Molecular Characterization of the Native (Non-Linked) CD160–HVEM Protein Complex Revealed by Initial Crystallographic Analysis
Abstract
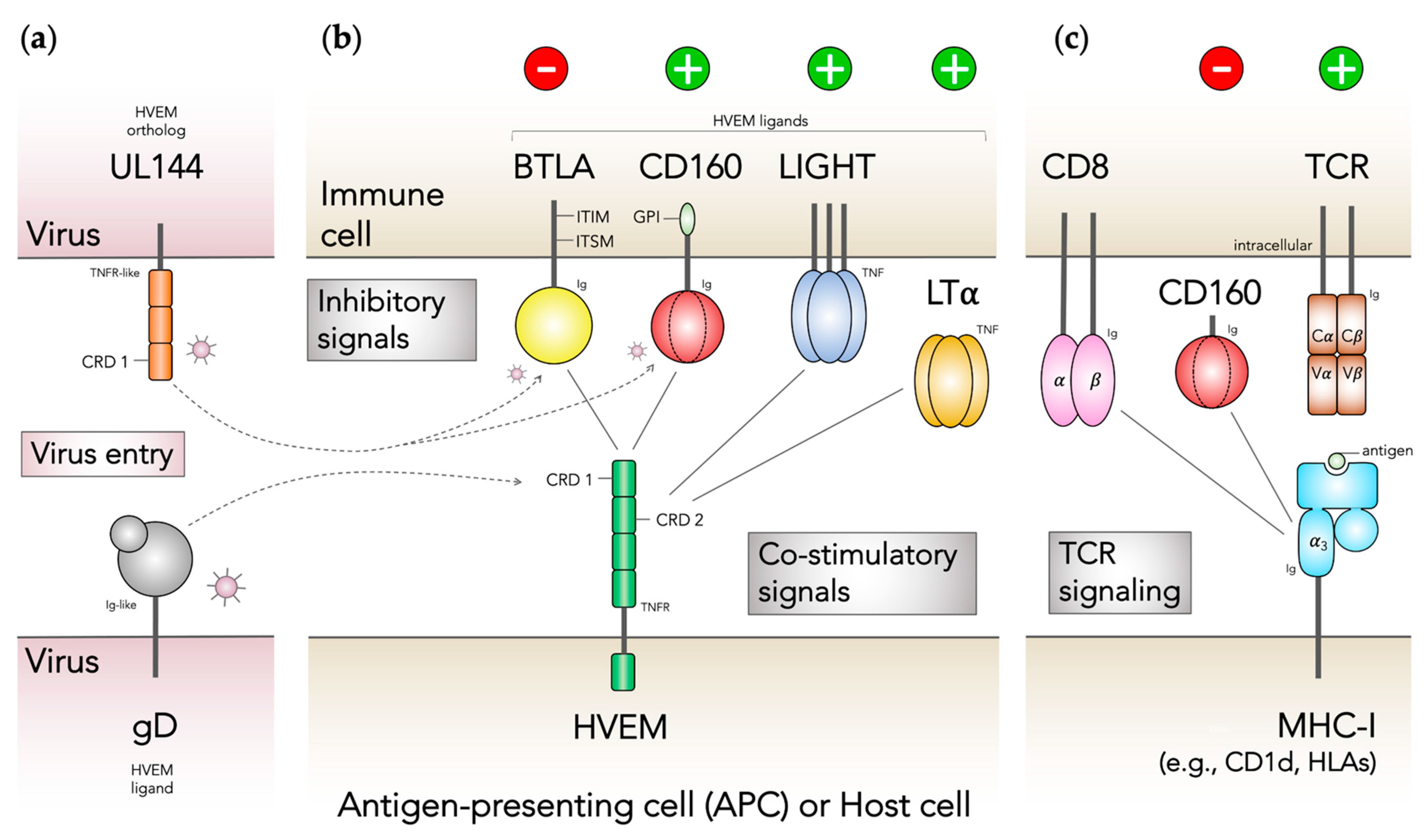
:1. Introduction
2. Materials and Methods
2.1. Macromolecule Production and Characterization
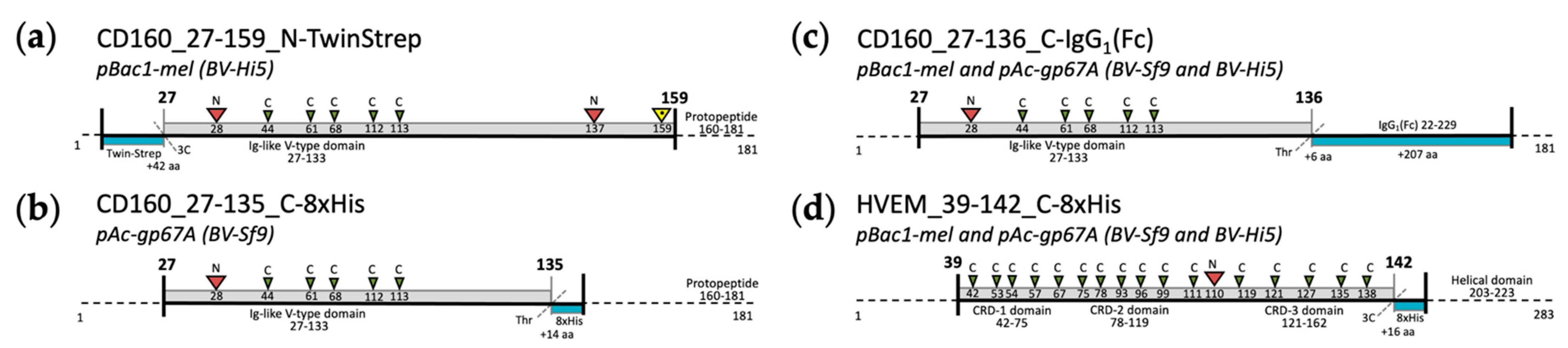
2.1.1. Molecular Cloning
2.1.2. Protein Production and Purification
2.2. Protein Peptides Identification
2.3. Crystallization
2.4. Data Collection and Processing
3. Results and Discussion
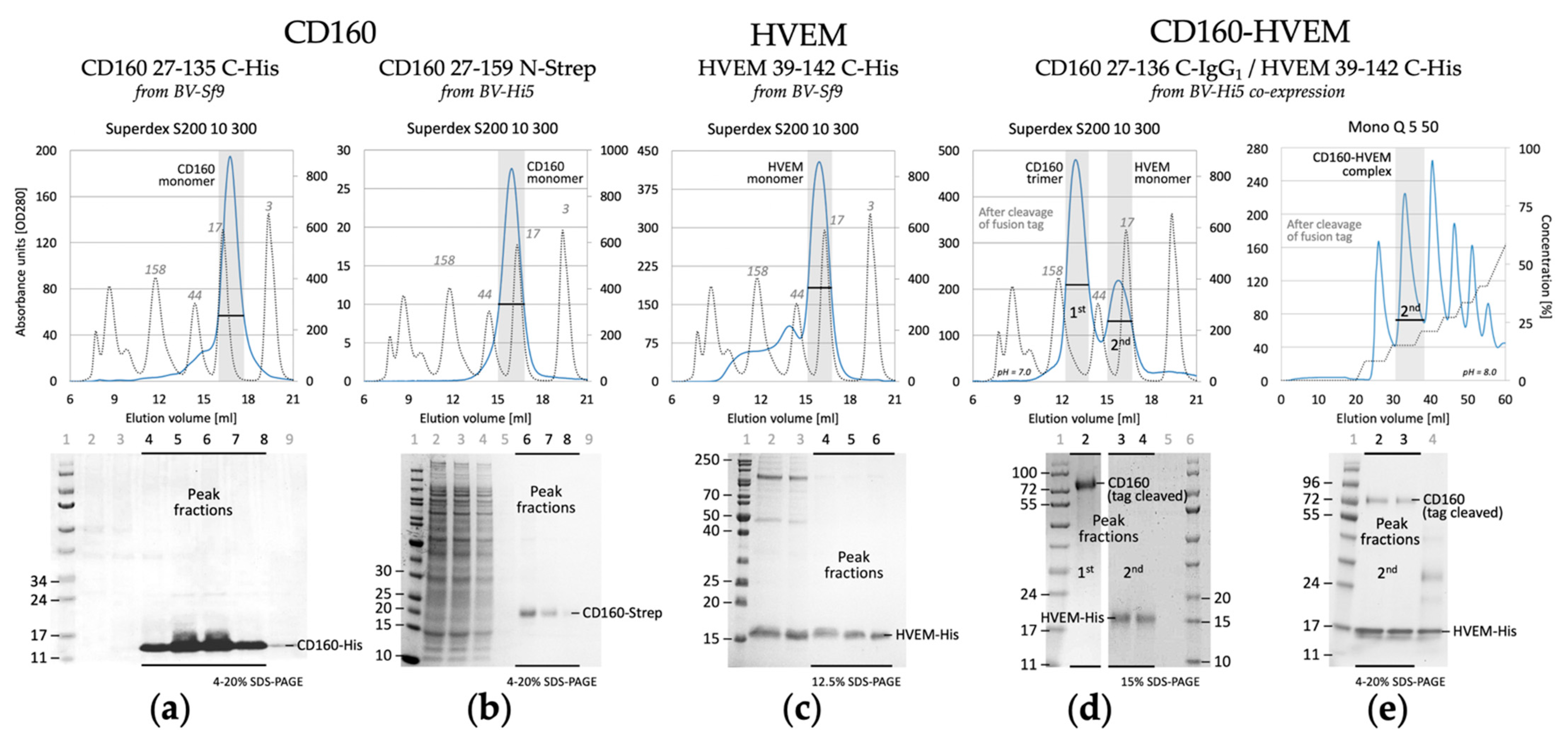
3.1. Purification and Characterization of CD160, HVEM and Their Complex
3.2. Peptide Identification of CD160 and HVEM by Mass Spectrometry
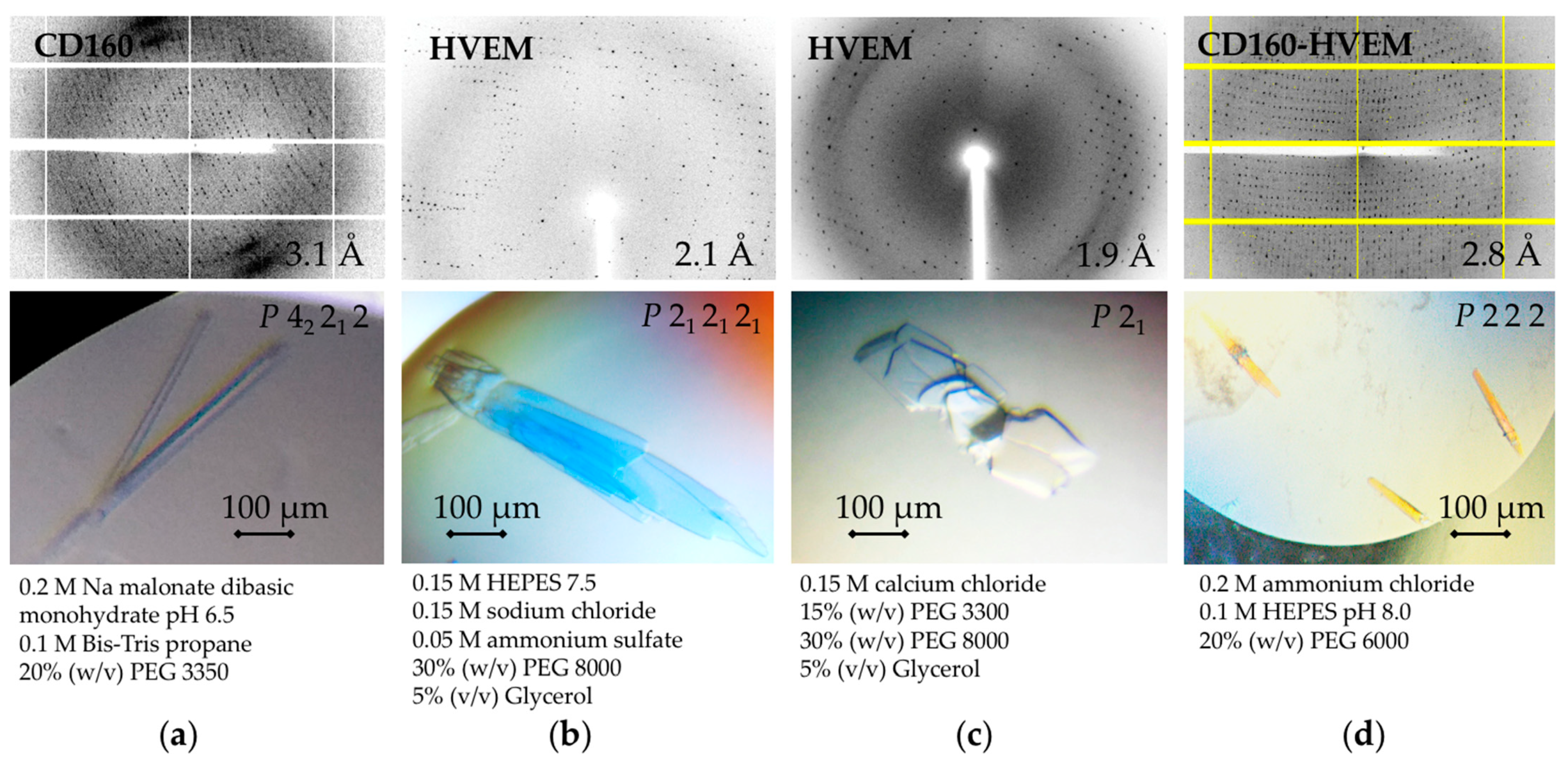
3.3. Crystallization of Native (Non-Linked) CD160–HVEM Complex and Both Proteins in the Absence of Their Binding Partner
3.4. Initial X-ray Analysis of the CD160, HVEM and CD160–HVEM Crystals
3.5. Functional Implications of Trimeric CD160 Occurring in Solution
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, W.; Garrett, S.C.; Fedorov, E.V.; Ramagopal, U.A.; Garforth, S.J.; Bonanno, J.B.; Almo, S.C. Structural basis of CD160: HVEM recognition. Structure 2019, 27, 1286–1295 e4. [Google Scholar] [CrossRef]
- Kojima, R.; Kajikawa, M.; Shiroishi, M.; Kuroki, K.; Maenaka, K. Molecular basis for herpesvirus entry mediator recognition by the human immune inhibitory receptor CD160 and its relationship to the cosignaling molecules BTLA and LIGHT. J. Mol. Biol. 2011, 413, 762–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiles, K.M.; Whitbeck, J.C.; Lou, H.; Cohen, G.H.; Eisenberg, R.J.; Krummenacher, C. Herpes simplex virus glycoprotein D interferes with binding of herpesvirus entry mediator to its ligands through downregulation and direct competition. J. Virol. 2010, 84, 11646–11660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anumanthan, A.; Bensussan, A.; Boumsell, L.; Christ, A.D.; Blumberg, R.S.; Voss, S.D.; Patel, A.T.; Robertson, M.J.; Nadler, L.M.; Freeman, G.J. Cloning of BY55, a novel Ig superfamily member expressed on NK cells, CTL, and intestinal intraepithelial lymphocytes. J. Immunol. 1998, 161, 2780–2790. [Google Scholar] [PubMed]
- Maiza, H.; Leca, G.; Mansur, I.G.; Schiavon, V.; Boumsell, L.; Bensussan, A. A novel 80-kD cell surface structure identifies human circulating lymphocytes with natural killer activity. J. Exp. Med. 1993, 178, 1121–1126. [Google Scholar] [CrossRef] [Green Version]
- Ward-Kavanagh, L.K.; Lin, W.W.; Sedy, J.R.; Ware, C.F. The TNF receptor superfamily in co-stimulating and co-inhibitory responses. Immunity 2016, 44, 1005–1019. [Google Scholar] [CrossRef] [Green Version]
- Bodmer, J.L.; Schneider, P.; Tschopp, J. The molecular architecture of the TNF superfamily. Trends Biochem. Sci. 2002, 27, 19–26. [Google Scholar] [CrossRef] [Green Version]
- Cai, G.; Anumanthan, A.; Brown, J.A.; Greenfield, E.A.; Zhu, B.; Freeman, G.J. CD160 inhibits activation of human CD4+ T cells through interaction with herpesvirus entry mediator. Nat. Immunol. 2008, 9, 176–185. [Google Scholar] [CrossRef]
- Sedy, J.R.; Bjordahl, R.L.; Bekiaris, V.; Macauley, M.G.; Ware, B.C.; Norris, P.S.; Lurain, N.S.; Benedict, C.A.; Ware, C.F. CD160 activation by herpesvirus entry mediator augments inflammatory cytokine production and cytolytic function by NK cells. J. Immunol. 2013, 191, 828–836. [Google Scholar] [CrossRef] [Green Version]
- Shui, J.W.; Larange, A.; Kim, G.; Vela, J.L.; Zahner, S.; Cheroutre, H.; Kronenberg, M. HVEM signalling at mucosal barriers provides host defence against pathogenic bacteria. Nature 2012, 488, 222–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolova, M.; Marie-Cardine, A.; Boumsell, L.; Bensussan, A. BY55/CD160 acts as a co-receptor in TCR signal transduction of a human circulating cytotoxic effector T lymphocyte subset lacking CD28 expression. Int. Immunol. 2002, 14, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Freeman, G.J. The CD160, BTLA, LIGHT/HVEM pathway: A bidirectional switch regulating T-cell activation. Immunol. Rev. 2009, 229, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Marquet, J.; Freeman, G.J.; Tawab, A.; Bouteiller, P.L.; Roth, P.; Bolton, W.; Ogg, G.; Boumsell, L.; Bensussan, A. Cutting edge: MHC class I triggering by a novel cell surface ligand costimulates proliferation of activated human T cells. J. Immunol. 1999, 162, 1223–1226. [Google Scholar] [PubMed]
- Barakonyi, A.; Rabot, M.; Marie-Cardine, A.; Aguerre-Girr, M.; Polgar, B.; Schiavon, V.; Bensussan, A.; Le Bouteiller, P. Cutting edge: Engagement of CD160 by its HLA-C physiological ligand triggers a unique cytokine profile secretion in the cytotoxic peripheral blood NK cell subset. J. Immunol. 2004, 173, 5349–5354. [Google Scholar] [CrossRef] [Green Version]
- Le Bouteiller, P.; Barakonyi, A.; Giustiniani, J.; Lenfant, F.; Marie-Cardine, A.; Aguerre-Girr, M.; Rabot, M.; Hilgert, I.; Mami-Chouaib, F.; Tabiasco, J.; et al. Engagement of CD160 receptor by HLA-C is a triggering mechanism used by circulating natural killer (NK) cells to mediate cytotoxicity. Proc. Natl. Acad. Sci. USA 2002, 99, 16963–16968. [Google Scholar] [CrossRef] [Green Version]
- Tsujimura, K.; Obata, Y.; Matsudaira, Y.; Nishida, K.; Akatsuka, Y.; Ito, Y.; Demachi-Okamura, A.; Kuzushima, K.; Takahashi, T. Characterization of murine CD160+ CD8+ T lymphocytes. Immunol. Lett. 2006, 106, 48–56. [Google Scholar] [CrossRef]
- Murayama, T.; Takegoshi, M.; Tanuma, J.; Eizuru, Y. Analysis of human cytomegalovirus UL144 variability in low-passage clinical isolates in Japan. Intervirology 2005, 48, 201–206. [Google Scholar] [CrossRef]
- Tu, T.C.; Brown, N.K.; Kim, T.J.; Wroblewska, J.; Yang, X.; Guo, X.; Lee, S.H.; Kumar, V.; Lee, K.M.; Fu, Y.X. CD160 is essential for NK-mediated IFN-gamma production. J. Exp. Med. 2015, 212, 415–429. [Google Scholar] [CrossRef] [Green Version]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Omasits, U.; Ahrens, C.H.; Muller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef] [Green Version]
- Kabsch, W. Xds. Acta Cryst. D Biol. Cryst. 2010, 66 Pt 2, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Vigdorovich, V.; Zhan, C.; Patskovsky, Y.; Bonanno, J.B.; Nathenson, S.G.; Almo, S.C. Increased heterologous protein expression in drosophila S2 cells for massive production of immune ligands/receptors and structural analysis of human HVEM. Mol. Biotechnol. 2015, 57, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Cryst. D Biol. Cryst. 2010, 66 Pt 1, 22–25. [Google Scholar] [CrossRef]
- Collaborative Computational Project. The CCP4 suite: Programs for protein crystallography. Acta Cryst. D Biol. Cryst. 1994, 50 Pt 5, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Giustiniani, J.; Bensussan, A.; Marie-Cardine, A. Identification and characterization of a transmembrane isoform of CD160 (CD160-TM), a unique activating receptor selectively expressed upon human NK cell activation. J. Immunol. 2009, 182, 63–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, A.; Boulagnon-Rombi, C.; Menguy, T.; Giustiniani, J.; Garbar, C.; Mascaux, C.; Labrousse, M.; Milas, C.; Barbe, C.; Bensussan, A.; et al. CD160 expression in retinal vessels is associated with retinal neovascular diseases. Invest. Ophthalmol. Vis. Sci. 2018, 59, 2679–2686. [Google Scholar] [CrossRef]
- Menguy, T.; Briaux, A.; Jeunesse, E.; Giustiniani, J.; Calcei, A.; Guyon, T.; Mizrahi, J.; Haegel, H.; Duong, V.; Soler, V.; et al. Anti-CD160, alone or in combination with Bevacizumab, is a potent inhibitor of ocular neovascularization in rabbit and monkey models. Invest. Ophthalmol. Vis. Sci. 2018, 59, 2687–2698. [Google Scholar] [CrossRef]
- Cheung, T.C.; Steinberg, M.W.; Oborne, L.M.; Macauley, M.G.; Fukuyama, S.; Sanjo, H.; D’Souza, C.; Norris, P.S.; Pfeffer, K.; Murphy, K.M.; et al. Unconventional ligand activation of herpesvirus entry mediator signals cell survival. Proc. Natl. Acad. Sci. USA 2009, 106, 6244–6249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proll, J.; Bensussan, A.; Goffin, F.; Foidart, J.M.; Berrebi, A.; Le Bouteiller, P. Tubal versus uterine placentation: Similar HLA-G expressing extravillous cytotrophoblast invasion but different maternal leukocyte recruitment. Tissue Antigens 2000, 56, 479–491. [Google Scholar] [CrossRef] [PubMed]
| Name of Protein | CD160 | HVEM |
|---|---|---|
| Source Organism | Homo sapiens (Human) | |
| DNA Source | Human cDNA | |
| Forward Primers † | 5′-ATTAACATCACCAGCTCAGCTTCCCAG-3′ | 5′-CTGCCGTCCTGCAAGGAGGACGAGTACCC-3′ |
| Reverse Primers † | 3′-TGTCTCTGTGAATAGAATGGAGAAAAA-5′ (27-135) | 3′-GGCGTAAGCGCGGCACGCGGCGCAGTGGTC-5′ (39-142) |
| 3′-CCCTGTCTCTGTGAATAGAATGGAGAA-5′ (27-136) | ||
| 3′-ACTGAGAGTGCCTTCATTATGGCTGA-5′ (27-159) | ||
| Cloning/Expression Vector | pBac1-mel and/or pAc-gp67A | |
| Expression Host | Insect Trichoplusia ni (Hi5) and/or Spodoptera frugiperda 9 (Sf9) | |
| NCBI Reference Sequence | NP_008984.1 | NP_003811.2 |
| CD160 | HVEM | CD160–HVEM | ||
|---|---|---|---|---|
| Diffraction source | Synchrotron DIAMOND BL-I04 | Synchrotron SSRL BL7-1 | Synchrotron DIAMOND BL-I04 | |
| Wavelength (Å) | 1.281 | 0.978 | 0.975 | 1.282 |
| Temperature (K) | 100 | |||
| Detector | PIXEL Dectris Eiger2 X 16M | CCD ADSC Quantum 315 | CCD ADSC Quantum 315 | PIXEL Dectris Eiger X 16M |
| Crystal-to-detector distance (mm) | 207 | 200 | 200 | 222 |
| Rotation range per image (°) | 0.1 | 1 | 1 | 0.15 |
| Total rotation range (°) | 360 | |||
| Exposure time per image (s) | 0.050 | 0.100 | 0.100 | 0.010 |
| Space group | P 42 21 2 | P 21 21 21 | P 21 | P 2 2 2 |
| a, b, c (Å) | 122.22, 122.22, 154.79 | 47.21, 62.42, 103.57 | 50.19, 47.51, 67.29 | 74.05, 94.14, 140.52 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 106.69, 90 | 90, 90, 90 |
| Mosaicity (°) | - | - | - | 0.14 |
| Resolution range (Å) | 3.12–75.46 (3.12–3.17) | 2.10–20.0 (2.10–2.16) | 1.90–20.00 (1.90–1.98) | 2.84–29.10 (2.84–2.91) |
| Redundancy | 26.4 | 7.6 | 8.6 | 19.8 |
| Unique reflections | 21562 | 17307 | 23026 | 23783 |
| Completeness (%) | 100.0 (100.0) | 98.8 (97.4) | 99.25 (96.22) | 99.10 (88.80) |
| CC half † | 1.0 (0.39) | 1.0 (0.63) | 1.0 (0.66) | 1.0 (0.80) |
| Mean I/σ(I) | 9.1 (0.3) | 8.1 (2.7) | 12.8 (2.2) | 20.9 (2.7) |
| R factor r.i.m. (%) ‡ | 0.334 (6.932) | 0.132 (0.976) | 0.182 (0.875) | 0.134 (0.981) |
| Overall B factor from Wilson plot (Å2) | 87.65 | 21.66 | 19.13 | 16.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lenhartová, S.; Nemčovič, M.; Šebová, R.; Benko, M.; Zajonc, D.M.; Nemčovičová, I. Molecular Characterization of the Native (Non-Linked) CD160–HVEM Protein Complex Revealed by Initial Crystallographic Analysis. Crystals 2021, 11, 820. https://doi.org/10.3390/cryst11070820
Lenhartová S, Nemčovič M, Šebová R, Benko M, Zajonc DM, Nemčovičová I. Molecular Characterization of the Native (Non-Linked) CD160–HVEM Protein Complex Revealed by Initial Crystallographic Analysis. Crystals. 2021; 11(7):820. https://doi.org/10.3390/cryst11070820
Chicago/Turabian StyleLenhartová, Simona, Marek Nemčovič, Radka Šebová, Mário Benko, Dirk M. Zajonc, and Ivana Nemčovičová. 2021. "Molecular Characterization of the Native (Non-Linked) CD160–HVEM Protein Complex Revealed by Initial Crystallographic Analysis" Crystals 11, no. 7: 820. https://doi.org/10.3390/cryst11070820
APA StyleLenhartová, S., Nemčovič, M., Šebová, R., Benko, M., Zajonc, D. M., & Nemčovičová, I. (2021). Molecular Characterization of the Native (Non-Linked) CD160–HVEM Protein Complex Revealed by Initial Crystallographic Analysis. Crystals, 11(7), 820. https://doi.org/10.3390/cryst11070820







