1. Discovering Crystals and Crystallography
My discovery of “crystallography” and “drug discovery” as research themes occurred in 1962. I was enrolled to do a degree in Natural Sciences with a focus on chemistry but my broad interests were in research underpinning new developments in medicine. I was also involved in radical politics and race and gender diversity issues. I had heard that Dorothy Hodgkin had a research team that was multidisciplinary, multinational and gender balanced in a way that I had not previously seen in Oxford. A colleague suggested that we apply for a multidisciplinary course that was run for undergraduate students as an option in the Laboratory of Crystallography where Dorothy was based. We were accepted and were not disappointed!
The discussions in the breaks and over lunch were about all elements of science, politics and life in general. The work in Dorothy’s laboratory had medical themes, including vitamin B12, penicillin and especially insulin, where her research was relevant to long-lasting insulins [
1]. However, she was a crystallographer and progress depended on obtaining crystals in order to define structures and understand functions. I decided this was a great training whether I ended up in academia, politics, science research or industry!
Two years later, I had to decide on my fourth-year research—equivalent to a Master’s degree. At the request of my tutor, Jack Barltrop, I visited Dyson Perrins Laboratory, the home of organic chemistry in Oxford for most of the past century. However, although I had enjoyed discussion in our weekly tutorials with Barltrop, I did not fall in love with the experimental organic chemistry in the lab. Having discovered the Laboratory of Crystallography in a one-term undergraduate project, with its amazing multidisciplinary and multinational culture, I ran down South Parks Road in Oxford to the Laboratory and pleaded with them to accept me there.
After a PhD with Tiny Powell learning the techniques, I moved into Dorothy Hodgkin’s laboratory in 1967 to study crystals of insulin, with the understanding that the knowledge would be useful in drug discovery. There, I became a member of her group involving Guy Dodson (New Zealand), Eleanor Dodson (Australia), and Margaret Adams (UK), to be joined by M Vijayan (India) the next year, Ted and Heather Baker (New Zealand), and Liang Dongcai (China), together with other visitors from the USA, Argentina, India, Japan, parts of Africa and elsewhere—a truly amazing environment. The insulin group relocated soon after to Zoology to join Professor David Phillips, later Lord David Phillips, who had moved from the Royal Institution to take up a new Chair of Molecular Biophysics in 1967.
Dorothy Hodgkin had left-wing views and her husband was a member of the Communist Party and author of some influential books on Africa that I had read earlier as a student. Nevertheless, she had very good relationships with pharmaceutical companies, including Novo in Denmark, Wellcome in the UK and Eli Lilly in the USA—all involved in insulin research for the treatment of diabetics. The companies provided us not only with valuable pure insulin samples, but also very exciting discussions about the underlying basic science of insulin crystallisation. The advantage to pharma companies of using the very small crystals of insulin for treatment of diabetes was that an injection once a day sufficed, as they had a very slow rate of dissolution in the bloodstream. Company scientists, in particular Jørgen Schlichtkrull at Novo in Denmark [
2], had spent a huge amount of time refining the conditions for crystallisation and I was amazed by this focus on what seemed to be a very academic approach to the subject, but it led to new long-lasting insulin forms such as 4Zn-insulin crystals. Thus, I was introduced to a laboratory in structural biology—the term was not used at that time—using crystals in my research. The environment was multidisciplinary, having physicists, chemists and biologists in the team, but also gender balanced and multinational. This became my model for how science should be. Crystals became the centre of my research life.
2. Solving the Crystal Structure of Insulin
Dorothy Hodgkin began researching on insulin after working with J.D. Bernal in the early 1930s for her PhD. She had finished her first degree in Chemistry in Oxford and then moved in 1932 to work in Cambridge, attracted by the research topic of crystallography but also by the charismatic multidisciplinary approach that J.D. Bernal had to science. In 1934, Bernal received beautiful crystals of pepsin from Nobel Laureate Theodor Svedberg in Uppsala, Sweden, to see whether they would diffract X-rays [
3]. The first experiments appeared negative but, after a few moments of depression, Bernal, who was also working on the structure of liquids including water, realised that the crystals should be kept in an aqueous environment where they would remain stable. He and Dorothy enclosed the crystals within a thin-walled capillary sitting in a small pool of mother liquor with a reservoir either side but sealed at each end of the capillary. To their surprise and pleasure, they found an amazing diffraction pattern, confirming that water was required for retention of protein architecture. Bernal phoned up the Editor of Nature and the paper was published within a couple of weeks [
4]. Dorothy and J.D. Bernal, known as “Sage”, wrote: “Now that a crystalline protein has been made to give X-ray photographs, it is clear that we have the means of checking them, by examining the structure of all crystalline proteins, arriving at far more detailed conclusions about protein structure than previous physical or chemical methods have been able to give”. The optimism was justified but the time scales were much longer than they had envisaged. Dorothy returned to Oxford in 1934 but obtained insulin crystals from Beecham Group plc, a British pharmaceutical company, who were producing the material for treatment of diabetics. Although Dorothy managed to get diffraction patterns of insulin [
5], the structure proved a challenge.
When I arrived fulltime in the Laboratory of Crystallography to start my PhD in 1964, it was announced that Dorothy Hodgkin had been awarded the Nobel Prize. This was in fact for the crystal structures of penicillin and vitamin B12, but there was a general recognition of the progress she had made with insulin and the expectation that the crystal structure would soon be solved. After Marjorie Harding, Beryl Rimmer and Sylvia Sheat in the 1950s, Guy Dodson, Eleanor Dodson and Margaret Adams were continuing to work on it in 1964 and produced some heavy atom derivatives to use in isomorphous replacement and anomalous scattering.
In 1965, there were already crystal structures from Cambridge including haemoglobin from Max Perutz [
6,
7] and myoglobin from John Kendrew and colleagues describing the iron-containing haem group which facilitates oxygen binding [
8]. Michael Rossmann, Richard Dickerson and David Davies from the Cambridge laboratory went on to make major breakthroughs in new labs in the US and Bror Strandberg in Scandinavia, while David Phillips and Tony North moved to solve the structure of hen egg-white lysozyme at the Royal Institution in London [
9]. This suggested a mechanism of catalytic activity, the first in three dimensions. Further insights into the mechanism of lysozyme were obtained from structures of some crystalline lysozyme-inhibitor complexes determined by X-ray analysis at 6 Å resolution by Louise Johnson and David Phillips [
10]. In 1966, I attended a presentation from David Phillips at the Royal Institution, where he had the sequence of hen egg-white lysozyme hanging from the ceiling, and the 3D model of the enzyme on the ground in front of him. However, the presentation by Louise Johnson showing inhibitor binding was equally stunning! It was a day that influenced my future career strongly!
Dorothy’s structure of vitamin B12 inspired me! My PhD from 1964 to 1967 on crystal structures of other metal complexes with Tiny Powell ensured that I was trained in the basic techniques. I joined Dorothy Hodgkin’s group that was next door to work on insulin in 1967. I immediately found myself thinking about which metal ions might be used to label the insulin more satisfactorily than had been achieved to that time. Already, lead ions (Pb
2+) had been used in an attempt to replace the zinc ions (Zn
2+) in insulin hexamers but this had led to some confusion, due to the fact that Pb
2+ acted as an A-type metal, binding oxygen ligands, whereas Zn
2+ was B-type. It was not surprising therefore that it had high occupancy at a further site near carboxylate sidechains close to the 3-fold axis of the 2 Zn insulin hexamer but half-way between the two zinc ions on the 3-fold axis of the rhombohedral crystals. I sought to find other metal ions that might bind without displacing the zinc ions. I decided to try uranyl ions, which I knew would bind to acid groups and see whether I could find a further useful derivative. This soon led to a low-resolution structure based on the uranyl lead and mercury derivatives [
11].
The low-resolution image excited me and showed how the two zinc ions coordinated three dimers in the zinc insulin hexamer. It led to an invitation to give a talk at a meeting on protein crystallography in the US later in 1969. I prepared slides of the low-resolution maps of the crystal structure but left Oxford soon after 2.8 Å resolution maps were calculated and as the modelling of the zinc-insulin hexameric structure was ongoing. I gave the talk on the low-resolution insulin crystal structure in Chicago and then moved to Long Island, where the 1969 International Congress of Crystallography was taking place at Stony Brook University, New York. I met with Dorothy and discussed the maps that had already been sent to me with the current molecular interpretation and modelling. As I walked with Dorothy from our hotel to the International Congress where several thousand participants were expected, the organiser of the conference came alongside us and started talking to Dorothy. He said that the plenary keynote lecturer, who was due to talk about the materials retrieved from the moon in a recent mission, had not been able to contribute due to the need to keep them safe-guarded to avoid infections that might arise from their release into the laboratory. He had heard that Dorothy’s group had just solved the crystal structure of insulin and then asked her to give the plenary lecture on insulin in place of the moon rock. Dorothy was aware that I had prepared slides on the low-resolution crystal structural information and had just given a talk on the new results. To my surprise she said, “I am happy to give a talk. I will say a few words and then Tom can talk about the structure in detail”.
This was a day or so before the lecture was due. I reviewed the slides that I had for a traditional kind of projector. I then very quickly used the material I had on the high-resolution structure to produce some new slides at the conference centre for a modern projector, and also some overlays of the electron density map that had been produced at high resolution for insulin crystals. This was an all-night job with no sleep. I required three kinds of projectors. I went back to the Conference Centre and persuaded them that, as it was such a huge lecture theatre and a very wide screen, they should allow me to use all three in parallel. I did not have time to discuss what I was going to say with Dorothy in detail but I confirmed that I was prepared to follow her. Dorothy went up onto the stage and introduced the topic and said who had been involved and how she had worked on this project since she first obtained crystals in 1934. She then passed over to me to give the main part of the presentation!
I began with a slide of the crystal electron density on one projector, another of the new insulin structure prepared overnight on a second projector and on the third overhead projector, I wrote some music—the tune that Dorothy hummed when she was excited by results. I then began my talk by announcing that this was the crystal structure of insulin derived from these beautiful electron density maps, some of the first structures ever defined by X-ray crystallography, and this is the tune that Dorothy always hummed, probably for the previous 35 years, when she saw the beautiful electron density maps. My talk was received well, with excitement and appreciation of what the Hodgkin group had achieved. However, when I met Dorothy afterwards, she said to me, “Tom I really didn’t know I hummed that tune when I was excited”.
The crystal structure defined the architecture of the insulin hexamer, with three dimers of insulin coordinated by two zinc ions on the 3-fold axis of the rhombohedral crystals in spacegroup R3 (
Figure 1). The dimers exhibited a pseudo 2-fold axis perpendicular to the 3-fold axis.
Soon after we had returned, Dorothy phoned the Editor of Nature to say that 35 years after starting work on insulin, the crystal structure was at last solved. A couple of weeks later, it was published in Nature [
12]. The insulin structure was further refined and the Hodgkin group produced a more detailed paper in 1971 [
13]. My own interests then were very much to understand the relationship of structure, chemistry and biological activity of insulin. Guy Dodson, Dan Mercola and I worked hard at a long review, where we related the structure to the biology in detail and this was published in a Advances in Protein Chemistry in 1972 [
14]. We discussed the role of the small crystals of zinc-insulin hexamers in biology as a storage mechanism of insulin in the beta-granules in evolution (
Figure 1). Related crystals had been used by pharma since the 1930s to treat diabetic patients. We also discussed the dissociation of the hexamer to allow the monomer to bind to its receptor, using conservation of residues in evolution to suggest where the binding site on insulin might be. For me, this was a further exciting development and finally convinced me that I ought to stay in crystallography but relate it firmly to biology.
3. Politics in the City of Oxford
In parallel to science and music, I had become, while an undergraduate in Oxford in 1963, Chair of the Joint Action Committee Against Racial Intolerance, and then later, when I had graduated and was doing my PhD, co-founding a City of Oxford organisation, known as Oxford Committee for Racial Integration. A few years before, very large numbers of West Indian and Bangladeshi immigrants were recruited to the Morris Motor and Pressed Steel companies in Oxford. There was an urgent need to have some activity in the city to reassure the immigrant population that they were welcome. With Michael Dummett, a very distinguished Professor of Philosophy, and Anne Dummett, his wife, who was politically active in the Liberal Party, several trade union leaders working in Morris Motors and Pressed Steel and a socially conscious clergyman, the new Oxford Committee for Racial Integration was established. This involved me heavily in politics of Oxford. I discovered that Oxford was much more than the centre where undergraduates lived in their colleges, did rowing on the river and multiple sports on the playing fields. There were also large working communities in East Oxford, with extensive housing estates that had grown to accommodate workers in the motor industry.
I was then asked by the Labour Party to stand for the City Council in St. Clements Ward, in East Oxford, close to the centre, where some immigrants had moved and which had previously been a Conservative stronghold. I campaigned on an environmental policy of preserving the centre of Oxford, making it a place where pedestrians and cyclists could be, and making sure that the traffic was not using it as a through route. In 1970, I was the only Labour councillor elected that year and I became a member of the City Council Planning Committee and interested in developing a new policy. This involved not only conserving the traditional centre but also developing communities with improved local environments. This became part of the Oxford City Labour Party manifesto and, in the following years, we won an increasing majority, so that we took control of Oxford in 1972. We were then able to pedestrianise the centre of Oxford and preserve many of the traditional areas. Cars were banned from going along the High Street, directly through the city centre, and instead routes were limited to a smaller number of cars directed alongside a road next to Magdalen College wall to go from East towards the North or West.
We had hoped to stop everything but buses, police and ambulances in terms of motorised vehicles going across Folly Bridge and Magdalen Bridge, but I was called down to Westminster to meet the Conservative Minister in charge of planning, appropriately named Keith Speed. He announced, “Blundell you may be in charge of the centre of Oxford planning, but I am in charge of road planning in the UK, including routes through city centres! You cannot block cars going over the Folly and Magdalen bridges”. However, we did manage to maintain the pedestrianisation policy and important conservation areas have remained restricted from motorised transport ever since. Nobody has changed it!
In general, I found huge support for the environmental changes that we developed in the Oxford City Centre from the general public and indeed from other parties as well as the Labour Party. However, politics is much more complicated than science and one can have completely objective arguments for making different decisions that are contradictory of each other. I found myself supporting environmental developments but also realising that working people in Oxford needed new housing near the centre. This demonstrated that, even within some areas of Oxford that I had hoped to preserve from further large building projects, we had to allow house building to avoid long travelling for Oxford working people. We did manage, however, to make North Oxford a Conservation Area, even though it had no listed buildings; rather we justified it on the basis of its impressive townscape. This policy controlled building in the large back gardens of many of the beautiful homes in North Oxford. However, the challenge between environment, industry, housing, travel and other aspects of people’s lives are real and I realised that politics was much more difficult than science.
4. World Travel for Crystallising My Ideas
Dorothy Hodgkin was very sympathetic to my doing something political but she was certainly worried that I was spending a lot of time from mid-afternoon onwards in politics and also still running a modern jazz group in the nights, albeit occasionally. My first wife, and mother of my son Ricky, was upset about the complexity of my life and our marriage broke up. I decided that I had to have a thinking period.
I, therefore, used several months in 1972 to leave Oxford, with an initial objective of attending a meeting in Japan. I first travelled on the train to Moscow in order to join the Trans-Siberian railway. I thought I would have time to think over the options in my life—to crystallise my ideas during travel to the Far East. This was quite a challenge, as I found myself in a small carriage with no skills in Russian language that would allow a conversation during the seven days of travel to Khabarovsk. I could not reach Vladivostok because it was closed to foreign travellers for military reasons. I eventually caught a plane travelling to Japan to attend and speak at the Ninth Congress of the International Union of Crystallography 26 August–7 September, 1972, in Kyoto.
I stayed for a while, making new Japanese friends before travelling on through Hong Kong, to Bangkok in Thailand to Calcutta in India. There, I was invited to give a talk on crystallography of insulin at one of the institutes before moving down to Bangalore to meet up with Vijayan, Siv Ramaseshan and other crystallographers whom I had met when they had worked with Dorothy Hodgkin in the Laboratory of Crystallography in Oxford over the previous eight years. The main features of my stay in Bangalore were multiple discussions and a lecture at the Indian Institute of Science focusing on the future uses of crystallography. I also took lessons on the veena and purchased one of these very large string instruments. I learnt a lot about Indian music but, after a few weeks, I decided I had better to return to Oxford. Travel was a challenge, as I had to find a further seat for my veena next to me, as it would have been clearly smashed if I had checked it in. On my return to Oxford, I found a veena player amongst the Indian community who continued to give me weekly lessons.
On my journey, I had decided to leave Oxford and to give up my politics and jazz. In discussion with Dorothy when I returned, she suggested that I should move to Sussex University, close to where I was brought up. Within one day, a University Lectureship was arranged and laboratory space allocated in the new building in the School of Biological Sciences just north of Brighton. I quickly applied for and was awarded a grant to work on insulin and glucagon structure, evolution and function from the Government Science Research Council. I was really lucky! There was already crystallography in Ron Mason’s laboratory in Inorganic Chemistry—where he had been appointed in 1971—but none in biology! I searched for the whereabouts of an ex-undergraduate student I had taught in Oxford, Ian Tickle, who had been one of my best tutees in Oxford. By then, he had become a well-trained crystallographer, moving for his PhD to the Netherlands. He agreed to return in 1973. Together with two very bright PhD students that I had been teaching as undergraduates in Oxford, Rod Pullen and John Jenkins, we moved to Sussex in 1973. I spent the time in between sorting out various policies and activities on the City Council. Steve Wood applied to do a PhD with me and joined us in Sussex as I set up a new group in an almost empty building that had just been constructed.
5. Crystallography in Biological Sciences at Sussex University
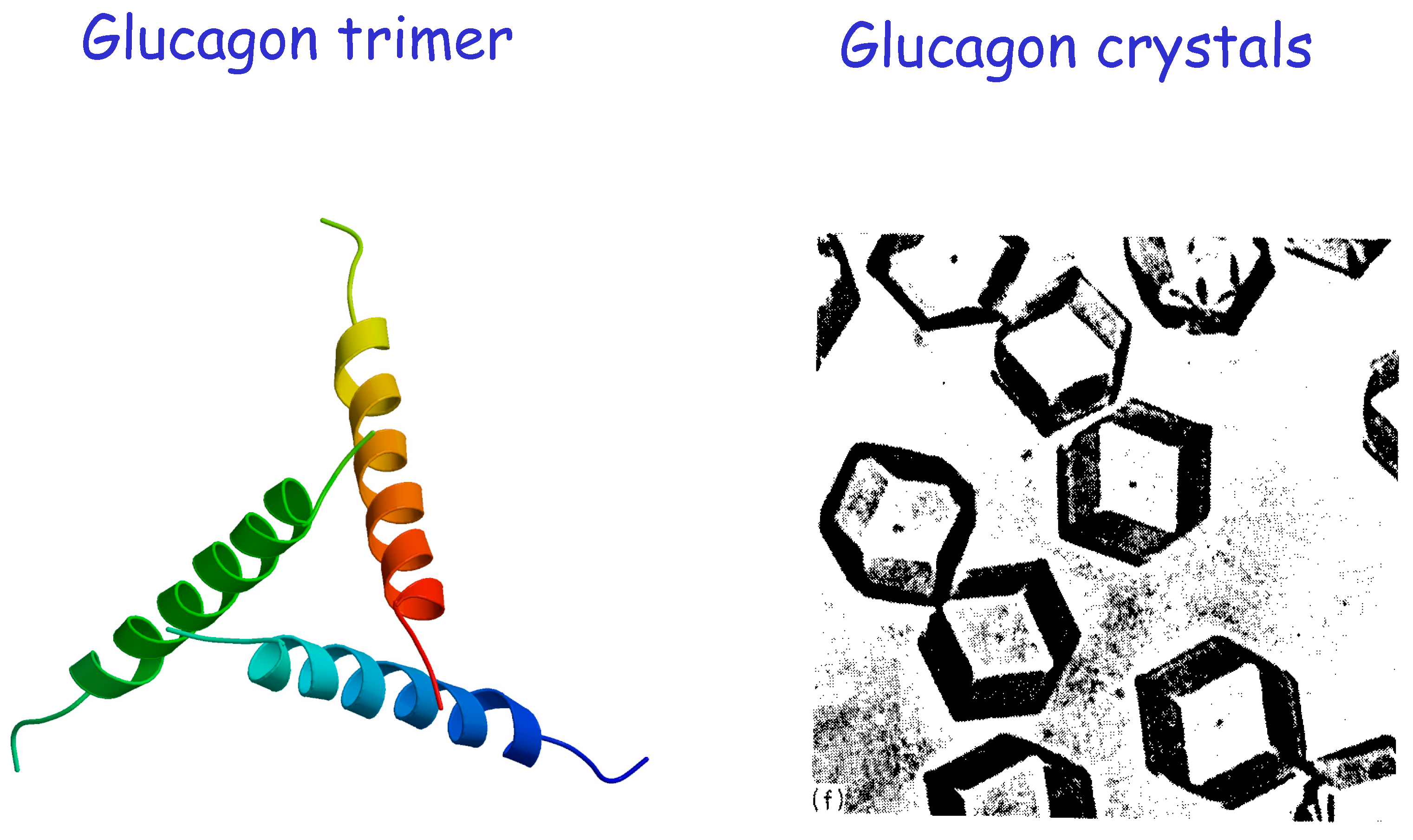
With a new focus on science and few other distractions as well as a superb young and enthusiastic research team, we got to work very quickly. I bought a house in Lewes, a few miles from Sussex University at Falmer on the outskirts of Brighton. I decided to start new biological crystallographic projects, the first of which was to look at the crystal structure of glucagon, where crystals were soon produced (
Figure 2).
The interest here was that whereas insulin puts down sugar levels in blood circulation, glucagon puts them up. They are the “yin and yang” of blood sugar control, very much exemplifying the ancient Chinese philosophy that opposite and contrary forces can act in complementary ways. Our paper was published in Nature in 1975 [
15]. Glucagon, unlike insulin, did not have a preformed structure in solution but rather assembled at the receptor. However, it formed trimers that assembled in crystals with cubic symmetry P2
13 as shown earlier by Murray Vernon King [
16], reflecting the stable storage form in the secretory granules of pancreatic α-cells. Our crystal structure demonstrated the arrangement of the trimers that also occur in solution (
Figure 2). The glucagon-receptor interaction inferred was the first structurally defined example of “concerted folding and binding” that I had come across. It triggered research on the two complementary ways of peptide–protein interactions exhibited by polypeptide hormones and growth factors: the first involving preformed structures but often with small conformational changes on binding; the other involving a preformed receptor with a disordered flexible polypeptide in solution requiring concerted folding and binding to crystallise. It was also very apparent even at that stage something had to compensate for the loss of entropy when the peptide bound the protein and if so it likely had a very well-defined binding site, suitable for drug discovery!
The second line of investigation was to understand the requirements of insulin binding to its receptor. We did this in collaboration with the group of Helmut Zahn and Dietrich Brandenburg in Aachen Germany, who synthesised novel insulins, and Jorgen Gliemann and Stefan Gammeltoft from Denmark, who did the receptor binding analyses. This multidisciplinary team tested the ideas we had established in Dorothy Hodgkin’s laboratory based on the crystal structure of insulin, bringing together chemistry, biochemistry, structural biology and biological assays to test our hypotheses. The objective of course was to find more efficient insulin-receptor binders that could be used clinically and may have greater affinity and longer half-lives. This was also published in Nature [
17].
The third manuscript to be published in Nature was a “think piece” focussing on whether the evolution of insulin was Darwinian or due to selectively neutral mutation [
18]. This exploited the extensive information about insulin, which was the first protein to be sequenced by Fred Sanger and for which there were in the 1970s many sequences from other species available. Our paper combined this with information of the 3D structures already defined by crystallography, in order to develop ideas about its evolution in terms of natural selection and multiple neutral amino-acid substitutions, building on ideas of Motoo Kimura about evolutionary rates at the molecular level [
19,
20]. The article depended very much on our detailed knowledge of insulin storage as crystals of zinc-insulin hexamers after processing of the proinsulin single chain to the A and B chains and the importance of the tertiary structure of the monomer in receptor binding. It illustrated how residues were selectively conserved not only to maintain and improve the three-dimensional structure of the protomer and interprotomer contacts in the dimer and hexamer, but also to facilitate processing of the proinsulin to insulin and interaction of insulin with its receptor. It demonstrated that much of the sequence change was likely selectively neutral, but conserved areas indicated evolutionary restraints in storage and receptor activation. The analysis provided one of the first detailed analyses of evolution of protein function from primitive fish species through to more recently evolved mammals including human. This paper arose very much from the discussions with John Maynard-Smith who had exciting ideas about evolutionary change and Sydney Shall, who was involved in understanding insulin function, at Sussex University, as well with Norman Lazarus who was involved in insulin research at The Wellcome Foundation Ltd in Kent. The insulin analyses guided the re-design of insulins in the pharmaceutical industry, indicating what was essential to activity and therefore what might be modified in drug discovery.
While still in Oxford, I had collaborated with Louise Johnson to write a review on protein crystallography. Since working with David Phillips on the crystal structure of lysozyme, Louise had moved to the laboratory of Fred Richards at Yale University for postdoctoral research in 1966. There, she had worked on the crystal structure of another enzyme, ribonuclease. After her postdoctoral year at Yale, she returned to the UK in 1967 and took up a post with David Phillips at Oxford in the new Laboratory of Molecular Biophysics that accommodated both the Phillips and Hodgkin protein crystallographic groups. Three years later, David Phillips asked Louise and myself to write a review on protein crystallography, which we did by bringing together our knowledge from the lysozyme and insulin structural work [
21]. In Sussex, being away from the Oxbridge environment, I realised that the knowledge of the protein crystallographic techniques to define structures was not fully recorded or widely disseminated. Louise and I decided to write a monograph on protein crystallography. This resulted in over 500 pages of detailed text, bringing together what we had learnt about the use of crystals in the groups of David Phillips and Dorothy Hodgkin in the previous decade, as well as from extensive collaborations not only with the US, but also Russia, China, India and elsewhere, where the subject was quickly evolving [
22].
6. Pepsin, Renin, HIV Protease and Structure-Based Drug Discovery
In the mid-1970s, I became aware that the original crystals of pepsin examined by Crowfoot and Bernal had not led to a 3D structure of the pepsin family. The sequence of pepsin itself was published by Tang et al. 1973 [
23]. In 1975, I decided to start with endothiapepsin, a pepsin homologue from the pathogenic fungus
Endothia parasitica, classed as an acid or aspartic protease and which we quickly crystallised and solved the structure [
24]. The structure had a deep active site cleft, and a symmetrical arrangement of the two catalytic aspartates located in the duplicated sequence Asp-Thr-Gly related by the pseudo 2-fold axis in the structure, later found in pepsin (
Figure 3). This led to further comparative structural analyses of the many pepsin-like proteases including the aspartic protease from Rhizopus chinensis (solved in the laboratory of David Davies in NIH) and the human chymosin [
24,
25].
In 1976, I had been appointed Professor and Head of the Crystallography Department in Birkbeck College. Another aspartic protease, renin, was identified as a target for inhibiting angiotensinogen cleavage leading to the hormone angiotensin, a peptide hormone that causes vasoconstriction and an increase in blood pressure [
26]. This led to discussions of using crystal structures in drug discovery of antihypertensives, both in academia and in industry.
However, in parallel, we decided to investigate the pseudo-2-fold axis in the crystal structures of these enzymes that extended beyond the active site residues shown in
Figure 3. This led first to a paper on their evolution through gene duplication from our group published in Nature in 1978 [
27], a collaboration with Jordan Tang, who had defined the pepsin sequence, and Mike James, another crystallographer who had worked in the Hodgkin group but also working on the aspartic proteases. We predicted that there must have been a dimeric evolutionary ancestor that consisted of two protomers, each equivalent to one half of the aspartic proteases/pepsins that had subsequently evolved by gene duplication and fusion.
In general, these developments led to an antagonistic response from some distinguished members of the academic community, who advised me not to focus on these “speculative” ideas but to think more about crystallography. Although I respected them and focused on experiment, I continued to look for the ancestral dimeric protease of the pepsins/aspartic proteases predicted in the 1978 Nature paper. It took six years before I realised in 1984 that it existed in the genome of HIV when the infectious agent was sequenced and AIDS became a pandemic.
Lynn Sibanda and I built models of HIV protease in the lab but I was reluctant to try to publish them in view of the criticism from colleagues. Rather, we started an experimental programme by producing crystals that came to fruition only in 1989 (
Figure 4). Between 1984 and 1989, several groups embarked on defining the crystal structures of the retroviral proteases with a focus on HIV protease as a drug target. A computational model was produced by colleagues Laurence Pearl and Taylor from our laboratory in Birkbeck [
28] based on the aspartic protease evolutionary relationship. Later, X-ray crystal structures were defined by Alex Wlodawer and coworkers for Rous sarcoma virus protease [
29] and for HIV protease by Navia et al., 1989 [
30], Wlodawer et al., 1989 [
31] and the Blundell laboratory [
32].
The resemblance of these putative ancestral dimers to renin suggested that inhibitors similar to those of renins and other pepsin-like enzymes might be effective [
34]. Further research in the Blundell laboratory was developed collaboratively with Pfizer [
35], with their work on the expression and characterisation of HIV protease and ours on the structure; this collaboration was an exercise in knowledge exchange and sharing between academia and industry and widely adopted by different companies! By 1997, four successful AIDS antivirals (saquinavir from Roche Pharmaceuticals, ritonavir from Abbot, indinavir from Merck and nelfinavir from Agouron) were on the market. They demonstrated the importance of understanding the genome not only in terms of the functions of gene products, but also of their crystal structures for use in structure-guided drug discovery, as recorded recently in an excellent history of macromolecular crystallography and its fruits [
36].
Our efforts at structure-guided drug discovery in academia in the 1980s included applications of protein crystallography and interactive computer graphics [
37]. We continued to focus on crystal structures of the inhibitor complexes of aspartic proteases using transition-state substrate analogues (
Figure 5) [
38,
39].
The approach found its most successful development with the renin inhibitor–aspartic protease complexes defined by high resolution crystallography in our laboratory and in a collaboration with Michael Szelke and published in Nature in 1987 [
40]. However, we continued to use endothiapepsin as an easier basis for crystal structural basis for drug discovery of renin inhibitors, for example in Sali A. et al. [
41,
42]. In parallel, we developed discussions of the antiviral agents for the treatment of AIDS using similar approaches [
43], which described the crystal structure of HIV protease and various antiviral agents for the treatment of AIDS.
7. Four Decades of Crystals and Drug Discovery in Large Pharma
Our work on crystallography of the aspartic protease inhibitors had already attracted attention in the pharmaceutical industry in the late 1970s. I was invited by Pfizer in Groton USA to discuss the renin–angiotensinogen structural work in relation to drug discovery, and was offered a contract as a scientific advisor in 1980. I soon discovered that there was also a very exciting drug discovery research programme in Pfizer at Sandwich in the UK. Simon Campbell had developed a very impressive, multidisciplinary and interactive group of scientists. I became a regular visitor to Pfizer at Sandwich and to the laboratories in Groton, Connecticut, which in the 1980s were the major research centres.
Simon Campbell’s radical approach to drug discovery in Sandwich led to very successful outcomes as recorded in an interview by Joanna Owens for Nature Reviews Drug Discovery much later in 2006 [
44]). Amlodipine, initially approved by the FDA in 1987, continues to be a widely used drug for the treatment of high blood pressure and angina; it sells for
$6 billion a year! Fluconazole, a broad-spectrum antifungal agent, active by both the oral and the intravenous routes, was developed for the treatment of superficial and systemic infections. Viagra and Revatio contain the same active ingredient, sildenafil, which was designed to treat pulmonary arterial hypertension. Simon Campbell noted what appeared at first to be a challenge to the use of the drug, when it was found to cause erections in male patients. He realised the opportunity and it became a major medication for use to treat erectile dysfunction, selling
$2.3 billion a year! Simon was one of the first industrial researchers to be elected to the Royal Society and he was knighted in 2015.
In parallel to my visits to Pfizer, I was invited to advise other companies including ICI in the UK, which later became Zeneca, and then merged with Sweden’s Astra to become AstraZeneca. I was also heavily involved in Celltech Group plc, which was a leading British-based biotechnology business based in Slough. It was founded by Gerard Fairtlough in 1980 with finance from the National Enterprise Board—a Labour Party Initiative of which I was very supportive. I was a scientific advisor in the 1980s, while it was focused mainly on antibody production in Slough, with a smaller effort in small-molecule drug discovery. Visits were always exciting, interactive and productive.
I then had a break while I ran research councils between 1991 and 1996, first the Director General of the Agricultural and Food Research Council (AFRC) with the plan to merge with the biological sciences funded within the Science and Engineering Research Council (SERC) to become the first Chief Executive of the BBSRC in 1994. The objective was to bring more basic bioscience into agricultural, food and biotechnology industries, in much the same way as MRC did for medical research.
In 1996, I returned to be a member of the main Board of CellTech. My time on the Board seemed to be completely focused on mergers, first with Chiroscience plc, to become Celltech Chiroscience before buying Medeva plc. In 2000, we bought other companies in Europe and eventually Oxford Glycosciences in July 2003. However, in 2004, Celltech was acquired by UCB, a Belgian small pharma, which I had not visited before and we become UCB Celltech. It was an interesting expansion but nothing quite equalled the science and innovation of the early days of Celltech, when it had an interactive and multidisciplinary culture not unlike the early days of Pfizer in Sandwich in the 1980s.
8. Software Underpinning Drug Discovery
In the 1980s, there was often no crystal structure available for new drug targets, on which to base an understanding of function and develop structure-guided drug development. Our academic laboratory, therefore, decided to develop modelling software for this purpose. The early papers focused on the use of computer graphics for interactive modelling of the human and mouse renins [
45], for which energy calculations were used to optimise protein–ligand interactions. These studies led to a highly cited review on prediction of protein structures and the design of novel molecules published in Nature by Lynn Sibanda and myself, along with Mike Sternberg and Janet Thornton who had been recruited to the Department in Birkbeck [
46]. This focused on knowledge-based approaches, depending on identification of analogies in secondary structures, motifs, domains or ligand interactions between a protein to be modelled and those of known three-dimensional structure, often described as comparative or homology modelling. Our approach underlined the importance of using multiple structures of close homologues with good crystallographic resolution to model the tertiary framework and then using rule based approaches to model insertions and deletions in loop regions, followed by energy minimisation and molecular dynamics approaches. Our new software, COMPOSER, was described in two papers. the first on the three-dimensional frameworks derived from a simultaneous superposition of multiple structures and knowledge-based building of insertions and deletions [
47], and the second on the rules for replacement of side chains [
48]. However, the development of these methods made us all aware of the need for protein sequence/structure information Peter Murray-Rust coordinated as a database published in Nature [
49].
A major challenge with respect to comparative/homology modelling is to select the templates to model the proteins, a process often called fold recognition. Andrej Sali had joined the laboratory to do experimental work for drug discovery, as described above, but his skills in computer programming made him keen to develop software as he became aware of the importance of good modelling procedures. In 1989, we published an approach to the definition of topological equivalence in homologous protein structures [
50]. In parallel, others developed software addressing this challenge, including David Eisenberg with a similar methodology involving inverse folding and local environments [
51], and software, using a new approach to protein fold recognition, known as THREADER by David Jones in the laboratories of Janet Thornton and Willie Taylor [
52].
In parallel, John Overington together with Andrej Sali in our laboratory investigated environment-specific substitution tables, based on analysis of crystal structures, to evaluate sequence and structure compatibility in evolution [
53]. We also devised methods using environment-specific amino acid substitution tables to assess tertiary templates for the prediction of tertiary folds [
54]. This was followed QSLAVE, software developed by Mark Johnson and John Overington for alignment and searching for common protein folds using a data bank of structural templates [
55]. However, the advance that has had the greatest impact from our laboratory was a further approach to comparative modelling. I had originally suggested we should exploit spatial restraints in a way that was similar to programs developed in our lab for use in the refinement of crystal structures. However, Andrej Sali developed ideas way beyond my first suggestion in his software MODELLER, which exploited spatial restraints derived from the structures of homologues [
56]. The original paper has at the time of writing been cited more than 12,000 times!
In the following decade, we focused on software and databases that were useful for modelling, for example JoY, a method for sequence-structure representation and analysis that facilitates the alignment of proteins and understanding the structural homology. It depends on annotating local environments of amino acids in the three-dimensional crystal structures and ensuring alignments are compatible with amino acids occurring at the equivalent positions [
57]. In parallel, we developed a further database, HOMSTRAD, comprising alignments of protein sequences and crystal structures for homologous families [
58]. Ji-Ye Shi in the group, an amazing PhD student from China, wrote FUGUE, which developed ideas, first exploited in QSLAVE, for sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties [
59]. We also used the local environment substitution tables developed earlier to devise software to predict the impacts of amino acid substitutions.
Of course software is difficult to maintain and its success depends on having excellent collaborators and students, but also on encouraging them to take away the software so as to develop and maintain it as a centre of their research. The most convincing aspect of this has been in the work of Andrej Sali who has maintained the Modeller software for nearly 30 years, ensuring its wide use and frequent citation.
9. Using Crystals to Explore Chemical Space for Drug Discovery
The multiple structures defined crystallographically allowed exploration of the chemical space of the small molecules that will bind to a potential drug target, both computationally and experimentally. In 1984, Peter Goodford founded a software company Molecular Discovery Ltd, working in the area of drug discovery. Its aim was to develop software, initially GRID, which uses a probe to explore the interaction of a small molecule with a protein of known structure [
60]. The energy values are computed at grid positions throughout and around the molecule. Various probes can be used including small molecules with aliphatic, nitrogen, aiming donors and carboxyl and hydroxyl acceptors. Molecular Discovery Ltd. developed a range of other software, which is widely used. This influenced a range of software such as LIGSITE that provides automatic detection of potential small-molecule-binding Sites in Proteins [
61], Fpocket which uses Voronoi tessellation [
62] and Pocket Depth [
63], a depth-based algorithm for identification of ligand binding sites in proteins. These and related approaches can recognise up to 95% of binding sites in known protein–ligand structures and have facilitated use of protein structures in drug discovery.
Another incredibly influential development in drug discovery was the work at Pfizer, Groton, Connecticut, USA, by Chris Lipinski in 1997, who defined criteria such as solubility and permeability for drug molecules in his Rule of 5 [
64]. This was influenced by analyses of successful drugs, then on the market, with respect to pharmacokinetics in the human body including ADME (Absorption Distribution Metabolism and Excretion). The Rule of 5 requires the drugs have no more than 5 hydrogen-bond donors or acceptors, no more than 5 rotatable bonds and a molecular mass less than 500 daltons as well as an octanol water partition co-efficient log P not greater than 5. These restraints together with a maximum of four rings led to a decrease from 10
80 possible molecules down to 10
63 compounds [
65]. They emphasised the huge challenges in exploring chemical space by searching for molecules with drug-like size of molecular weight 500 daltons. Awareness of the Rule of 5 in the industry has led to very efficient drug discovery, particularly for human targets.
10. Crystallography and Fragment-Based Drug Discovery
In the 1990s, drug developers became increasingly aware that, even after adopting the Rule of 5, a huge number of molecules of drug-like size remained in order to explore chemical space. This led to the idea that smaller molecules of around 300 molecular weight, known as fragments, might be used, as this would lead to increased promiscuity and require smaller libraries.
A fragment is around the minimum size required to bind to a so-called “hotspot”. A hotspot is usually in a deep cavity on the protein surface, where there is a juxtaposition of polar and non-polar regions, making water molecules “unhappy”. This arises from the fact that bound water molecules in the apo-form will have lost rotational and translational entropy compared with their being in a water environment. If a fragment binds at the hotspot the loss of entropy in binding in one orientation is compensated by the gain of rotational and translational entropy, usually of several liberated waters. We, together with collaborators from the Crystallographic Data Centre in Cambridge, have developed more recently computational approaches to defining hot spots [
66,
67]. Once the fragment is bound in a way that it has a clear orientation, it can be elaborated to establish further interactions within the pocket or closely surrounding region without increasing the loss of translation and rotational entropy. Fragments usually have millimolar binding and it is possible to maximise interactions of each step in the elaboration of the fragment in order to gain nanomolar binding with a drug-like molecule of approximately 500 daltons molecular weight.
This approach, now known as fragment-based drug discovery, was originally developed using NMR approaches to monitor the fragment binding [
68]. However, structure-guided methods using crystallography are most efficient as they define the position of the fragment before growing it or linking to other fragments. The use of X-ray crystallography in fragment screening (
Figure 6) has been pioneered in Astex [
69,
70]. Astex was founded by Harren Jhoti, Chris Abell and myself in 1999 and funded by Abingworth Investments, which I had advised for many years before. The laboratory was established with three postdocs in the Blundell and Abell laboratories in the Departments of Biochemistry and Chemistry in Cambridge University, where it was demonstrated that X-ray analysis could define the fragment positions accurately with high-resolution X-ray methods. In parallel, Harren Jhoti, who had been at GSK, established himself initially in the Grafton Centre, in the centre of Cambridge to recruit his team, before moving to the Cambridge Science Park to establish the early stage drug discovery effectively. Tim Haines, an entrepreneur funded by Abingworth, joined as CEO and Harren initially was Chief Scientific Officer, but after five years became CEO.
Fragment-based drug discovery usually now involves an initial screen using fluorescence-based thermal-shift measurements, ligand-based NMR, surface plasmon resonance and X-ray screening of crystals [
71]. The fragment is first shown to bind the protein, and the structure of the protein-chemical fragment complex defined by X-ray or NMR. The kinetics are investigated using surface plasmon resonance and the thermodynamics using isothermal calorimetry. Fragment-based screening is widely used and many of the X-ray screening has been established at synchrotrons, in the UK by Dr Frank von Delft and his colleagues at the Diamond Light Source. Frank had been in our laboratory in Cambridge but not involved in fragment-based approaches. His method combines a high-throughput synchrotron data collection from crystals with some very nice software called PanDDA [
72], in which the electron density for the fragment bound state is calculated taking into account the occupancy of the fragment and weighting in the apo- and bound-states properly.
Astex established funding not only from venture capital but also through collaborations initially with each of AstraZeneca, Novartis, Janssen, and GSK. This allowed drugs to be moved through clinical trials. By 2012 Astex had eight candidate drugs in clinical trials, some internally developed and others in the collaborations. The cost of the in-house clinical trials is huge and it was clear we needed a major investment. In 2013, Astex was sold to a Japanese Pharma giant, Otsuka, in a $886 million deal, but the Cambridge laboratories have been maintained and expanded.
In 2016, Astex achieved a milestone with a US FDA filing of a new drug known as Ribociclib, in a collaboration with Novartis, finally gaining FDA approval in March, 2017. The drug is used in combination therapy for advanced breast cancer with Letrozole as first-line treatment. Ribociclib was followed in April, 2019 by FDA approval of a further drug, Erdafitinib, with Janssen for treatment of metastatic urothelial carcinoma. Astex remains very much as it was on the Science Park as part of Otsuka, but now focusing on fragment-based drug discovery not only in cancer but also in diseases of the central nervous system. Harren Jhoti continues to play a role in Astex and in the larger Otsuka family of companies in Japan and the United States, as well as UK. I continue to be on the Board of Astex Pharma and also to chair the Science Advisory Board.
11. Crystals and Antimycobacterial Discovery for Infectious Disease
In 2006, Ken Duncan of the Gates Foundation contacted Astex to see if there was an interest in collaboration with the Bill and Melinda Gates Foundation as part of the Integrated Methods for Tuberculosis Drug Discovery (IM TB) to use fragment-based discovery for the design of new treatments for tuberculosis. At that time Astex was working on oncology targets and was aware that it needed to maintain this focus in order to establish its position in drug discovery and get further investment for its early development and clinical trials. Furthermore, the market for therapeutics for tuberculosis was likely to be in developing countries, where the population cannot afford the costs of drugs that are used in economically developed countries. It seemed sensible therefore that this was done in a university context. Therefore, Chris Abell and I volunteered to develop a parallel approach using crystal structures and fragment-based drug discovery in our laboratories in the University of Cambridge.
In Africa, the position was that over 50% of tuberculosis cases had often not been diagnosed, the BCG vaccine provided only marginal protection for a few years and drug therapy was long and complicated, taking six to nine months for drug sensitive tuberculosis and as long as two years for drug-resistant tuberculosis. The first line drugs were Rifampicin, Ethambutol, Pyrazinamide and Isoniazid [
73], but multiple drug-resistant tuberculosis was on the rise with only two new tuberculosis drugs in 30 years.
We had already developed some early work in the University on targeting protein–protein interactions, which was another area of greater challenge than targeting the active sites of protein kinases and other druggable targets favoured by big pharma (see below). We were therefore able to establish collaboration with the Gates Foundation fairly quickly. There were two initial objectives—one was to substantially improve the ability to discover, identify and validate targets linked with TB persistence and secondly to find and optimise small-molecule inhibitors of validated targets.
The focus on tuberculosis, nevertheless, required new thinking, as the cell wall of
Mycobacterium tuberculosis provides a challenge that differs from that of human cells, there are many efflux pumps in
M. tuberculosis and enzymes that can degrade candidate molecules that do reach the intracellular region. These challenges, however, are well suited to fragment-based discovery, which allows the design of molecules that meet new requirements [
74]. We followed the fragment-based approach taken for the cancer targets in Astex (see
Figure 7), but introducing further selection criteria as the fragments were elaborated. In order to understand the challenges and to select suitable targets Bernardo Ochoa-Montano in our group set out to develop a database of crystallographic and modelled structures of the
M. tuberculosis proteome, called CHOPIN [
75]. We considered this to be a suitable name, given the fact that CHOPIN probably died of tuberculosis. The database has models of around 80% of the gene products; it first accepts any crystal structures and then exploits those of homologues using the software developed in the Blundell over the past three decades in particular FUGUE [
59] and MODELLER [
56]. The higher oligomeric states were modelled as well as the individual protomers. In addition, the binding sites were defined and hotspots used to identify those that were likely to be druggable.
The team set off to select targets for which there were crystal structures and to apply the methods developed for fragment-based discovery. We considered some well-established targets in tuberculosis including the enoyl-acyl carrier protein reductase, which is involved in the synthesis of mycolic acids and is the target of the first and second-line tuberculosis drugs isoniazid and ethionamide. We also looked at KatG, which is a heme enzyme catalase peroxidase, which is involved in activation of isoniazid. Although we have defined crystal structures for all proteins that we have targeted, most recently we have used cryo-EM, which has undergone a resolution revolution. This combined approach has been applied to follow the drug discovery against KatG, cryo-EM providing images at 3 Å resolution, work developed by Asma Munir and Amanda Chaplin in our group [
76]. Our work has led to low-micromolar and nanomolar candidate molecules. The real challenge with tuberculosis, however, is persuading companies to take any candidate drug through clinical trials. Even for wealthy charitable foundations like the Bill and Melinda Gates Foundation it is a challenge.
Bill Gates himself visited the group in Cambridge a few years later when the work was ongoing. He requested that his visit was not advertised and he arrived alone to be greeted by the combined Chemistry and Biochemistry teams. He spoke with each member of our structural biology and chemistry teams, who introduced themselves by name and country/language—members were truly international, with over thirty languages between them. Bill Gates recounted how Melinda had seen a programme about tuberculosis in Africa and had encouraged him to be involved in this area, one of the major steps in establishing the Gates Foundation. I discussed with him how tuberculosis was a real challenge to my own family on my wife’s side, who live in Matabeleland Zimbabwe and where a number of family members had contracted the disease, mainly as they moved down to work in South Africa.
In parallel to these developments, we have also targeted other mycobacteria. One of these is
Mycobacerium abscessus, which is an infectious agent that infects many Cystic Fibrosis patients. Professor Andres Floto from the Clinical School leads the research programme in Cambridge for the Cystic Fibrosis Trust. With targeting
M. abscessus for cystic fibrosis, the challenge is that patients are few in number making it financially it less attractive for pharmaceutical companies [
77]. Nevertheless, successful development of drugs for the market has been achieved by Vertex, a company that is in both the United States and the UK. Our work has focused always on obtaining crystal structures but as there are many fewer structures for the
M. abscessus proteome, we have again developed a database, in this case called Mabellini, to extend the structural knowledge experimentally defined by crystallography [
78]. With Dr Sherine Thomas in the lead in our laboratory we have targeted several enzymes, including
M. abscessus TrmD (tRNA-(N(1)G37) 4 methyltransferase). TrmD is an essential tRNA modification enzyme in bacteria that prevents errors in the reading frame during protein translation and represents an attractive potential target for the development of new antibiotics. We reproduced the crystal structure and used fragment-based drug discovery to the design a new class of inhibitors with low-micromolar in vitro TrmD inhibitory activity [
79]. Several of these compounds exhibit activity against planktonic
M. abscessus and
M. tuberculosis as well as against intracellular
M. abscessus and
M. leprae, indicating their potential as the basis for a novel class of broad-spectrum mycobacterial drugs.
After a lecture in 2016 at the International Mycobacterial Congress in Fort Collins, Colorado, USA, I was approached by the American Leprosy Mission to see whether we would collaborate with them, using the techniques we had developed for M. tuberculosis and abscessus, but focusing targets from the closely related M. leprae. The stigma of leprosy has meant that, even in countries such as India or Brazil, where the disease is rampant, it is not widely known that it is contracted by~200,000 people each year. Leprosy, which causes chronic infections of the skin and peripheral nerves, is treated using the same multidrug therapy as tuberculosis including Dapsone, Rifampicin and Clofazimine.
In 2016, Dr Sundeep Chaitanya Vedithi, Director of Drug Discovery for the American Leprosy Mission, joined our laboratory in Cambridge and began educate us about the challenges of leprosy [
80]. We had many new things to learn including the fact that the genome of
M. leprae has undergone reductive evolution and has only 1615 genes, although there are 1293 pseudo genes. This means that the mycobacterium is very host dependent and difficult to culture. Sundeep has developed the programme both by developing the database known as HANSEN in which around 70% of the models are good quality and most of the rest have some reasonable structures built in the usual way [
81]. We have also used computational saturation mutagenesis to predict structural consequences of systematic mutations in the beta subunit of RNA polymerase in
M. leprae [
82]. In parallel, the American Leprosy Mission is funding fragment-based discovery in the laboratory, with Dr. Marta Acebron focusing on the experimental crystallographic and drug discovery programme.
12. Multicomponent Systems as Drug Targets in Cancer
Most biological systems involve multicomponent systems in order to introduce sensitive regulation into the cell and whole organism. Fifty years ago most researchers assumed that regulatory and signalling processes involved binary interactions, for example through a single receptor and its ligand. In the 1960s and 1970 when we defined the structures of insulin and glycogen, we were active at inferring which regions of the structures were important for activity but little was known about the sequence or structure of the receptors, or indeed about other components of the signalling systems.
Our first attempts to define structures of complexes came much later in our work on nerve growth factor in 1991 when our 2.3 Å resolution structure demonstrated a new protein fold comprising three antiparallel β-strands forming a flat surface facilitating dimerisation [
83]. We later described the mouse 7S NGF, a complex of nerve growth factor with four binding proteins (two α-NGF and two γ-NGF), which assembles as a symmetrical hetero-hexamer organised around the β-NGF dimer [
84]. The crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor was defined in 1999 demonstrating a similar assembly to that of the 7S NGF complex [
85]. A further six years later led to the characterisation of a symmetric dimer binding the extracellular domains of two receptor molecules in a similar way, so dimerising the receptor [
86].
In parallel to these studies, we examined the structure of FGF paralogues interacting with the FGF receptors. In 2000, we defined the structure of fibroblast growth receptor extracellular domain bound to ligand FGF and heparin sulphate glycosaminoglycan polysaccharide (
Figure 8) [
87]. The molecular association of heparin with FGF and its receptor was known to be essential for biological activity, but the interactions defined in our crystal structure indicated a multicomponent complex. Our contention that this complex structure was biologically relevant caused extensive controversy!
However, another group had suggested that a 2FGF: 2FGFR: 2heparin ternary complex existed on the basis of a 3 Å resolution crystal structure by colleagues [
88]. This was inconsistent with the results obtained in collaboration with Luca Pellegrini and Ashok Venkitaraman [
89], so we proceeded in the following years to check the stoichiometry by other biophysical methods including size-exclusion chromatography [
90] and isothermal titration calorimetry [
91]. These and other experiments led to the conclusion that the earlier crystal structure that appeared to indicate a 2:2:2 complex of the FGF-FGFR-heparin probably arose from two sites are partially occupied in a way that would be generated by disorder of two 2:2:1 complexes around the crystallographic 2-fold axis
Most efforts at drug discovery on FGF receptors were focused on the intracellular kinase domain, giving rise to useful molecules. Multiple small-molecule inhibitors targeting this family of kinases have been developed, and some of them are in clinical trials. The pan-FGFR inhibitor erdafitinib, originally discovered by Astex Pharmaceuticals and licensed to Janssen Pharmaceuticals as JNJ-42756493 for further development, has recently been approved by the U.S. Food and Drug Administration (FDA) for the treatment of metastatic or unresectable urothelial carcinoma [
92].
An attempt to improve selectivity by targeting the protein–protein interactions in the extracellular region of FGF receptors was developed in a collaborative exercise involving sixteen different groups coordinated by Marc Herbert from Sanofi-Aventis, from Montpellier, France. [
93]. The new FGFR inhibitor, SSR128129E (SSR), binds to the extracellular part of the receptor. It does not compete directly with FGF for binding to FGFR but inhibits in an allosteric manner. SSR was the first reported small-molecule allosteric inhibitor of FGF/FGFR signalling, acting via binding to the extracellular part of the FGFR. This orally deliverable, small-molecule multiFGFR inhibitor showed promising therapeutic anti-cancer efficacy
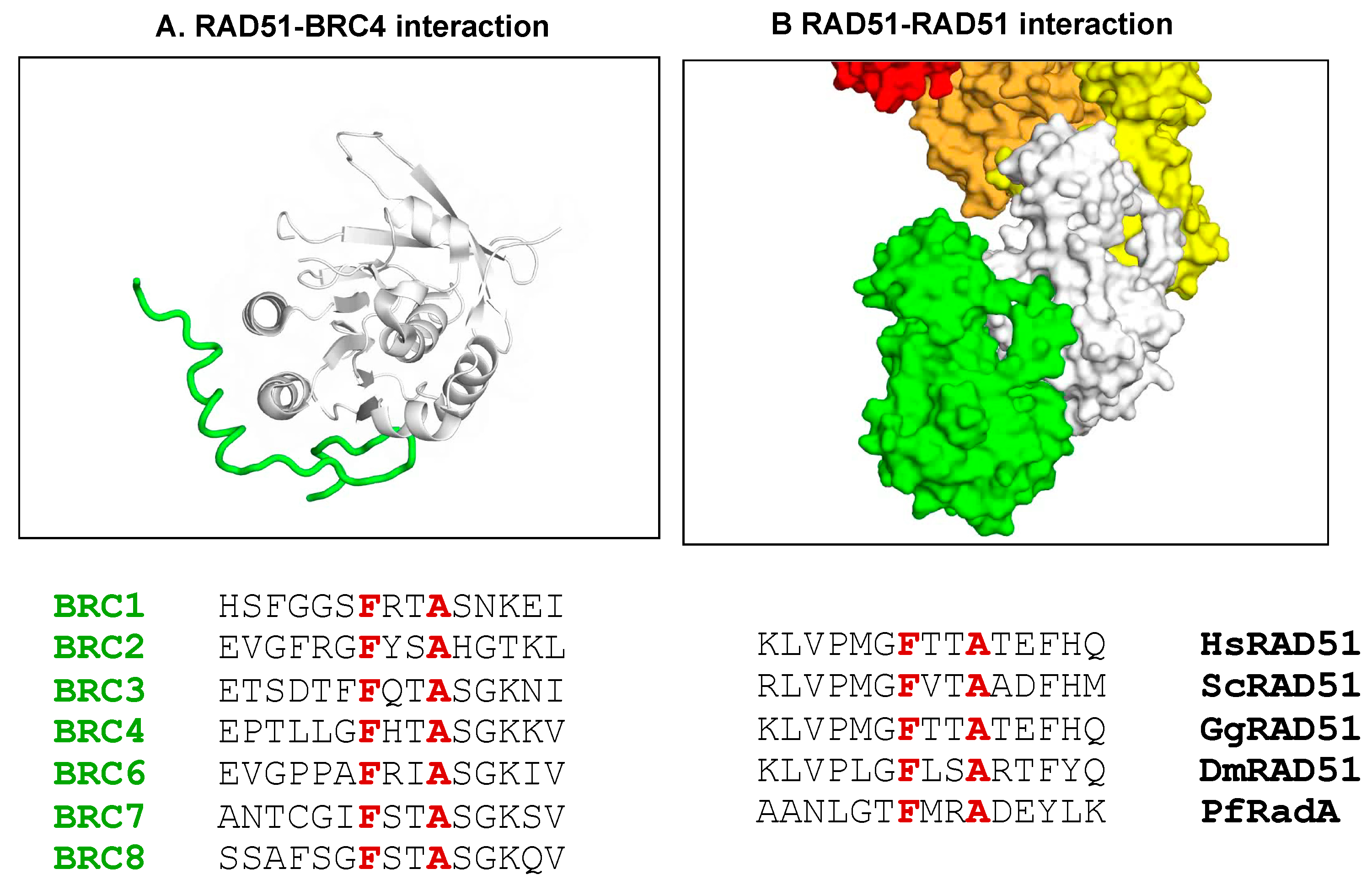
These early experiments emphasised the challenge for targeting drugs at protein–protein interactions. In general, the protein–protein interfaces were relatively flat and the number of druggable sites very few. However, it was evident from other studies in our laboratory of protein–protein interactions that when one component was involved in concerted folding and binding onto a pre-defined site then the pockets were generally deeper. Our initial studies, involving Luca Pellegrini, of such a system involved the interaction of Rad51 with BRCA2. The flexible region of BRCA2 involved in concerted folding and binding with the Rad51 involved a conserved sequence of FxxA, in which the phenylalanine (F) initially docks into a deep pocket (
Figure 9). Pellegrini et al. (2002) demonstrated that the intervening residues, which are variable in evolution, then fold to allow the conserved alanine to dock into a second pocket ([
94]).
In a collaboration with Ashok Venkitaraman from the Clinical School in Cambridge and Marko Hyvonen and May Marsh in our group in the Department of Biochemistry, we screened using the fragment based approach producing multiple micromolar fragment binding (
Figure 10) including merging of fragment and peptide data to eventually give inhibitors with nanomolar binding [
95,
96]. However, even when we obtained nanomolar compounds targeting the Rad51 BRCA2 system and obtained good nanomolar binding, it proved impossible to persuade any companies to join us.
We have also targeted protein–protein interactions in the alternative DNA repair system, Non-Homologous End Joining (NHEJ). This involves the recognition of DNA double-strand breaks by Ku70/80, the recruitment of a huge DNA protein kinase (DNA-PKcs) that interacts with Ku, and multiple components that form scaffolds and strings that regulate the assembly as the DNA ligase is recruited to repair the assembled ends [
97]. Individual components, even the largest molecules such as DNA-PKcs with 4128 amino acids have been defined by X-ray analysis at 4.3 Å [
98]. Furthermore, such systems often change over time, providing major challenges to crystallography, and require dissection using novel single-molecule forceps and related techniques [
99]. These structures are proving useful in the design of new cancer therapeutics, not only through targeting the DNA-PKcs kinase active site [
100], but also at protein–protein interfaces, especially where polypeptide with an intrinsically disorder region folds and binds, such as the binding of the Artemis tail to a site on the DNA-Ligase IV in NHEJ [
101,
102].
13. Crystals and Drug Discovery: The International Scene
This personal history has shown that for more than five decades, the use of crystals and X-ray analysis to define structures has contributed to the discovery of new medicines in a very significant way. As it is a personal history, I have mainly described developments in the UK, particularly in Oxford, London and Cambridge, where Max Perutz, John Kendrew, David Phillips and Dorothy Hodgkin made very impressive contributions to haemoglobin, myoglobin, lysozyme and insulin.
However, these developments in structural biology and drug discovery were international, having strong parallels not only in the United States, in particular in Yale, Harvard, UCLA and UCSF, but also in Russia, China, India, Brazil, South Africa and elsewhere. For example, when I first visited Russia for the International Crystallography Meeting in 1966, Boris Vainshtein, who was Head of the Institute of Crystallography, was already strongly supporting the work on biological systems. At this time, Natalia Andreeva work on pepsin, the structure of which was eventually published in 1976 at high resolution [
103,
104].
Developments in China were more complex. A key factor in establishing protein crystallography in China was the impact of Fred Sanger on Chinese scientists who had been in Cambridge in the early 1950s, while Sanger produced the first sequence of a protein, insulin. When back in China and challenged to do something for China during the Great Leap Forward in 1958, they decided to follow the advice of Frederick Engels a century before: “if you know the formula of something, then synthesise it.” They realised that they knew the formula of insulin, so why not follow the radical Frederick Engels, who was much respected in China, and synthesise it. Soon after, encouraged by the visit of Dorothy Hodgkin to Shanghai in 1960, they were inspired to crystallise synthetic insulin and check that it was identical to bovine insulin. Liang Dong Cai was subsequently sent to Oxford in 1966 to learn protein crystallography. He returned in 1967, apparently for the Cultural Revolution, but worked on the crystal structure of insulin in the Institute of Biophysics in Beijing, where he eventually became Director [
105].
The incorporation of crystallographic structural work into drug discovery in US large pharma including Pfizer occurred soon after in the late 1970s and early eighties, in Belgium in Janssen Pharmaceutica, in Denmark in Novo, and in many other countries throughout the world, including China. This has been a truly international translation of basic science with an impressive impact on an applied area.
14. Recent Developments and Perspectives in Crystals, Crystallography and Drug Discovery
Over recent years, new technologies have improved our knowledge of crystals and crystallography to contribute new insights. Amongst these have been a new generation of light sources [
106]. First-generation rings were built for high-energy physics research and only the second generation were designed from the start as light sources. The third-generation rings came online in 1992 with straight sections for insertion devices and lower electron beam emittance. Undulators gave further impressive gains in brightness. Since then, fourth-generation light sources exceed the performance of previous sources by an order of magnitude or more, not only in brightness but also coherence and pulse duration with an impressive wavelength range from the vacuum UV to hard X-rays. Linac-based Free-Electron Lasers [
107] offer subpicosecond pulses and huge technological opportunities for the future analysis of molecules and crystals.
Other developments have arisen from computer analysis, often using molecular dynamics or normal mode analysis based on time averaged structures derived from X-ray crystallography. These have had major impacts on understanding cryptic pocket formation in protein targets [
108]. Cryptic pockets are often exposed on protein targets when drugs bind and so provide alternatives to classical binding sites for drug development. Simulation-based approaches demonstrate that cryptic sites do not correspond to local minima in the computed conformational free-energy landscape of the unliganded proteins. Temperature-based enhanced sampling approaches also do not help, although simulations with fragments can stabilise cryptic sites and help in defining them in a way than can be used for drug discovery [
108].
However, the major impact in structural biology over recent years has been electron microscopy, where the resolution revolution depended on the development of new cryo-EM machines, new detectors and new software [
109,
110]. These developments were recognised in the award of the 2017 Nobel Prize for Chemistry to Richard Henderson, Jacques Dubochet and Joachim Frank “for developing cryo-electron microscopy for the high-resolution structure determination of biomolecules in solution”. Our own laboratory has been impacted by this revolution. As described in the previous section, the structure of the very large enzyme DNA-PKcs was solved by the X-ray analysis at 4.3 Å [
98], but recently, structures have been obtained of DNA-PKcs by cryo-EM at 2.8 Å and its complex with Ku70/80 and DNA at 3.8 Å resolution (previously 6.6 Å) [
111], examples of how the resolution revolution has changed structural biology over the past decade. It has even allowed Harren Jhoti and Pamela Williams and the team at Astex along with collaborators at Isohelio Ltd, to develop fragment-based drug discovery using cryo-EM, demonstrating that fragment-sized molecules can be accurately described when bound to large proteins, allowing cryo-EM to contribute even further to drug discovery [
112]).
The next decade will be a very exciting time for many academics and pharma companies as they follow the trend to use cryo-EM for structural biology and drug discovery. Nevertheless, crystals and crystallography will continue to play a major role in understanding protein function and drug discovery.