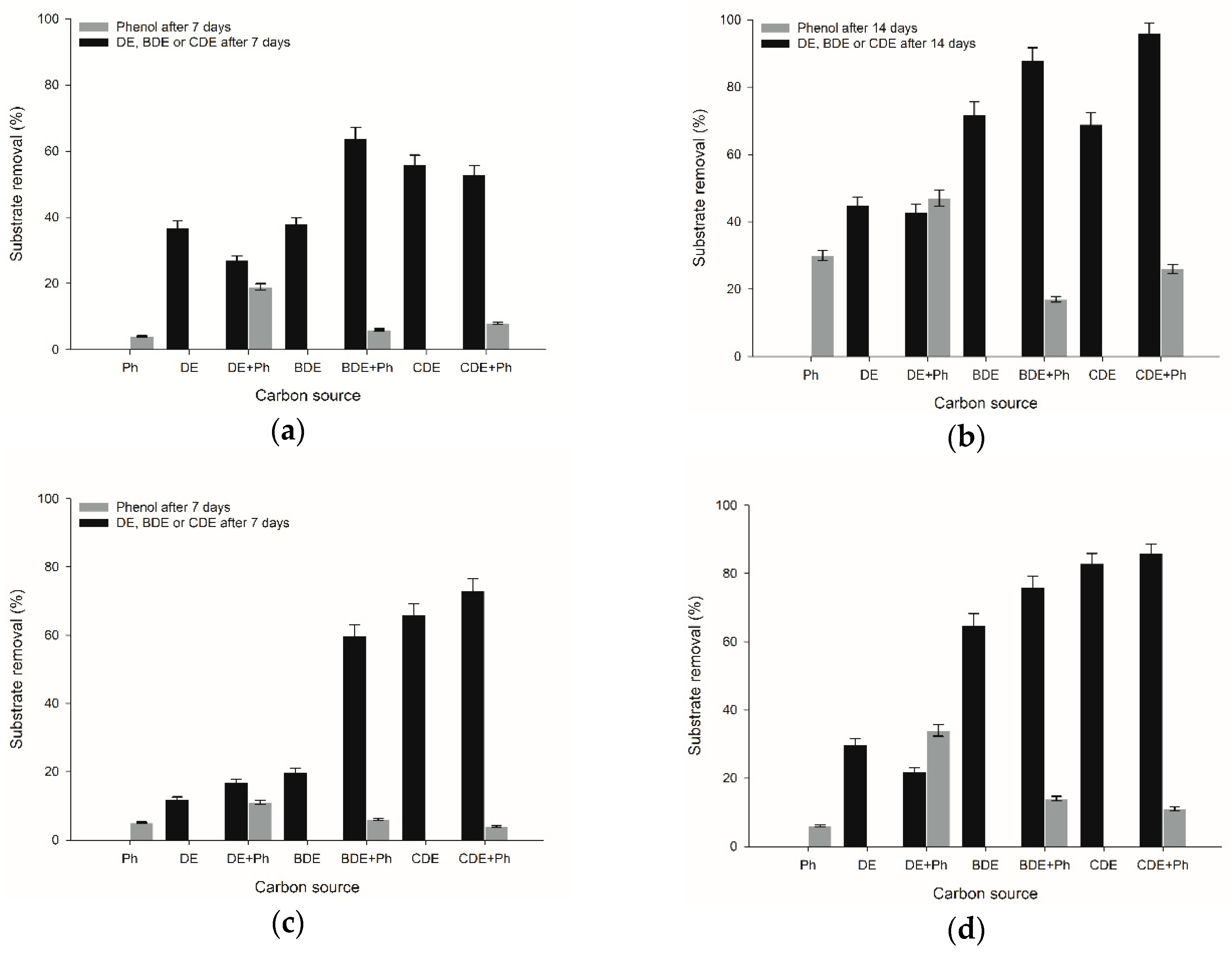
2.1. Biodegradation of (4-Monohalogenated) Diphenyl Ethers
2.2. Cell Metabolic Activity
2.3. Enzymatic Activity
2.4. Cell Membrane Permeability and Surface Hydrophobicity
3. Materials and Methods
3.1. Chemicals
3.2. Preparation of Bacterial Suspensions and Their Treatment
3.3. Biodegradation
3.4. Cells’ Metabolic Activity
3.5. Crude Extract Preparation and Enzyme Activity Evaluation
3.6. Membrane Permeability
3.7. Cell Surface Hydrophobicity
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Yu, H.; Yang, H.; Cui, D.; Lv, L.; Li, B. Synthesis and herbicidal activity of diphenyl ether derivatives containing unsaturated carboxylates. J. Agric. Food Chem. 2011, 59, 11718–11726. [Google Scholar] [CrossRef] [PubMed]
- McGrath, T.J.; Ball, A.S.; Clarke, B.O. Critical review of soil contamination by polybrominated diphenyl ethers (PBDEs) and novel brominated flame retardants (NBFRs); concentrations, sources and congener profiles. Environ. Pollut. 2015, 230, 741–757. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Feng, M.; Zhang, X.; Wang, L.; Wang, Z. Occurrence of polychlorinated diphenyl ethers in Nanjing section of the Yangtze River: Level and distribution pattern. Environ. Sci. Pollut. Res. 2015, 22, 9224–9232. [Google Scholar] [CrossRef] [PubMed]
- Sjödin, A.; Patterson, D.G.; Bergman Åke, Å. A review on human exposure to brominated flame retardants—Particularly polybrominated diphenyl ethers. Environ. Int. 2003, 29, 829–839. [Google Scholar] [CrossRef]
- Kelly, B.C.; Ikonomou, M.G.; Blair, J.D.; Gobas, F.A. Bioaccumulation behaviour of polybrominated diphenyl ethers (PBDEs) in a Canadian Arctic marine food web. Sci. Total Environ. 2008, 401, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, M.A.; Laessig, R.H.; Reed, K.D. Polybrominated diphenyl ethers (PBDEs): New pollutants-old diseases. Clin. Med. Res. 2003, 1, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Koistinen, H.; Koistinen, R.; Dell, A.; Morris, H.R.; Easton, R.L.; Patankar, M.S.; Oehninger, S.; Clark, G.F.; Seppälä, M. Glycodelin from seminal plasma is a differentially glycosylated form of contraceptive glycodelin-A. Mol. Hum. Reprod. 1996, 2, 759–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domingo, J.L. Polychlorinated diphenyl ethers (PCDEs): Environmental levels, toxicity and human exposure: A review of the published literature. Environ. Int. 2006, 32, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Mrozik, A.; Piotrowska-Seget, Z.; Łabużek, S. Bacteria in bioremediation of hydrocarbon-contaminated environment. Postępy Mikrobiol. 2005, 44, 227–238. [Google Scholar]
- Hiraishi, A. Biodiversity of dioxin-degrading microorganisms and potential utilization in bioremediation. Microbes Environ. 2014, 18, 105–125. [Google Scholar] [CrossRef]
- Lv, Y.; Li, L.; Chen, Y.; Tang, Z.; Hu, Y. Effects of glucose and biphenyl on aerobic cometabolism of polybrominated diphenyl ethers by Pseudomonas putida: Kinetics and degradation mechanism. Int. Biodeterior. Biodegrad. 2016, 108, 76–84. [Google Scholar] [CrossRef]
- Seo, J.S.; Keum, Y.S.; Li, Q.X. Bacterial Degradation of Aromatic Compounds. Int. J. Environ. Res. Public Health 2009, 6, 278–309. [Google Scholar] [CrossRef] [Green Version]
- Dvořák, P.; Nikel, P.I.; Damborský, J.; de Lorenzo, V. Bioremediation 3.0: Engineering pollutant-removing bacteria in the times of systemic biology. Biotechnol. Adv. 2017, 35, 845–866. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, B.; Wang, C.P.; Fan, J.Z.; Sun, H.W. Aerobic degradation of trichloroethylene by co-metabolism using phenol and gasoline as growth substrates. Int. J. Mol. Sci. 2014, 15, 9134–9148. [Google Scholar] [CrossRef] [PubMed]
- Domaradzka, D.; Guzik, U.; Hupert-Kocurek, K.; Wojcieszyńska, D. Cometabolic Degradation of Naproxen by Planococcus sp. Strain S5. Water. Air. Soil Pollut. 2015, 226, 297. [Google Scholar] [CrossRef] [PubMed]
- Hedegaard, M.J.; Deliniere, H.; Prasse, C.; Dechesne, A.; Smets, B.F.; Albrechtsen, H.J. Evidence of co-metabolic bentazone transformation by methanotrophic enrichment from a groundwater-fed rapid sand filter. Water Res. 2018, 129, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Khodaei, K.; Nassery, H.R.; Asadi, M.M.; Mohammadzadeh, H.; Mahmoodlu, M.G. BTEX biodegradation in contaminated groundwater using a novel strain (Pseudomonas sp. BTEX-30). Int. Biodeterior. Biodegrad. 2017, 116, 234–242. [Google Scholar] [CrossRef]
- Derakhshan, Z.; Mahvi, A.H.; Ehrampoush, M.H.; Mazloomi, S.M.; Faramarzian, M.; Dehghani, M.; Yousefinejad, S.; Ghaneian, M.T.; Abtahi, S.M. Studies on influence of process parameters on simultaneous biodegradation of atrazine and nutrients in aquatic environments by a membrane photobioreactor. Environ. Res. 2018, 161, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Escola Casas, M.; Ooi, G.T.H.; Kaarsholm, K.M.S.; Bester, K.; Andersen, H.R. Influence of humic acid addition on the degradation of pharmaceuticals by biofilms in effluent wastewater. Int. J. Hyg. Environ. Health 2017, 220, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Baggi, G.; Bernasconi, S.; Zangrossi, M. 3-Chloro-, 2,3- and 3,5-dichlorobenzoate co-metabolism in a 2-chlorobenzoate-degrading consortium: Role of 3,5-dichlorobenzoate as antagonist of 2-chlorobenzoate degradation—Metabolism and co-metabolism of chlorobenzoates. Biodegradation 2005, 16, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Priya, V.S.; Philip, L. Biodegradation of dichloromethane along with other VOCs from pharmaceutical wastewater. Appl. Biochem. Biotechnol. 2013, 169, 1197–1218. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tay, J.H. Toxic and inhibitory effects of trichloroethylene aerobic co-metabolism on phenol-grown aerobic granules. J. Hazard. Mater. 2015, 286, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Mamma, D.; Kalogeris, E.; Papadopoulos, N.; Hatzinikolaou, D.G.; Christrakopoulos, P.; Kekos, D. Biodegradation of phenol by acclimatized Pseudomonas putida cells using glucose as an added growth substrate. J. Environ. Sci. Health. A. Tox. Hazard. Subst. Environ. Eng. 2004, 39, 2093–2104. [Google Scholar] [CrossRef] [PubMed]
- Bakhshi, Z.; Najafpour, G.; Kariminezhad, E.; Pishgar, R.; Mousavi, N.; Taghizade, T. Growth kinetic models for phenol biodegradation in a batch culture of Pseudomonas putida. Environ. Technol. 2011, 33, 1835–1841. [Google Scholar] [CrossRef] [PubMed]
- Perpetuo, E.A.; Silva, D.N.; Avanzi, I.R.; Gracioso, L.H.; Baltazar, M.P.G.; Nascimento, C.A.O. Phenol biodegradation by a microbial consortium: Application of artificial neural network (ANN) modelling. Environ. Technol. 2012, 33, 1739–1745. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-M.; Nam, I.-H.; Murugesan, K.; Schmidt, S.; Crowley, D.E.; Chang, Y.-S. Biodegradation of diphenyl ether and transformation of selected brominated congeners by Sphingomonas sp. PH-07. Appl. Microbiol. Biotechnol. 2007, 77, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Xie, M.; Li, Y.; Gao, G.; Bartlam, M.; Wang, Y. Biodegradation of decabromodiphenyl ether (BDE 209) by a newly isolated bacterium from an e-waste recycling area. AMB Express 2018, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Heidari, H.; Sedighi, M.; Zamir, S.M.; Shojaosadati, S.A. Bisphenol A degradation by Ralstonia eutropha in the absence and presence of phenol. Int. Biodeterior. Biodegradation 2017, 119, 37–42. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Wang, C.-K.; Shih, Y.-H. Microbial degradation of 4-monobrominated diphenyl ether in an aerobic sludge and the DGGE analysis of diversity. J. Environ. Sci. Heal. Part B 2010, 45, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, V.; Balakrishnan, V.; Philip Robinson, J.; Ramakrishnan, M. Anticancer and apoptosis-inducing effects of Moringa concanensis using hepG2 cell lines. Bangladesh J. Pharmacol. 2014, 9, 604–609. [Google Scholar] [CrossRef]
- Vollár, M.; Gyovai, A.; Szűcs, P.; Zupkó, I.; Marschall, M.; Csupor-Löffler, B.; Bérdi, P.; Vecsernyés, A.; Csorba, A.; Liktor-Busa, E.; et al. Antiproliferative and antimicrobial activities of selected bryophytes. Molecules 2018, 23, 1520. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Sun, G.-D.; Jin, J.-H.; Liu, Y.; Luo, M.; Zhong, Z.-P.; Liu, Z.-P. Successful bioremediation of an aged and heavily contaminated soil using a microbial/plant combination strategy. J. Hazard. Mater. 2014, 264, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Scelza, R.; Antonietta Rao, M.; Gianfreda, L. Effects of compost and of bacterial cells on the decontamination and the chemical and biological properties of an agricultural soil artificially contaminated with phenanthrene. Soil Biol. Biochem. 2007, 39, 1303–1317. [Google Scholar] [CrossRef]
- Xu, Y.; Lu, M. Bioremediation of crude oil-contaminated soil: Comparison of different biostimulation and bioaugmentation treatments. J. Hazard. Mater. 2010, 183, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.N.; Kumari, B.; Upadhyay, S.K.; Mishra, S.; Kumar, D. Bacterial degradation of pyrene in minimal salt medium mediated by catechol dioxygenases: Enzyme purification and molecular size determination. Bioresour. Technol. 2013, 133, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Nowak, A.; Mrozik, A. Degradation of 4-chlorophenol and microbial diversity in soil inoculated with single Pseudomonas sp. CF600 and Stenotrophomonas maltophilia KB2. J. Environ. Manage. 2018, 215, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Wojcieszyńska, D.; Hupert-Kocurek, K.; Guzik, U. Factors affecting activity of catechol 2,3-dioxygenase from 2-chlorophenol-degrading Stenotrophomonas maltophilia strain KB2. Biocatal. Biotransformation 2013, 31, 141–147. [Google Scholar] [CrossRef]
- Guzik, U.; Greń, I.; Wojcieszyńska, D.; Łabużek, S. Isolation and characterization of a novel strain of Stenotrophomonas maltophilia possessing various dioxygenases for monocyclic hydrocarbon degradation. Brazilian J. Microbiol. 2009, 40, 285–291. [Google Scholar] [CrossRef]
- Wang, S.; Bai, N.; Wang, B.; Feng, Z.; Hutchins, W.C.; Yang, C.-H.; Zhao, Y. Characterization of the molecular degradation mechanism of diphenyl ethers by Cupriavidus sp. WS. Environ. Sci. Pollut. Res. 2015, 22, 16914–16926. [Google Scholar] [CrossRef] [PubMed]
- Solyanikova, I.P.; Golovleva, L.A. Bacterial degradation of chlorophenols: Pathways, biochemica, and genetic aspects. J. Environ. Sci. Health. B. 2004, 39, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Sikkema, J.; de Bont, J.A.; Poolman, B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 1995, 59, 201–222. [Google Scholar] [PubMed]
- Zhao, C.; Yan, M.; Zhong, H.; Liu, Z.; Shi, L.; Chen, M.; Zeng, G.; Song, B.; Shao, B.; Feng, H. Biodegradation of polybrominated diphenyl ethers and strategies for acceleration: A review. Int. Biodeterior. Biodegradation 2018, 129, 23–32. [Google Scholar] [CrossRef]
- Hao, O.J.; Kim, M.H.; Seagren, E.A.; Kim, H. Kinetics of phenol and chlorophenol utilization by Acinetobacter species. Chemosphere 2002, 46, 797–807. [Google Scholar] [CrossRef]
- Marqués, S.; Ramos, J.L. Transcriptional control of the Pseudomonas putida TOL plasmid catabolic pathways. Mol. Microbiol. 1993, 9, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, L.; Wang, R.; Wang, X.; Ye, J.; Long, Y. Biosorption and degradation of decabromodiphenyl ether by Brevibacillus brevis and the influence of decabromodiphenyl ether on cellular metabolic responses. Environ. Sci. Pollut. Res. 2016, 23, 5166–5178. [Google Scholar] [CrossRef] [PubMed]
- Heipieper, H.J.; Keweloh, H.; Rehm, H.J. Influence of phenols on growth and membrane permeability of free and immobilized Escherichia coli. Appl. Environ. Microbiol. 1991, 57, 1213–1217. [Google Scholar] [PubMed]
- Chen, X.; Song, D.; Xu, J.; Li, E.; Sun, G.; Xu, M. Role and mechanism of cell-surface hydrophobicity in the adaptation of Sphingobium hydrophobicum to electronic-waste contaminated sediment. Appl. Microbiol. Biotechnol. 2018, 102, 2803–2815. [Google Scholar] [CrossRef] [PubMed]
- Kaczorek, E.; Sałek, K.; Guzik, U.; Dudzińska-Bajorek, B. Cell surface properties and fatty acids composition of Stenotrophomonas maltophilia under the influence of hydrophobic compounds and surfactants. New Biotechnol. 2013, 30, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Jafarirad, S.; Mehrabi, M.; Divband, B.; Kosari-Nasab, M. Biofabrication of zinc oxide nanoparticles using fruit extract of Rosa canina and their toxic potential against bacteria: A mechanistic approach. Mater. Sci. Eng. C 2016, 59, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Devi, K.P.; Sakthivel, R.; Nisha, S.A.; Suganthy, N.; Pandian, S.K. Eugenol alters the integrity of cell membrane and acts against the nosocomial pathogen Proteus mirabilis. Arch. Pharm. Res. 2013, 36, 282–292. [Google Scholar] [CrossRef] [PubMed]

| Relative Enzymatic Activity (mU mg−1) | P. plecoglossicida IsA | P. fluorescens B01 | ||
|---|---|---|---|---|
| 1,2-DO | 2,3-DO | 1,2-DO | 2,3-DO | |
| SS | 277.72 ± 13.89 | 268.63 ± 13.43 | 0.01 ± 0.00 | 19.26 ± 0.96 |
| Ph | 127.10 ± 6.35 | 119.59 ± 5.98 | 0.05 ± 0.02 | 19.90 ± 0.99 |
| DE | 255.28 ± 12.76 | 110.64 ± 5.53 | 0.02 ± 0.01 | 12.44 ± 0.62 |
| DE + Ph | 0.66 ± 0.03 | 0.09 ± 0.02 | 494.50 ± 24.72 | 26.22 ± 1.31 |
| BDE | 83.28 ± 4.16 | 49.22 ± 2.46 | 8.47 ± 0.42 | 0.01 ± 0.00 |
| BDE + Ph | 339.27 ± 16.96 | 157.05 ± 7.58 | 126.71 ± 6.34 | 0.07 ± 0.03 |
| CDE | 0.08 ± 0.02 | 0.07 ± 0.02 | 2.11 ± 0.11 | 0.85 ± 0.04 |
| CDE + Ph | 2.19 ± 0.11 | 0.02 ± 0.00 | 237.70 ± 11.89 | 0.02 ± 0.01 |
| DE | DE + Ph | BDE | BDE + Ph | CDE | CDE + Ph | ||
|---|---|---|---|---|---|---|---|
| P. plecoglossicida IsA | MP (μmol L−1·min) | 51.0 ± 2.6 | 46.0 ± 2.3 | 53.0 ± 2.7 | 49.0 ±2.5 | 54.0 ± 2.7 | 51.0 ± 2.6 |
| CSH (%) | 27.0 ± 1.4 | 31.0 ± 1.6 | 65.0 ± 3.3 | 36.0 ±1.8 | 54.0 ± 2.7 | 54.0 ± 2.5 | |
| P. fluorescens B01 | MP (μmol L−1·min) | 34.0 ±1.7 | 45.0 ± 2.3 | 33.0 ± 1.7 | 37.0 ± 1.9 | 45.0 ± 2.3 | 54.0 ± 2.7 |
| CSH (%) | 44.0 ± 2.2 | 51.0 ± 2.6 | 56.0 ± 2.8 | 30.0 ± 1.5 | 6.0 ± 0.3 | 16.0 ± 0.8 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pacholak, A.; Smułek, W.; Zdarta, A.; Zgoła-Grześkowiak, A.; Kaczorek, E. Bacterial Biodegradation of 4-Monohalogenated Diphenyl Ethers in One-Substrate and Co-Metabolic Systems. Catalysts 2018, 8, 472. https://doi.org/10.3390/catal8100472
Pacholak A, Smułek W, Zdarta A, Zgoła-Grześkowiak A, Kaczorek E. Bacterial Biodegradation of 4-Monohalogenated Diphenyl Ethers in One-Substrate and Co-Metabolic Systems. Catalysts. 2018; 8(10):472. https://doi.org/10.3390/catal8100472
Chicago/Turabian StylePacholak, Amanda, Wojciech Smułek, Agata Zdarta, Agnieszka Zgoła-Grześkowiak, and Ewa Kaczorek. 2018. "Bacterial Biodegradation of 4-Monohalogenated Diphenyl Ethers in One-Substrate and Co-Metabolic Systems" Catalysts 8, no. 10: 472. https://doi.org/10.3390/catal8100472
APA StylePacholak, A., Smułek, W., Zdarta, A., Zgoła-Grześkowiak, A., & Kaczorek, E. (2018). Bacterial Biodegradation of 4-Monohalogenated Diphenyl Ethers in One-Substrate and Co-Metabolic Systems. Catalysts, 8(10), 472. https://doi.org/10.3390/catal8100472









