Abstract
ω-Aminododecanoic acid is considered as one of the potential monomers of Nylon 12, a high-performance member of the bioplastic family. The biosynthesis of ω-aminododecanoic acid from renewable sources is an attractive process in the polymer industry. Here, we constructed three artificial self-sufficient P450s (ArtssP450s) using CYP153A13 from Alcanivorax borkumensis and cytochrome P450 reductase (CPR) domains of natural self-sufficient P450s (CYP102A1, CYP102A5, and 102D1). Among them, artificial self-sufficient P450 (CYP153A13BM3CPR) with CYP102A1 CPR showed the highest catalytically activity for dodecanoic acid (DDA) substrate. This form of ArtssP450 was further co-expressed with ω-TA from Silicobacter pomeroyi and AlkJ from Pseudomonas putida GPo1. This single-cell system was used for the biotransformation of dodecanoic acid (DDA) to ω-aminododecanoic acid (ω-AmDDA), wherein we could successfully biosynthesize 1.48 mM ω-AmDDA from 10 mM DDA substrate in a one-pot reaction. The productivity achieved in the present study was five times higher than that achieved in our previously reported multistep biosynthesis method (0.3 mM).
1. Introduction
Production of biochemicals from vegetable oil derivatives (e.g., free fatty acids (FFAs)) have drawn great attention as an alternative means to develop sustainable and green production processes, known as biorefinery [1,2,3,4]. Fatty acids and fatty acid derivatives are employed for the production of various polymer intermediates and precursors with broad commercial and pharmaceutical implications, including cosmetics, adhesives, lubricants, surfactants, coatings, biofuels, and anticancer agents [5,6,7]. In particular, long chain ω-hydroxy fatty acids (ω-OHFAs) contain two functional groups—hydroxy and carboxyl groups—at their ends, and they can therefore be further oxidized to fatty aldehydes, dicarboxylic acids, and oxo-fatty acids. Recently, it was reported that dodecanoic acid (DDA) can be efficiently transformed to ω-amino dodecanoic acid (ω-AmDDA) by utilizing different enzymes like Cytochrome P450 monooxygenases, alcohol dehydrogenases (AlkJ), alkane hydroxylase AlkBGT, Baeyer–Villiger monooxygenases (BVMOs), esterases, and ω-transaminases (ω-TAs) [8,9,10]. ω-AmDDA is a very important monomer for the synthesis of Nylon 12, along with other aliphatic polyamides. Owing to its extraordinary heat-, abrasion-, chemical-, UV-, and scratch-resistance capabilities, Nylon 12 is frequently used as a coating agent on fuel and braking systems in most passenger cars [9,10]. Usually, Nylon 12 is industrially produced from the monomer ω-laurolactam through ring opening polymerization, the chemical synthesis of which is initiated by the trimerization of 1,3-butadiene originating from steam cracking (200–300 °C) of crude oil [9,10]. However, this method has raised many environmental concerns. Considering the “green” alternatives, synthesis of ω-amino dodecanoic acid from DDA is a potential approach to address this issue. Moreover, DDA shows relatively high solubility in water compared to other medium- to long-chain fatty acid substrates. Earlier, our group reported that CYP153A13 from Alcanivorax borkumensis SK2 efficiently transformed DDA to ω-hydroxy dodecanoic acid (ω-OHDDA) in an in vivo reaction [10,11,12].
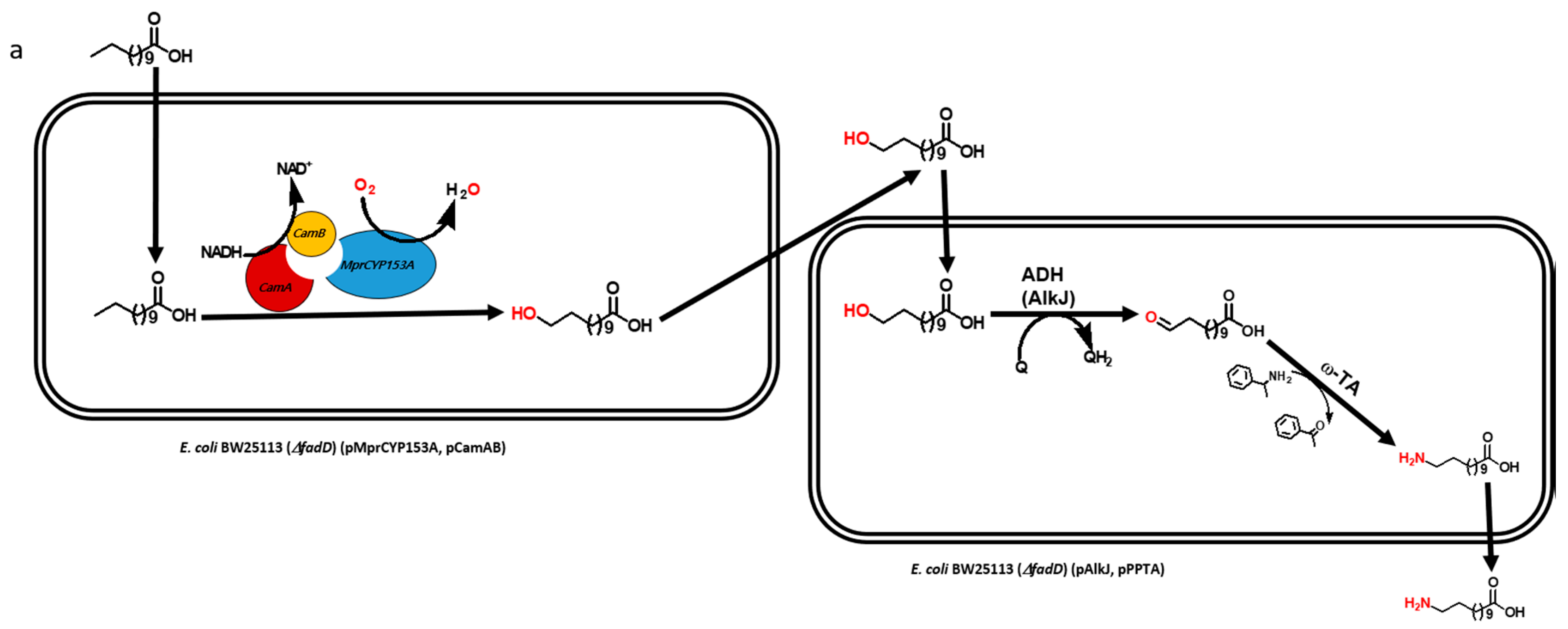
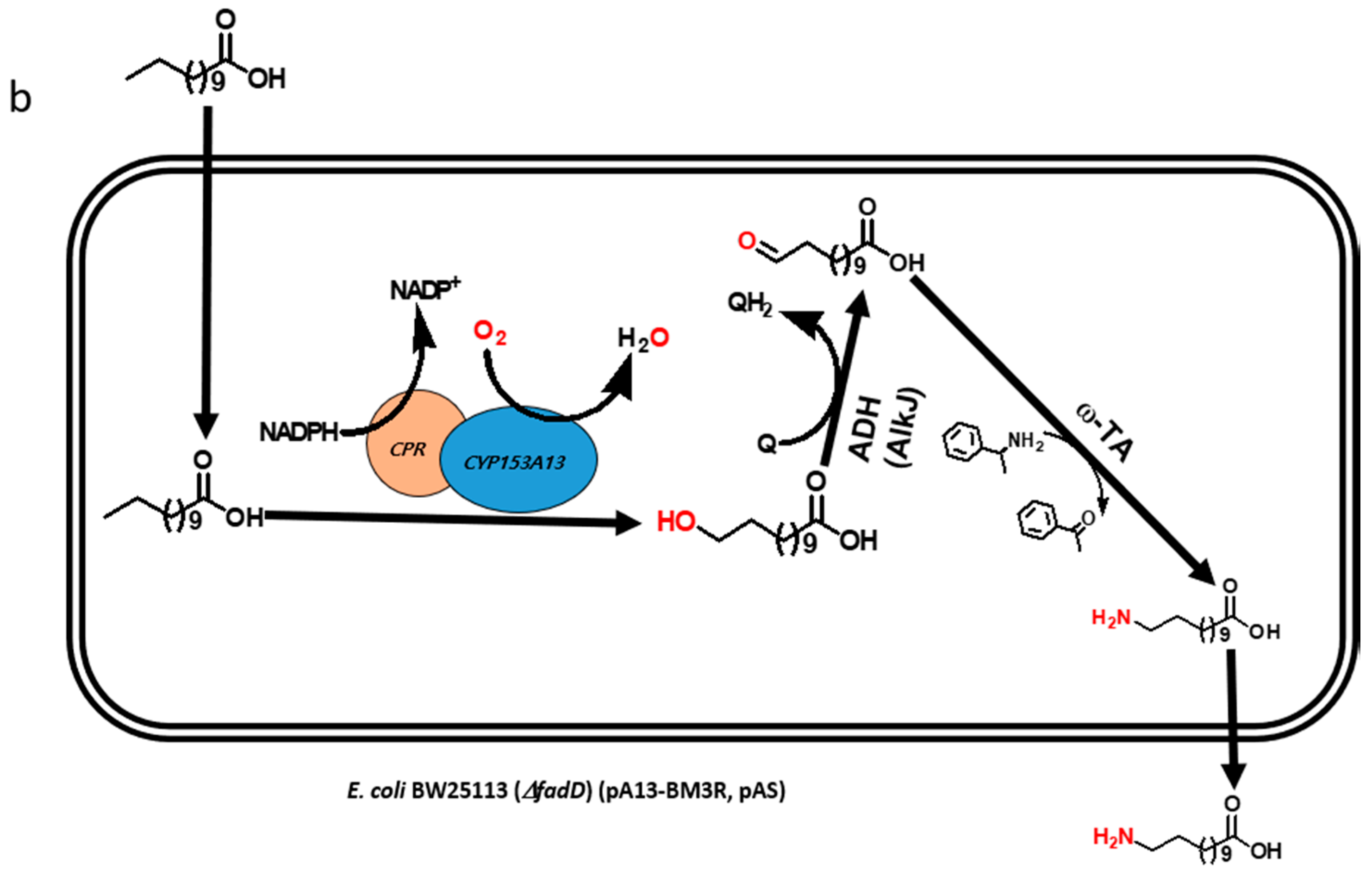
Multistep biocatalytic synthesis often negatively impacts practical industrial applications [13,14]. With increasing number of steps in biocatalytic reactions, the number of generated by-products also increases, many of which may be toxic to cells and make it difficult to separate desirable products. Moreover, the expected expression of multiple recombinant proteins and efficient bioconversion from multistep enzymatic reaction itself is a challenging task [10,15]. Hence, the use of lesser number of recombinant proteins is of immense importance for the biosynthesis of industrially important monomers in a one-pot reaction [16]. Recently, we reported the biosynthesis of ω-AmDDA using the cascade of novel CYP153A, AlkJ, and ω-TA enzymes [10]. In that study, we could achieve more than 87% conversion of ω-OHDDA to ω-AmDDA in a sequential biocatalytic reaction. However, the conversion was negligible in a one-pot reaction (Figure 1a). We therefore envisioned to biosynthesize ω-AmDDA in a single-cell, one-pot cascade reaction. In the present study, we changed the reaction pattern of CYP153A13 from a three-protein system to a single-protein system for ω-hydroxylation of DDA. As CYP153A belongs to the class I CYPs, three component systems are required for the activation of the enzyme (Figure S1). Two additional proteins—putidaredoxin reductase (CamA) and putidaredoxin (CamB)—are related to electron transfer system [10,11,12]. Self-sufficient P450s, belonging to Class VIII CYPs, are catalytically self-sufficient monooxygenase that contain a heme domain (P450 domain) and a flavin reductase domain (cytochrome P450 reductase (CPR) domain) on a single polypeptide chain ([11,12,17,18,19]; Figure S1). Recently published findings have shown that the fusion construct of CYP153A with the CPR domain of natural self-sufficient P450s results in the generation of the active form of the enzyme and successfully catalyzes a biocatalytic reaction [11,12,17]. In the present study, we also co-expressed the artificial self-sufficient P450s (ArtssP450s) with an alcohol dehydrogenase (AlkJ) from Pseudomonas putida and an omega transaminase (ω-TA) from Silicobacter pomeroyi for the synthesis of ω-AmDDA from DDA in a one-pot single-cell reaction (Figure 1b).
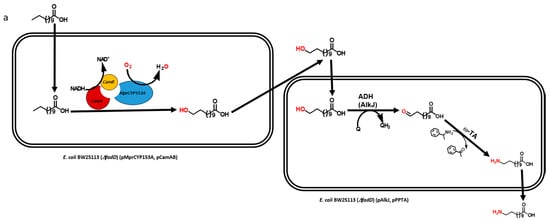
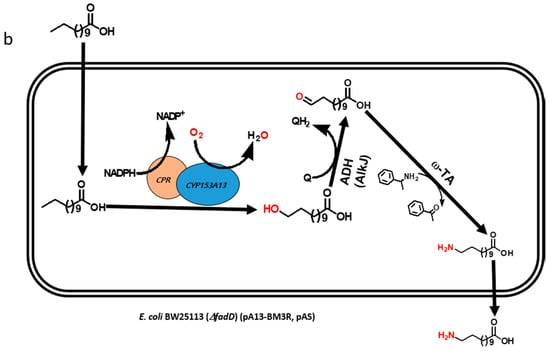
Figure 1.
Schematic diagram showing the biosynthesis of ω-amino dodecanoic acid (ω-AmDDA) from dodecanoic acid (DDA) using (a) three-protein system (previous study [10]) and (b) a single-protein system (this study) for ω-hydroxylation of DDA.
2. Results and Discussion
2.1. Construction and Expression of Artificial Self-Sufficient P450s (ArtssP450s)
CYP153A13 of Alcanivorax borkumensis (Abk) was cloned into NdeI and HindIII restriction sites of pET24ma plasmid vector. The HindIII restriction site was chosen to make it a common restriction site to fuse CYP153A13 with CPR domains of CYP102A1, CYP102A5, and CYP102D1 (Figures S2 and S3). After successful cloning of CYP153A13, CYP153A13-containing plasmid was used again to make artificial self-sufficient fusion construct of CYP153A13 and CPRs in the same vector. All three CPR domains were amplified; RE digested and ligated into HindIII and NotI sites for CYP102A5 and CYP102D1 and into HindIII and XhoI sites for CYP102A1. To remove HindIII sites within the CPR domain of CYP102A1, the 243th alanine residue DNA codon was changed from GCT to GCA by utilizing overlap extension PCR method. Next, all the ArtssP450s were confirmed by sequencing and nomenclatured newly as AbkCYP153A13BM3CPR (for CYP102A1 CPR), AbkCYP153A13Bc21CPR (for CYP102A5 CPR), and AbkCYP153A13SavCPR (for CYP102D1 CPR) on the basis of their gene source (See Materials and Method). All three different CPRs were chosen from three different natural self-sufficient P450s based on previous reports. Among them, BM3 is a well reported self-sufficient P450, and its CPR domain is frequently used for making artificial constructs [11,12,17]. The self-sufficient P450 of Bc21 (CYP102A5) has been reported to possess the highest turnover rates among P450s [19]. In addition, a codon-optimized Sav (CYP102D1) has been reported to show similar catalytic activity for saturated and unsaturated fatty acids as that of other members of the CYP102A family [20].
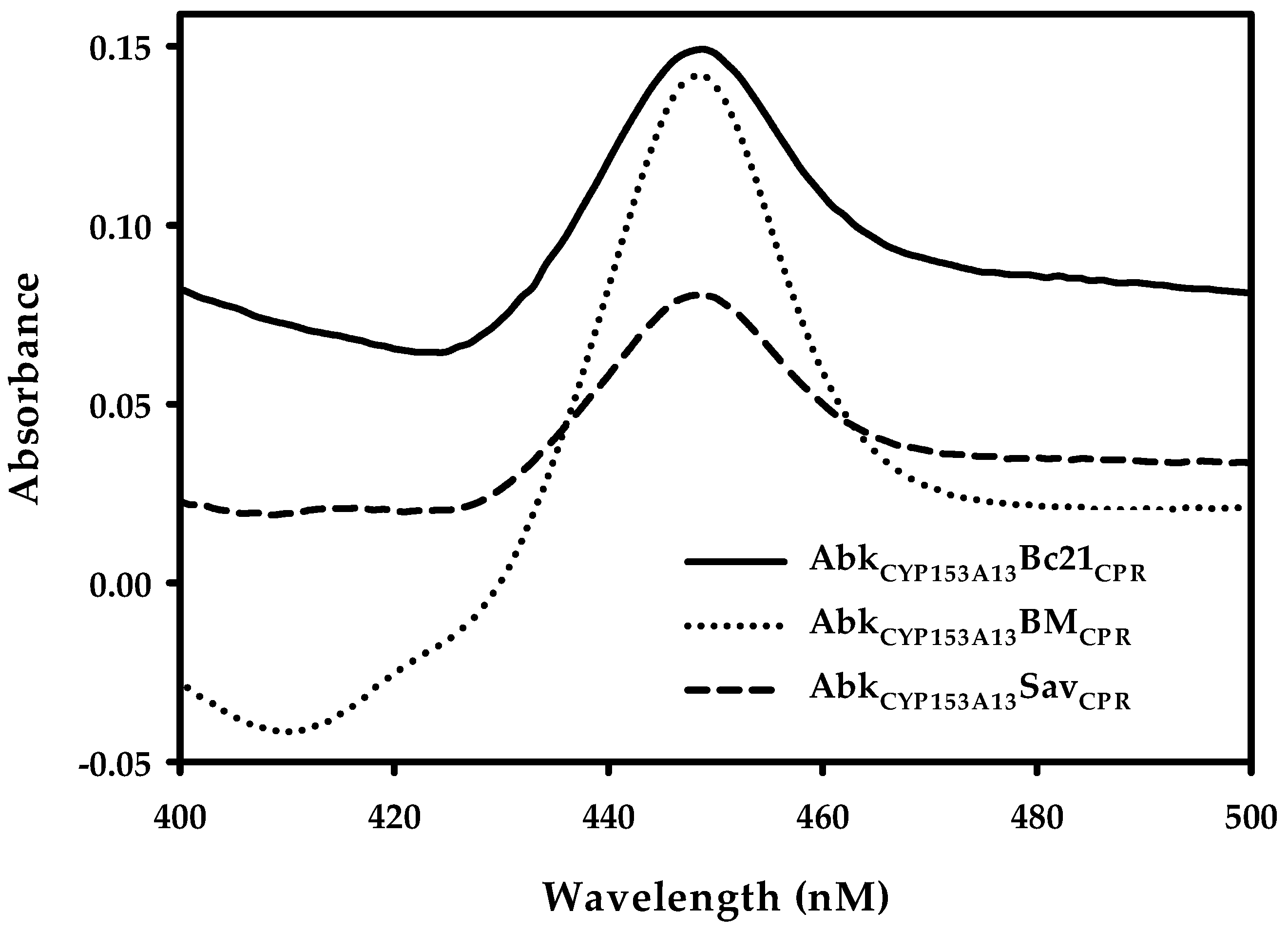
After successful construction of ArtssP450s, the expression of all recombinant proteins was tested at three different temperature (20, 25, and 30 °C) and three different isopropyl-thio-β-d-galactopyranoside (IPTG) concentrations—0.01, 0.1, and 0.5 mM—in terrific broth (TB) bacterial expression media. All three proteins showed better expression at 20 °C with 0.1 mM IPTG concentration. The protein expression was confirmed through SDS-PAGE gel analysis (Figure S4). SDS-PAGE analysis demonstrated clear protein expression band at expected size (~120 kDa). Additionally, the conventional carbon monoxide (CO)-differential spectral assay with sodium dithionite was used in cell-free extracts as a standard method to determine the concentration of active P450. The characteristic absorbance at 450 nm for CO complexed to the reduced ferrous state of P450 was observed (Figure 2), corroborating their active expression. The amount of active P450 was 1.51, 2.25, and 0.75 nmol mL−1 for AbkCYP153A13BM3CPR, AbkCYP153A13Bc21CPR, and AbkCYP153A13SavCPR, respectively. These results were well correlated with the reported titer range of the active P450 expression (0.6–2.33 nmol mL−1) in E. coli system [18,19,20,21]. It has also been reported that, in many cases, the co-expression of CPRs lead to formation of high titers of active P450 [21]. In the CO-binding analysis, AbkCYP153A13Bc21CPR construct seemed to be the most active, being nearly three times higher than AbkCYP153A13SavCPR and two times higher than AbkCYP153A13BM3CPR.
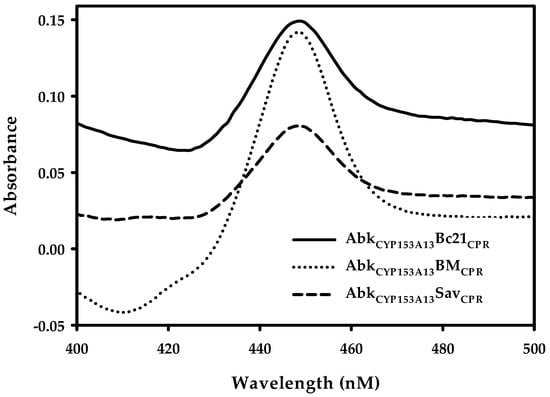
Figure 2.
Carbon monoxide (CO)-binding analysis of ArtssP450s. AbkCYP153A13, Alcanivorax borkumensis CYP153A13; AbkCYP153A13BM3CPR, AbkCYP153A13 fused with BM3 (CYP102A1) cytochrome P450 reductase (CPR); AbkCYP153A13Bc21CPR, AbkCYP153A13 fused with Bacillus cereus (CYP102A5) CPR; and AbkCYP153A13SavCPR, AbkCYP153A13 fused with Streptomyces avermitilis (CYP102D1) CPR.
2.2. Production of ω-OHDDA Using ArtssP450s
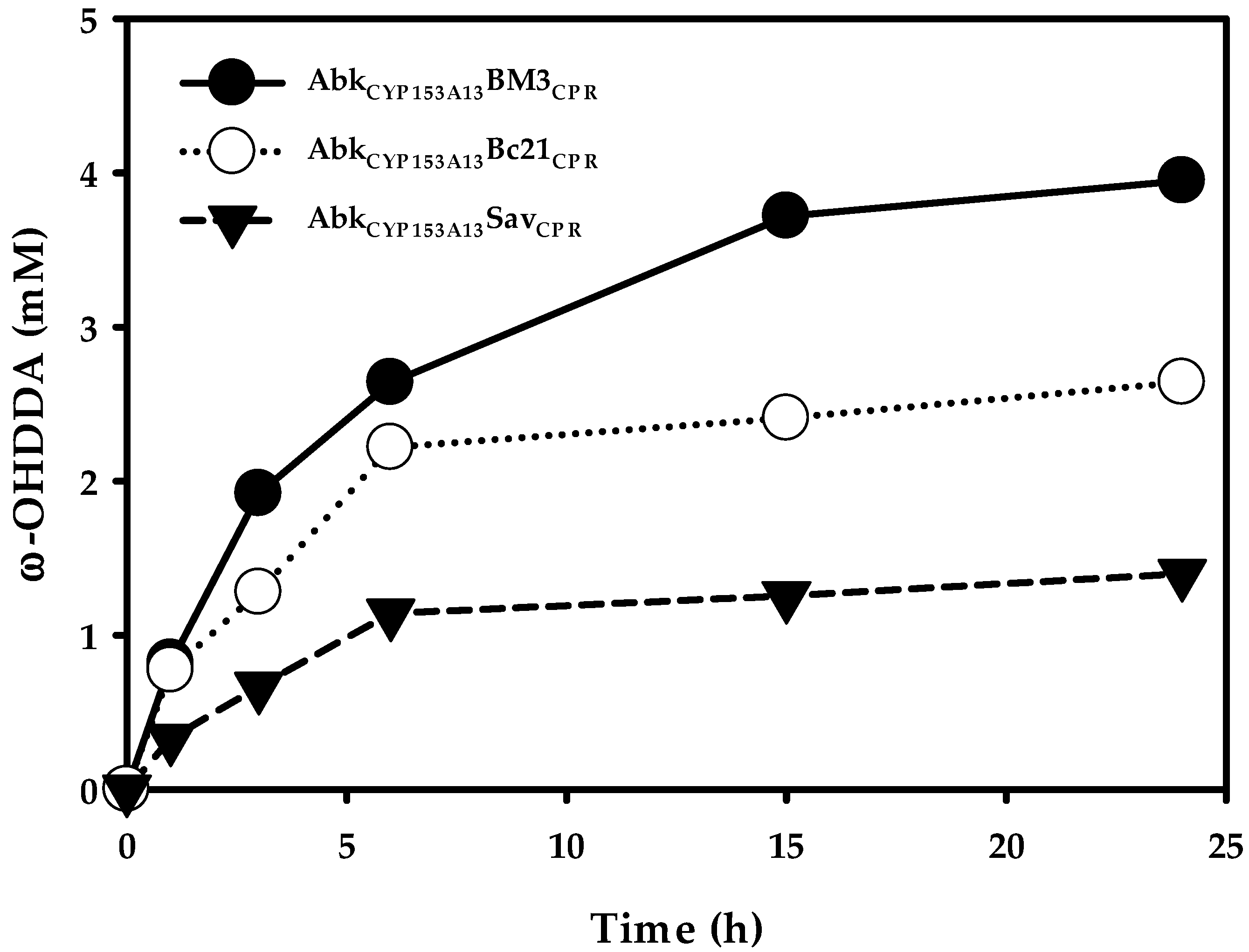
It has been reported that AbkCYP153A13 exhibits better ω-OHDDA productivity among CYP153A13, CYP153A33, and CYP153A35 in an in vivo biocatalytic reaction employing three protein system [11]. In order to know the ω-OHDDA productivity of newly constructed ArtssP450s in a single protein system, all three enzymes were tested with 10 mM DDA substrate in a whole-cell reaction. Additionally, a control whole-cell reaction was performed using AbkCYP153A13 in CamAB system. The initial specific rates of ω-hydroxylation were 0.10, 0.09, and 0.037 mmol/gCDW/h for AbkCYP153A13BM3CPR, AbkCYP153A13Bc21CPR, and AbkCYP153A13SavCPR, respectively. Accordingly, 24-h reactions resulted in 3.95, 2.64, and 1.40 mM ω-OHDDA, respectively (Figure 3). Although the initial specific rates of ω-hydroxylation and production of ω-OHDDA after 24 h were better for AbkCYP153A13 in CamAB system (0.16 mmol/gCDW/h and 6.21 mM, respectively) (Data not shown), we used single-protein system due to its obvious advantages. Among ArtssP450s, the initial specific rate of AbkCYP153A13BM3CPR and AbkCYP153A13Bc21CPR were very close. Unexpectedly, the highest productivity (3.95 mM) was observed in AbkCYP153A13BM3CPR. As AbkCYP153A13Bc21CPR showed the highest amount of active P450 in CO-binding spectrum analysis, it was estimated that the productivity would also be the highest for AbkCYP153A13Bc21CPR (Figure 2). However, the published findings have demonstrated that BM3 natural self-sufficient P450 (CYP102A1) shows the highest catalytic activity among all P450s [20]. Moreover, CamAB system showed better ω-OHDDA productivity than ArtssP450 systems developed in the present study. Despite these findings, the use of ArtssP450s for ω-OHDDA biosynthesis was of utmost importance in consideration of the one-pot biocatalytic cascade reaction in order to get the final product of this study, i.e., ω-AmDDA (Figure 1b). As AbkCYP153A13BM3CPR system showed the highest ω-OHDDA productivity among all the ArtssP450 constructs, it was further used to produce ω-AmDDA from DDA (Figure 1).
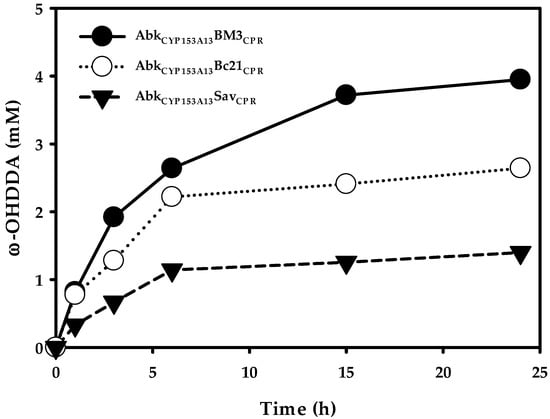
Figure 3.
ω-hydroxy dodecanoic acid (ω-OHDDA) production by ArtssP450s. Reaction conditions: substrate concentration, 10 mM; volume, 10 mL in 100 mL flask; temperature, 30 °C; cell type, BW25113 (ΔfadD, DE3) containing pA13-BM3R, pA13-Bc21R, pA13-SavR; cell OD600: 30 and buffer, 100 mM potassium phosphate buffer (pH 7.5) with 1% (w/v) glucose.
2.3. Biosysnthesis of ω-AmDDA in One-Pot Reaction
2.3.1. Co-expression of AbkCYP153A13BM3CPR with AlkJ and Sp ω-TA
The production of ω-AmDDA from DDA in a one-pot reaction requires co-expression of AbkCYP153A13BM3CPR with AlkJ and ω-TA. In this reaction, the DDA substrate is initially converted to ω-OHDDA by AbkCYP153A13BM3CPR. AlkJ then catalyzes the transformation of ω-OHDDA to ω-OxoDDA, which is further transaminated by ω-TA to yield ω-AmDDA. Therefore, AbkCYP153A13BM3CPR was co-expressed with AlkJ and ω-TA at 20 °C using 0.1 mM IPTG induction in TB media. The expression of all three proteins in the soluble form was confirmed by SDS-PAGE gel analysis (Figure 4). Next, the co-expressed cells were used in a whole-cell reaction to produce ω-AmDDA from DDA in a one-pot reaction. The reaction was initiated by adding 5 mM DDA (stock in dimethyl sulfoxide (DMSO), 5% (v/v) final concentration), 0.1 mM pyridoxal-5′-phosphate (PLP), and 40 mM benzylamine using co-expressed cells (0.3 gDCW/mL) in 100 mM Tris-HCl buffer. The reaction mixture was incubated at 35 °C and 200 rpm. After 6 h, 0.8 and 0.3 mM ω-OHDDA and ω-AmDDA, respectively, were detected (data not shown). This can be explained by the fact that AbkCYP153A13BM3CPR transformed DDA to ω-OHDDA. AlkJ converted this ω-OHDDA to ω-OxoDDA, which was utilized by Sp ω-TA to form ω-AmDDA. Albeit with low conversion percentage, the successful biotransformation of DDA to ω-AmDDA encouraged us to further improve the productivity by optimizing different parameters.
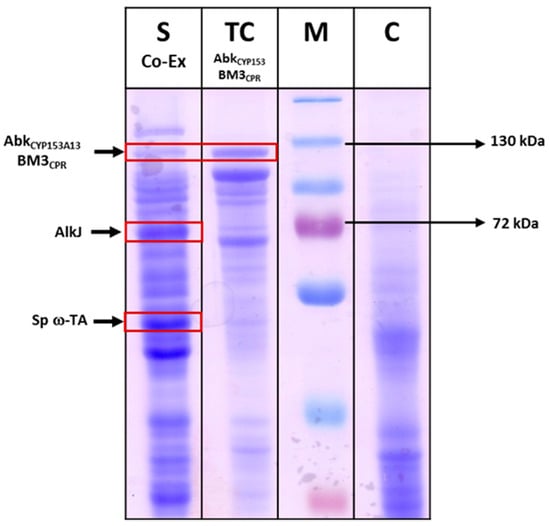
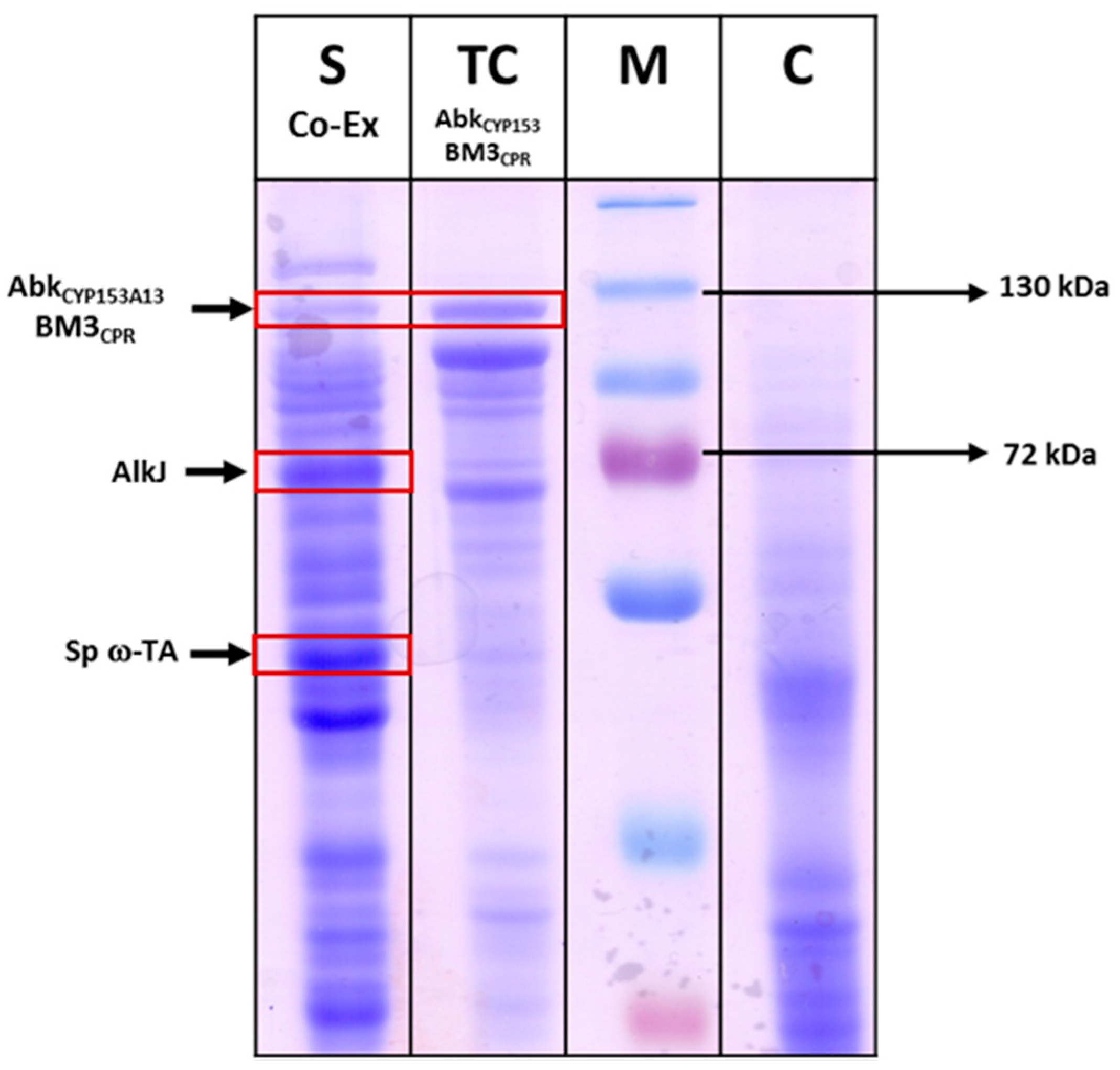
Figure 4.
SDS-PAGE analysis of recombinant E. coli expressing AbkCYP153A13BM3CPR (~120 kDa), AlkJ (~61.2 kDa), and Sp ω-TA (~52 kDa) co-expression. C: control (E. coli BW25113(DE3) ΔfadD cells without any plasmid were used as control); M: Marker; TC: total cells; SP: soluble proteins. Protein expression was carried out using 0.1 mM isopropyl-thio-β-d-galactopyranoside (IPTG) at 20 °C and 200 rpm for 16 h.
2.3.2. Effect of Benzylamine (BA) on ω-OHDDA Productivity
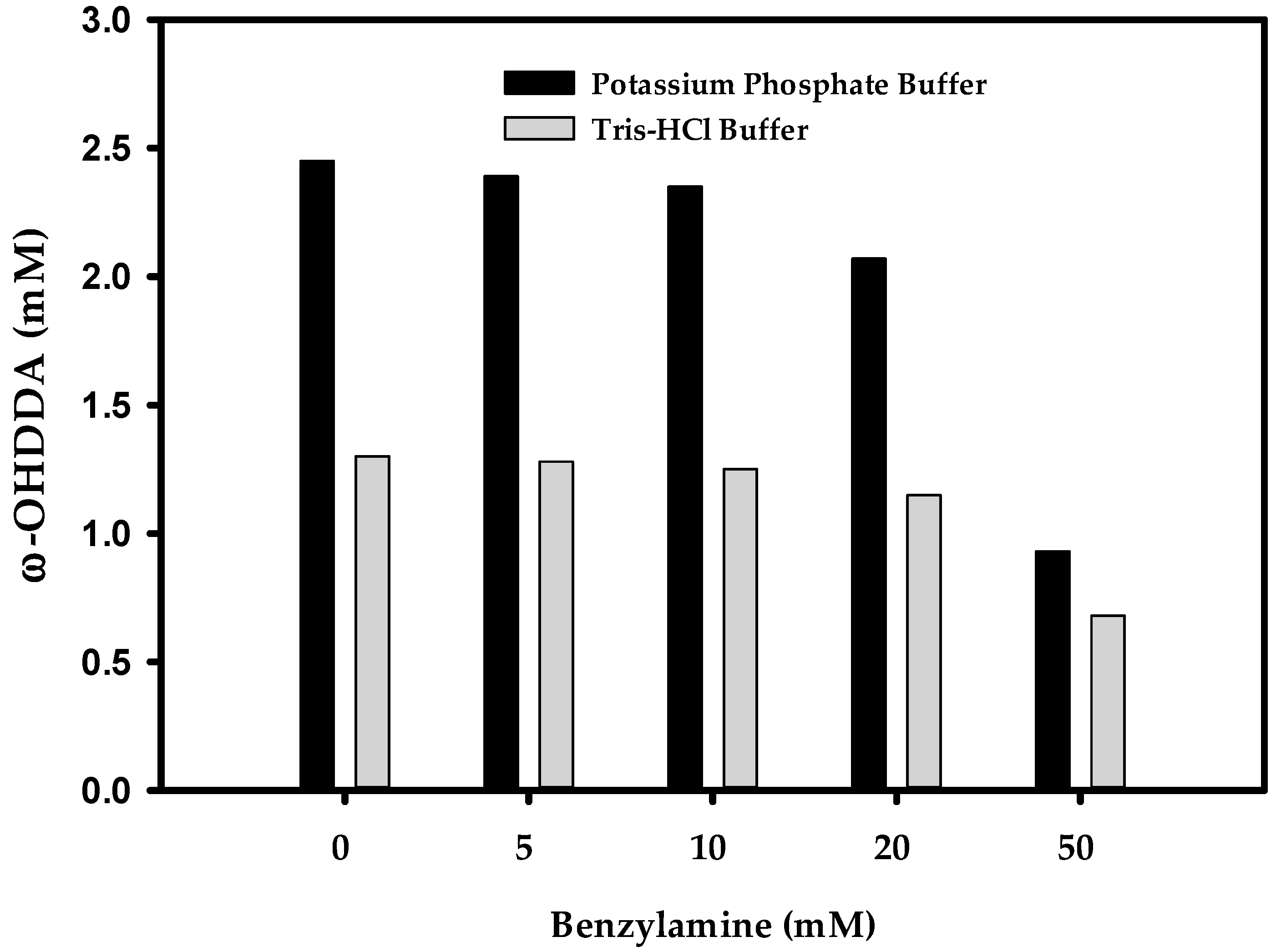
The selection of a suitable amino donor is of immense importance to enhance the yield of ω-TA reaction. Organic syntheses by ω-TAs may suffer from unfavorable reaction equilibrium due to insufficient amount of amino donor [16]. To maximize the productivity of ω-TAs, shifting the equilibrium to the product using higher concentration of amino donor is extremely important [22]. Previously, while synthesizing ω-AmDDA from ω-OHDDA using AlkJ and Sp ω-TA, we found benzylamine (BA) as the best amino donor [10]. It has been reported that the performance of P450 employed in the amination cascade is influenced by the amino donor used [23]. Therefore, it was presumed that the productivity of ω-OHDDA might be hindered due to excess amount of BA as amino donor for ω-TA in the one-pot reaction demonstrated herein. Before optimizing ω-AmDDA productivity, the biosysnthesis of ω-OHDDA by AbkCYP153A13BM3CPR was tested in 100 mM (pH 7.5) potassium phosphate buffer and Tris-HCl buffer using five different concentrations of BA. It was demonstrated that 0–5 mM BA had negligible effect on the production of ω-OHDDA in both types of buffers, but the productivity was better in the phosphate buffer. Moreover, 50 mM BA had remarkable effect on ω-OHDDA in both the cases (Figure 5). This could be explained by the interception of the hydrogen-borrowing step (NADP+/NADPH) of the cofactors in endogenous E. coli enzyme systems, which may lead to imbalanced regeneration of the cofactors [23].
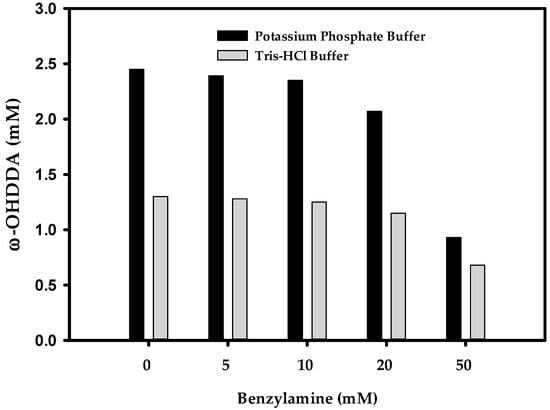
Figure 5.
Effect of BA on ω-OHDDA productivity. Reaction conditions: substrate concentration: 10 mM; volume: 10 mL in 100 mL flask; temperature: 35 °C; cell type: BW25113 (ΔfadD, DE3) containing pA13-BM3R; cell OD600: 30 and buffer, 100 mM (pH 7.5) with 1% (w/v) glucose; reaction time: 5 h.
We have previously reported that ω-TA enzyme is more active in Tris-HCl buffer with alkaline pH [10]. In the present study, it was revealed that ArtssP450s did not show any activity for ω-hydroxylation of DDA in an alkali pH (>8) (data not shown). Therefore, further optimization of ω-AmDDA production in a one-pot reaction was continued in 100 mM Tris-HCl buffer (pH 7.5), which seemed to be a convenient reaction condition for all the enzymes.
2.3.3. Optimization of ω-AmDDA Biosynthesis
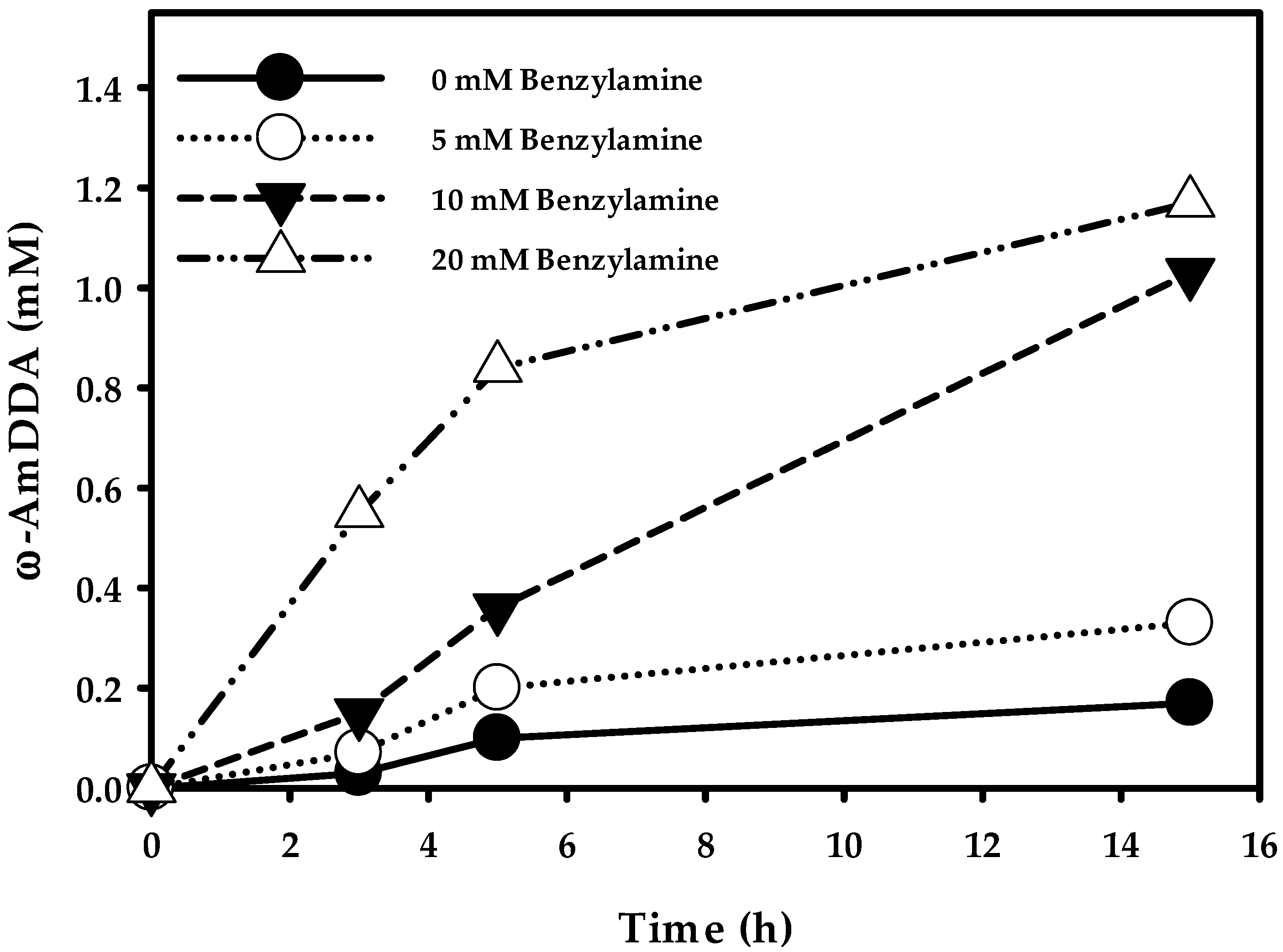
Finally, in order to check the optimum productivity of ω-AmDDA in a one-pot reaction, cells (0.3 gCDW/mL) co-expressing AbkCYP153A13BM3CPR, AlkJ, and Sp ω-TA were used in various concentration of BA (0–50 mM) in a whole-cell reaction. The reaction was initiated by adding 10 mM DDA (stock in DMSO, 5% (v/v) final concentration), 0.1 mM PLP, and 20 mM benzylamine. The reactants were incubated at 35 °C and 200 rpm. After 3 h of whole-cell reaction, the amount ω-AmDDA was 0.03, 0.07, 0.15, and 0.55 mM when the BA was used in the reaction was 0, 5, 10, and 20 mM, respectively. In particular, it was observed that the amount of ω-OHDDA after 3 h was 1.21, 1.10, 0.95, and 0.82 mM, respectively, with the subsequent concentration of used BA. The production of both ω-AmDDA and ω-OHDDA increased up to 5 h, and the amount of ω-AmDDA was 0.10, 0.20, 0.36, and 0.84 mM, respectively, whereas the amount of ω-OHDDA was 2.35, 2.22, 1.98, and 1.64 mM, respectively, for the corresponding amount of BA used. Interestingly, the biosynthesis of ω-AmDDA was increasing over the 15 h reaction period (Figure 6), while the production of ω-OHDDA dropped after 5 h. The whole-cell reaction yielded 0.17, 0.33, 1.03, and 1.17 mM of ω-AmDDA and 1.87, 1.61, 1.12, and 0.56 mM of ω-OHDDA after 15 h when 0, 5, 10, and 20 mM BA was used, respectively (Figure 6). The present study demonstrated that the use of 20 mM BA as an amino donor in a one-pot whole-cell reaction gave the best productivity for the synthesis of ω-AmDDA from DDA substrate. Moreover, the best whole-cell reaction condition was continued up to 24 h. It was found that 1.48 mM ω-AmDDA was biosynthesized from 10 mM DDA, which is five times more than our last report [10]. Whole-cell reaction of E. coli cells expressing MprCYP153A/CamAB for ω-hydroxylation of DDA and E. coli cells expressing AlkJ/mll1207 ω-TA resulted in the generation of ~0.3 mM ω-AmDDA from DDA in a 24 h reaction [10].
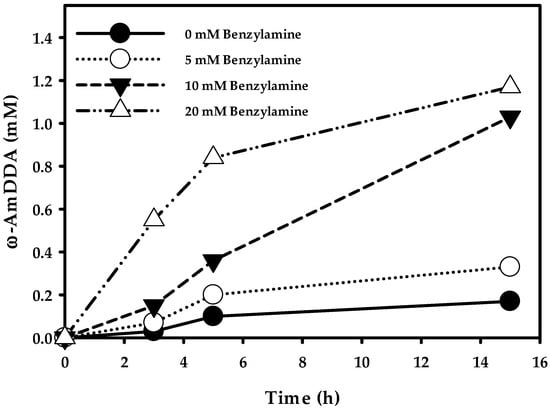
Figure 6.
Biosysnthesis of ω-AmDDA from DDA using co-expressed AbkCYP153A13BM3CPR, AlkJ, and Sp ω-TA enzymes. Reaction conditions: substrate concentration: 10 mM; volume: 10 mL in 100 mL flask; temperature: 35 °C; cell type, BW25113 (ΔfadD, DE3) containing pA13-BM3R and pAS; cell OD600: 30; cofactor: 0.1 mM pyridoxal-5′-phosphate (PLP); amino donor: 20 mM benzylamine and buffer, 100 mM Tris-HCl buffer (pH 7.5) with 1% (w/v) glucose.
It has been reported that AlkJ from Pseudomonas putida GPo1 shows thermal denaturation at moderate temperatures, and Tstab—the temperature where the enzyme shows 50% residual activity—was found to be 34 °C [24]. Thus, biotransformations carried out at lower temperatures could have resulted in improved overall yield in the present study. However, the major challenge in one-pot biotransformations employing more than one protein—as demonstrated in the present study—is the satisfactory performance of all proteins involved. In these multiprotein biotransformations, not every protein can be optimally functional in the given set of reaction parameters. Hence, there is always a compromise involved in yield of final product and the optimal performance of the proteins employed in the multiprotein biotransformations. In the addition, the final outcome of multiprotein biotransformations is severely compromised when proteins exhibit differential performance at specific reaction conditions, such as temperature and/or reaction pH. It should also be noted that whole-cell biocatalysts are more stable than their purified counterparts. Nevertheless, in our previously reported study [10], it was observed that all of the initially added ω-OHDDA (5 mM) was consumed within 1 h when whole-cell reaction was performed at 35 °C, implying that AlkJ activity was enough to carry out transformation of ω-OHDDA.
It has been well reported that the fadL is required for the transport of long chain fatty acids in E. coli [25,26]. Although overexpression of fadL was one of the possible approaches to improve the mass transfer of DDA substrate across cell membrane in the present study, it could have adversely affected the expression of other proteins. As expression of a recombinant protein may impart a metabolic burden on the host microorganism [27], the primary object of multiprotein, one-pot biotransformations should be the use of lesser number of recombinant proteins to achieve the synthesis of a desired product. Recently, Janßen et al. [28] reported the improved transfer of long-chain fatty acids in fadL-overexpressed E. coli cells. However, a recently reported study by our group [10] demonstrated that E. coli cells can satisfactorily uptake DDA without overexpression of fadL.
3. Materials and Methods
3.1. Chemicals and Media
All chemicals such as DDA, ω-OHDDA, ω-AmDDA, dimethyl sulfoxide (DMSO), IPTG, 5-aminolevulinic acid (5-ALA), N,O-Bis(trimethylsilyl)-trifluoroacetamide (BSTFA), N-Methyl-N-(trimethylsily) trifluoroacetamide (MSTFA), PLP, benzylamine, and pyridine were purchased from Sigma-Aldrich (St. Louis, MO, USA). Chloroform was obtained from Junsei (Tokyo, Japan). Bacteriological agar, Luria-Bertani (LB) broth, and terrific broth (TB) media were bought from BD Difco (Franklin Lakes, NJ, USA). All chemicals used in this study were of analytical grade.
3.2. Artificial Self-Sufficient P450 Construction and Gene Manipulation
All the bacterial strains and plasmid vectors used in this study are listed in Table 1. Alcanivorax borkumensis SK2 (KACC no. 12864) was procured from the Korean Agricultural Culture Collection (KACC, Jeonju, Korea). Genomic DNA was extracted from lyophilized commercial cell stock using a kit (G-Spin™ Genomic DNA Extraction Kit for bacteria (iNtRON Biotechnology, Suwon, Korea)). The gene encoding CYP153A13 (GI: CAL15649.1) from A. borkumensis SK2 was amplified by PCR with oligonucleotides (Table S1). After restriction digestion and ligation with T4 DNA ligase, the plasmid was utilized to transform competent E. coli DH5α cells. Successful cloning was confirmed by DNA sequencing. Construction of artificial self-sufficient fusion proteins (pA13-BM3R, pA13-Bc21R, and pA13-SavR) with CYP153A13 and the reductase domains (CPRs) of self-sufficient P450s (CYP102A1 from Bacillus megaterium, CYP102A5 from Bacillus cereus, and CYP102D1 from Streptomyces avermitilis) were performed following previously described procedures [18]. CPR domains of self-sufficient P450s were analyzed using online bioinformatics tools Pfam 31.0 site (http://pfam.xfam.org/). Gene synthesis and codon optimization for E. coli codon preferences was performed by Cosmo Genetech (Cosmo Genetech, Seoul, Korea).

Table 1.
Plasmids and strains used in this study.
3.3. Expression of Enzymes
E. coli BW25113 (DE3) ∆fadD strain [10] was utilized for the biotransformation studies, wherein fatty acid degrading β-oxidation pathway was blocked. Plasmid DNA were transformed into host strains using standard heat shock method. Transformants were selected based on their antibiotic resistance [30]. Transformants were grown overnight at 37 °C in 10 mL LB medium containing 50 μg/mL of kanamycin (for pA13-BM3R, for pA13-Bc21R, and for pA13-SavR) and/or 100 μg/mL ampicillin (for pAS, pAlkJ, and pPPTA). The seed cultures were added to expression flask into 1/3 ratio. All artificial self-sufficient P450 protein expression were carried out in 200 mL of Terrific Broth in a 1 L flask. They were cultured at 37 °C until cell concentration reached an OD600 of 0.6–0.8. The induction was performed by adding 0.1 mM IPTG, 0.5 mM 5-ALA as heme precursor, and 0.1 mM FeSO4, and cells were grown at 20 °C for 16 h. The expression of Sp ω-TA/AlkJ with or without artificial self-sufficient P450 was carried out in 200 mL of LB medium in 1 L flasks. Induction was performed by adding 0.1 mM IPTG, and cells were allowed to grow for 16 h at 20 °C.
After 16 h, cells were harvested by centrifugation (4000 rpm, 20 min, 4 °C), washed with phosphate-buffered saline (PBS), and resuspended in 100 mM potassium phosphate buffer (pH 7.5) containing 1% (w/v) glucose.
3.4. CO-Binding Assay and Gel Electrophoresis
The expressed proteins were subjected to SDS-PAGE and spectrophotometric analysis in order to measure the CO-binding activity. UV absorption spectra of CO-bound artificial self-sufficient P450 proteins after sodium dithionite reduction were measured by Cary 100 UV-Vis spectrometry (Agilent Technologies, CA, USA) by wavelength scan from 400 to 500 nm. The concentration of P450 was measured by CO-binding affinity using an extinction coefficient of 91.9 mM−1 cm−1 at 450 nm. SDS-PAGE (Bio-Rad Laboratories, Inc, Hercules, CA, USA.) was carried out with 12% polyacrylamide gel as described elsewhere [31]. Proteins were visualized by Coomassie® brilliant blue R-250 staining.
3.5. Biotransformation
Biotransformation of FFAs to ω-OHFAs was performed according to the previously reported method [10]. For the biotransformation of ω-OHDDA acid to ω-AmDDA, E. coli BM-AS (Table 1) cotransformed strain was grown and harvested as described above. The cell pellets were resuspended in 100 mM Tris-HCl buffer (pH 7.5 and 8.0), and biotransformation (300 mgCDW/mL) was performed at 35 °C at 150 rpm in a shaking incubator following our previous published protocols [10]. The whole-cell reaction was started by the addition of ω-OHDDA (stock in DMSO, 5% (v/v) final concentration).
To assess the biotransformation of DDA to ω-OHDDA by ArtssP450s, whole cell reaction was performed according to the previously reported method [10]. Briefly, induced cells expressing ArtssP450s in a single protein system were grown for 16 h at 20 °C. Cells were harvested by centrifugation (4000 rpm, 20 min, 4 °C) and washed with PBS. To initiate the biotransformation, resting cells (300 mgCDW/mL), DDA (10 mM; stock in DMSO, 5% (v/v) final concentration) and buffer (Potassium phosphate; 100 mM, pH 7.5) with 1% (w/v) glucose were added to a 100 mL flask to a final volume of 10 mL. This reaction mixture was incubated at 30 °C and 200 rpm for 24 h. For one-pot biotransformation of DDA to ω-AmDDA, E. coli BM-AS (Table 1) cotransformed strain was grown and harvested as described above. Biotransformation was initiated by adding resting cells (300 mgCDW/mL), DDA (10 mM; stock in DMSO, 5% (v/v) final concentration) and buffer (Tris-HCl; 100 mM, pH 7.5 and 8.0) to a 100 mL flask to a final volume of 10 mL. This reaction mixture was incubated at 35 °C and 150 rpm for 24 h.
3.6. Product Identification and Quantification
Whole-cell reactions of both the biotransformations, i.e., DDA to ω-OHDDA and DDA to ω-AmDDA, were stopped and acidified with 6 M HCl to pH 2.0. The substrates and products of DDA biotransformation were extracted with an equal volume of chloroform (200 µL) after vigorous vortexing for 1 min (Table S2). After centrifugation, the extracted sample in chloroform (bottom layer) was transferred to a new microcentrifuge tube for derivatization. These samples were transformed to their trimethylsilyl (TMS) derivatives by incubation with an excess of BSTFA at 50 °C for 20 min. In the case of biotransformation generating ω-AmDDA, the acidified reaction samples (with 6 M HCl) were centrifuged, and the supernatant was dried in a vacuum concentrator. After complete drying, the supernatant was dissolved again to the original volume with pyridine. The sample was then mixed with an equal volume of MSTFA by vigorous vortexing for 1 min and converted to the TMS derivatives by incubation at 50 °C for 20 min.
Analytical conditions for free fatty acids and their derivatives using gas chromatography are well established and reported in our previously published reports [4,7,10,11,12,20]. Quantitative analysis was performed using a gas chromatography instrument with a flame ionization detector (GC/FID) fitted with an AOC-20i series auto sampler injector (GC 2010 plus Series, Shimadzu Scientific Instruments, Kyoto 604-8511, Japan). Two-microliter samples were inserted by split mode (split ratio 20:1) and examined by means of a nonpolar capillary column (5% phenyl methyl siloxane capillary 30 m × 320 μm i.d., 0.25-μm film thickness, HP-5). The oven temperature program for fatty acid analysis was 50 °C for 1 min, an increase by 10 °C/min to 250 °C, and hold for 10 min. The inlet temperature was 250 °C, and the detector temperature was 280 °C. The flow rate of the carrier gas (He) was 1 mL/min, and the flow rates of H2, air, and He in FID were 45 mL/min, 400 mL/min, and 20 mL/min, respectively. For the analysis of DDA and ω-OHDDA, commercial decanoic acid was used as internal standard. Internal standard was added after stopping the reaction and before the centrifugation step of chloroform extraction. For ω-AmDDA analysis, the oven temperature program was modified. The initial oven temperature was 90 °C, which was then increased by 15 °C/min to 250 °C, holding at this temperature for 5 min. Internal standard was not used. As chloroform extraction was not carried out in this analysis, there was presumably no loss of product and substrate as in the chloroform extraction. Pyridine was used for dilution of the product after vacuum evaporation of buffer–water, and dilution factor was considered while quantifying the product yield. Products of the biotransformations were confirmed by comparing the GC chromatograms with authentic references (Figures S5–S8).
4. Conclusions
In summary, we constructed artificial self-sufficient P40s by fusing CYP153A13 with various CPR domains—CYP102A1, CYP102A5, and CYP102D1—and tested them for the hydroxylation of DDA to ω-OHDDA. Furthermore, a single-cell system was generated for the biosynthesis of Nylon 12 monomer ω-AmDDA in a one-pot reaction by co-expressing the best artificial self-sufficient P40 generated (i.e., AbkCYP153A13BM3CPR), AlkJ, and Sp ω-TA enzymes. This highly efficient biocatalytic cascade produced ω-AmDDA in an easy and cost-effective way. It is worth emphasizing that the productivity of the biocatalytic cascade reported herein was five times higher than that of our previously reported sequential cascade method [10].
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4344/8/9/400/s1, Figure S1: Electron transportation system bacterial CYP. A. class I, and B. class VIII; Figure S2: Co-expression plasmid diagram for the synthesis of ω-AmDDA; Figure S3: Schematic diagram of ArtssP450s construction strategy using restriction sites into pET24ma vector; Figure S4: SDS-PAGE analysis of recombinant E. coli expressing ArtssP450s (~120 kDa). C: control; M: marker; TC: total cells; SP: soluble proteins. Protein expression was carried out using 0.1 mM IPTG, 0.1 mM FeSO4, and 0.5 mM 5-ALA at 20 °C and 170 rpm; Figure S5: GC chromatogram of the chemical standard of DDA. The peak at 13.5 min is the internal standard decanoic acid; Figure S6: GC chromatogram of the chemical standard of ω-AmDDA. The peak at 13.5 min is the internal standard decanoic acid; Figure S7: GC chromatogram of authentic ω-AmDDA and reaction mixture of DDA to ω-AmDDA; Figure S8: GC/MS analysis of authentic ω-AmDDA and reaction mixture of DDA to ω-AmDDA; Table S1: Primers used for the construction of ArtssP450s; Table S2: Retention time of the substrates and products by gas chromatography.
Author Contributions
H.Y., and T.C. designed the experiments of the project. M.M.A. carried out the research works as part of his PhD project. H.Y. supervised the whole studies reported in the manuscript. M.M.A. wrote the manuscript. M.D.P. revised this manuscript. H.J., and S.S. assisted in experimental tools.
Funding
This research was funded by the Ministry of Trade, Industry and Energy of South Korea (MOTIE, Korea) under the Industrial Technology Innovation Program, Grant Nos. 10062550 and 10076343.
Conflicts of Interest
The authors declare no financial or commercial conflict of interest.
References
- Steen, E.J.; Kang, Y.; Bokinsky, G.; Hu, Z.; Schirmer, A.; McClure, A.; del Cardayre, S.B.; Keasling, J.D. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 2010, 463, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Yang, J.E.; Ha, J.Y.; Chae, T.U.; Shin, J.H.; Gustavsson, M.; Lee, S.Y. Bio-based production of monomers and polymers by metabolically engineered microorganisms. Curr. Opin. Biotechnol. 2015, 36, 73. [Google Scholar] [CrossRef] [PubMed]
- Zorn, K.; Oroz-Guinea, I.; Brundiek, H.; Bornscheuer, U.T. Engineering and application of enzymes for lipid modification, an update. Prog. Lipid Res. 2016, 63, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, M.; Sung, S.; Jeon, H.; Patil, M.D.; Chung, T.; Yun, H. Biosynthesis of medium- to long-chain α,ω-diols from free fatty acids using CYP153A monooxygenase, carboxylic acid reductase, and E. coli endogenous aldehyde reductases. Catalysts 2018, 8, 4. [Google Scholar] [CrossRef]
- Metzger, J.O.; Bornscheuer, U. Lipids as renewable resources: Current state of chemical and biotechnological conversion and diversification. Appl. Microbiol. Biotechnol. 2006, 71, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Ness, J.E.; Xie, W.; Zhang, X.; Minshull, J.; Gross, R.A. Biosynthesis of monomers for plastics from renewable oils. J. Am. Chem. Soc. 2010, 132, 15451–15455. [Google Scholar] [CrossRef] [PubMed]
- Durairaj, P.; Malla, S.; Nadarajan, S.P.; Lee, P.-G.; Jung, E.; Park, H.H.; Kim, B.-G.; Yun, H. Fungal cytochrome P450 monooxygenases of Fusarium oxysporum for the synthesis of ω-hydroxy fatty acids in engineered Saccharomyces cerevisiae. Microb. Cell Fact. 2015, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-W.; Lee, J.-H.; Bornscheuer, U.T.; Park, J.-B. Microbial Synthesis of Medium-Chain α,ω-Dicarboxylic Acids and ω-Aminocarboxylic Acids from Renewable Long-Chain Fatty Acids. Adv. Synth. Catal. 2014, 356, 1782–1788. [Google Scholar] [CrossRef]
- Ladkau, N.; Assmann, M.; Schrewe, M.; Julsing, M.K.; Schmid, A.; Bühler, B. Efficient production of the Nylon 12 monomer ω-aminododecanoic acid methyl ester from renewable dodecanoic acid methyl ester with engineered Escherichia coli. Metab. Eng. 2016, 36, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, M.M.; Jeon, H.; Nadarajan, S.P.; Chung, T.; Yoo, H.-W.; Kim, B.-G.; Patil, M.D.; Yun, H. Biosynthesis of the Nylon 12 monomer, ω-aminododecanoic acid with novel CYP153A, AlkJ, and ω-TA enzymes. Biotechnol. J. 2018, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.; Park, B.G.; Ahsan, M.M.; Kim, J.; Yun, H.; Choi, K.Y.; Kim, B.G. Production of ω-hydroxy palmitic acid using CYP153A35 and comparison of cytochrome P450 electron transfer system in vivo. Appl. Microbiol. Biotechnol. 2016, 100, 10375–10384. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.; Park, B.G.; Yoo, H.-W.; Kim, J.; Choi, K.-Y.; Kim, B.-G. Semi-rational engineering of CYP153A35 to enhance ω-hydroxylation activity toward palmitic acid. Appl. Microbiol. Biotechnol. 2018, 102, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Patil, M.D.; Grogan, G.; Yun, H. Biocatalyzed C-C bond formation for the production of alkaloids. ChemCatChem 2018. [Google Scholar] [CrossRef]
- Patil, M.D.; Dev, M.J.; Shinde, A.S.; Bhilare, K.D.; Patel, G.; Chisti, Y.; Banerjee, U.C. Surfactant-mediated permeabilization of Pseudomonas putida KT2440 and use of the immobilized permeabilized cells in biotransformation. Process Biochem. 2017, 63, 113–121. [Google Scholar] [CrossRef]
- Lin, B.; Tao, Y. Whole-cell biocatalysts by design. Microb. Cell Fact. 2017, 16, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Patil, M.D.; Grogan, G.; Bommarius, A.; Yun, H. Recent advances in ω-transaminase-mediated biocatalysis for the enantioselective synthesis of chiral amines. Catalysts 2018, 8, 254. [Google Scholar] [CrossRef]
- Honda Malca, S.; Scheps, D.; Kuhnel, L.; Venegas-Venegas, E.; Seifert, A.; Nestl, B.M.; Hauer, B. Bacterial CYP153A monooxygenases for the synthesis of omega-hydroxylated fatty acids. Chem. Commun. 2012, 48, 5115–5117. [Google Scholar] [CrossRef] [PubMed]
- Scheps, D.; Honda Malca, S.; Richter, S.M.; Marisch, K.; Nestl, B.M.; Hauer, B. Synthesis of ω-hydroxydodecanoic acid based on an engineered CYP153A fusion construct. Microb. Biotechnol. 2013, 6, 694–707. [Google Scholar] [PubMed]
- Chowdhary, P.K.; Alemseghed, M.; Haines, D.C. Cloning, expression and characterization of a fast self-sufficient P450: CYP102A5 from Bacillus cereus. Arch. Biochem. Biophys. 2007, 468, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.Y.; Jung, E.; Yun, H.; Yang, Y.H.; Kim, B.G. Engineering class I cytochrome P450 by gene fusion with NADPH-dependent reductase and S. avermitilis host development for daidzein biotransformation. Appl. Microbiol. Biotechnol. 2014, 98, 8191. [Google Scholar] [CrossRef] [PubMed]
- Hausjell, J.; Halbwirth, H.; Spadiut, O. Recombinant production of eukaryotic cytochrome P450s in microbial cell factories. Biosci. Rep. 2018, 38, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mathew, S.; Jeong, S.-S.; Chung, T.; Lee, S.-H.; Yun, H. Asymmetric synthesis of aromatic β-amino acids using ω-transaminase: Optimizing the lipase concentration to obtain thermodynamically unstable β-keto acids. Biotechnol. J. 2016, 11, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Tavanti, M.; Mangas-Sanchez, J.; Montgomery, S.L.; Thompson, M.P.; Turner, N.J. A biocatalytic cascade for the amination of unfunctionalised cycloalkanes. Org. Biomol. Chem. 2017, 15, 9790–9793. [Google Scholar] [CrossRef] [PubMed]
- Kirmair, L.; Skerra, A. Biochemical analysis of recombinant AlkJ from Pseudomonas putida reveals a membrane-associated, FAD-dependent dehydrogenase suitable for the biosynthetic production of aliphatic aldehydes. Appl. Environ. Microbiol. 2014, 80, 2468–2477. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Black, W.; Yoon, J.M.; Shanks, J.V.; Jarboe, L.R. Improving Escherichia coli membrane integrity and fatty acid production by expression tuning of FadL and OmpF. Microb. Cell Fact. 2017, 16, 38. [Google Scholar] [CrossRef] [PubMed]
- Jeon, E.Y.; Song, J.W.; Cha, H.J.; Lee, S.M.; Lee, J.; Park, J.B. Intracellular transformation rates of fatty acids are influenced by expression of the fatty acid transporter FadL in Escherichia coli cell membrane. J. Biotechnol. 2018, 281, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [PubMed]
- Janßen, H.; Steinbüchel, A. Fatty acid synthesis in escherichia coli and its applications towards the production of fatty acid based biofuels. Biotechnol. Biofuels 2014, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Sambrook, J. Molecular Cloning: A Laboratory Manual/joseph Sambrook, David w. Russell; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Bollag, D.M.; Rozycki, M.D.; Edelstein, S.J. Protein Methods, 2nd ed.; Wiley-Liss: Hoboken, NJ, USA, 1996; ISBN 978-0-471-11837-4. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).