Abstract
In this paper, we describe the synthesis and structural characterization of the 1-(cyclohex-1-en-1-yl)-3-(prop-2-yn-1-yl)-1,3-dihydro-2H-benzimidazol-2-one (2) via IR, NMR (1H and 13C), and HRMS. The crystal structure of the isolated organic compound 2 was confirmed through single-crystal X-ray diffraction analysis. The experimental results regarding the molecular geometry and intermolecular interactions within the crystal are in accordance with the DFT calculations and Hirshfeld surface analysis.
1. Introduction
Aromatic compounds with heterocyclic structures incorporating nitrogen and oxygen play an important role in organic chemistry synthesis. Benzimidazoles containing nitrogen are well known for their wide range of biological activities [1,2,3,4]. In addition, they have been studied extensively for their anticancer activity [5], antifungal agents [1,6], antiviral activity [7,8], and antidiabetic properties [9,10]. Moreover, the benzimidazolone nucleus is often used as a scaffold for the development of therapeutic molecules with medicinal and biological applications. Benzimidazolone derivatives display noticeable pharmacological applications such as antimicrobial [11,12], anti-inflammatory [13], and antitumor agents [7,14]. Additionally, the use of benzimidazole and its derivatives as corrosion inhibitors for mild steel in an acidic medium has been reported [15,16]. Several methodologies can be employed for the synthesis of benzimidazolone derivatives, with one common approach involving the condensation reaction between o-phenylenediamine and ketoester derivatives [17,18].
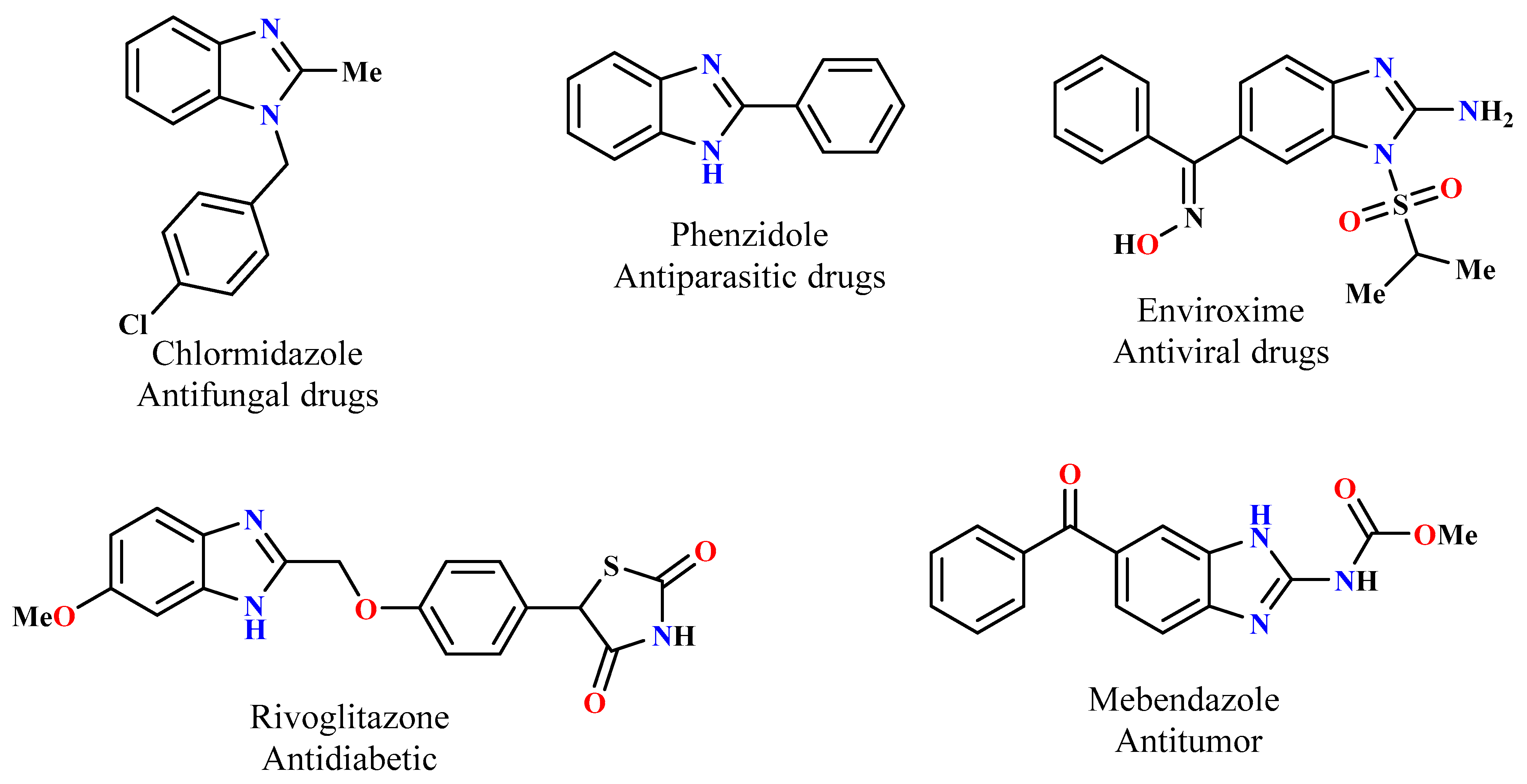
Within the various categories of heterocycles, benzimidazole derivatives have gained notable recognition in medicinal chemistry [19,20,21]. As a result, significant interest has been directed toward the creation of new benzimidazole systems. Since the N-alkylation of benzimidazole derivatives generates rigid analogs, it is highly desirable to develop such scaffolds [22,23,24]. Recently, alkylating agents have been extensively studied, which has led to the development of many biologically active benzimidazole compounds, particularly molecules that are based on a triazolic moiety [25,26]. Figure 1 shows some different compounds reported in the literature that have been prepared for their biological activities from benzimidazole and its derivatives.
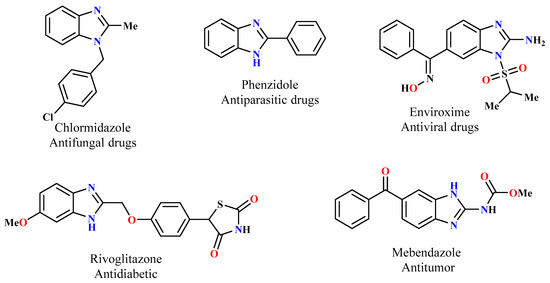
Figure 1.
Examples of biologically active benzimidazole derivatives.
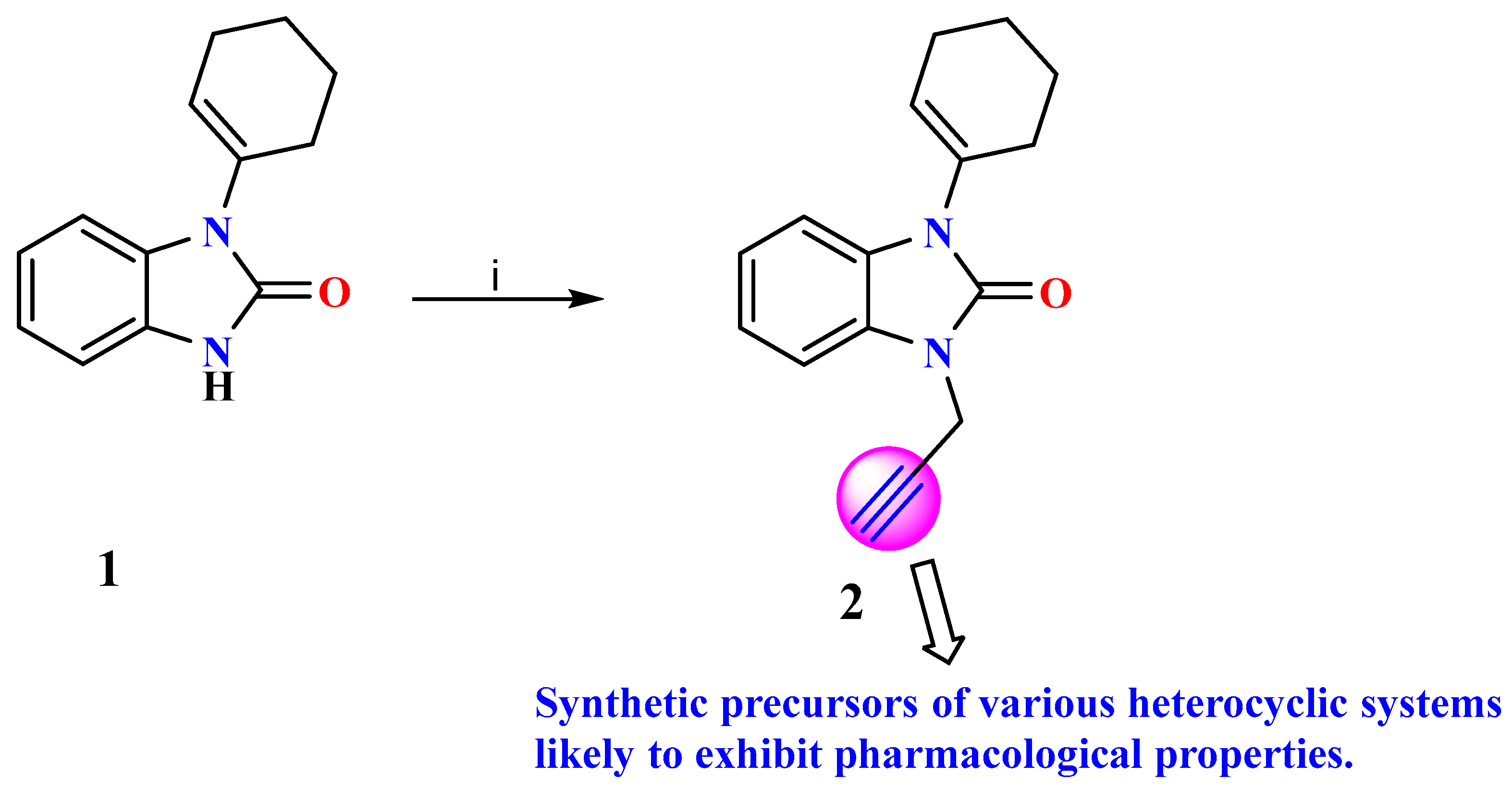
In this work, we approached the synthesis of benzoimidazol-2-one 2 through the solid–liquid phase-transfer alkylation of benzimidazolone 1. The structure of (2) was established using FT-IR and NMR methods and confirmed through a single-crystal X-ray structure determination. Furthermore, Hirshfeld surface (HS) analysis was used to reveal the different intramolecular contacts, their percentage contribution, the contour and surface plots (dnorm, di, and de), and the major as well as minor contributions of the contacts for compound 2. Furthermore, the chemical activity parameters determined via the molecular electrostatic potential (MEP) were investigated, demonstrating the nucleophilic and electrophilic character, which is related to the charge distribution around the crystal molecule (2).
2. Results and Discussion
2.1. Synthesis and Structural Characterization
The condensation reaction, a valuable synthetic process in organic chemistry, leads to the formation of benzimidazole and benzodiazepine derivatives. Continuing our previous investigations on the synthesis of benzimidazolone derivatives, we report the synthesis of 1-(cyclohex-1-en-1-yl)-3-(prop-2-yn-1-yl)-1,3-dihydro-2H-benzimidazol-2-one (2) in one step using a direct alkylation reaction under phase-transfer catalysis conditions (Scheme 1). The title compound (2) was synthesized through the direct N-alkylation of benzimidazolone 1 using a previously described process [18,22]. Compound 1 was reacted with propargyl bromide which acted as a catalyst. The reaction was carried out at room temperature for 6 h. Then, the reaction mixture was purified with a hexane/ethyl acetate mixture as an eluent, and then we filtered off the precipitated solid, dried well, and recrystallized from ethyl acetate to prepare 2 with a good yield.
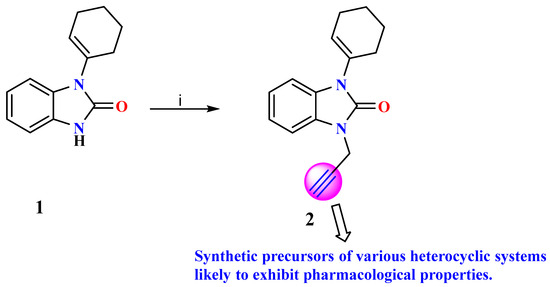
Scheme 1.
Reagent and condition: (i) R-X, K2CO3 and DMF/CH3CN, T.A, 6 h.
The structure of 2 was determined using FT-IR and NMR spectroscopy methods, with extensive validation accomplished by cross-referencing the acquired spectroscopic data with the available literature [18]. Furthermore, the geometric arrangement of the chemical was validated using XRD analysis, providing additional confirmation of its structural characteristics.
2.2. FT-IR Analysis
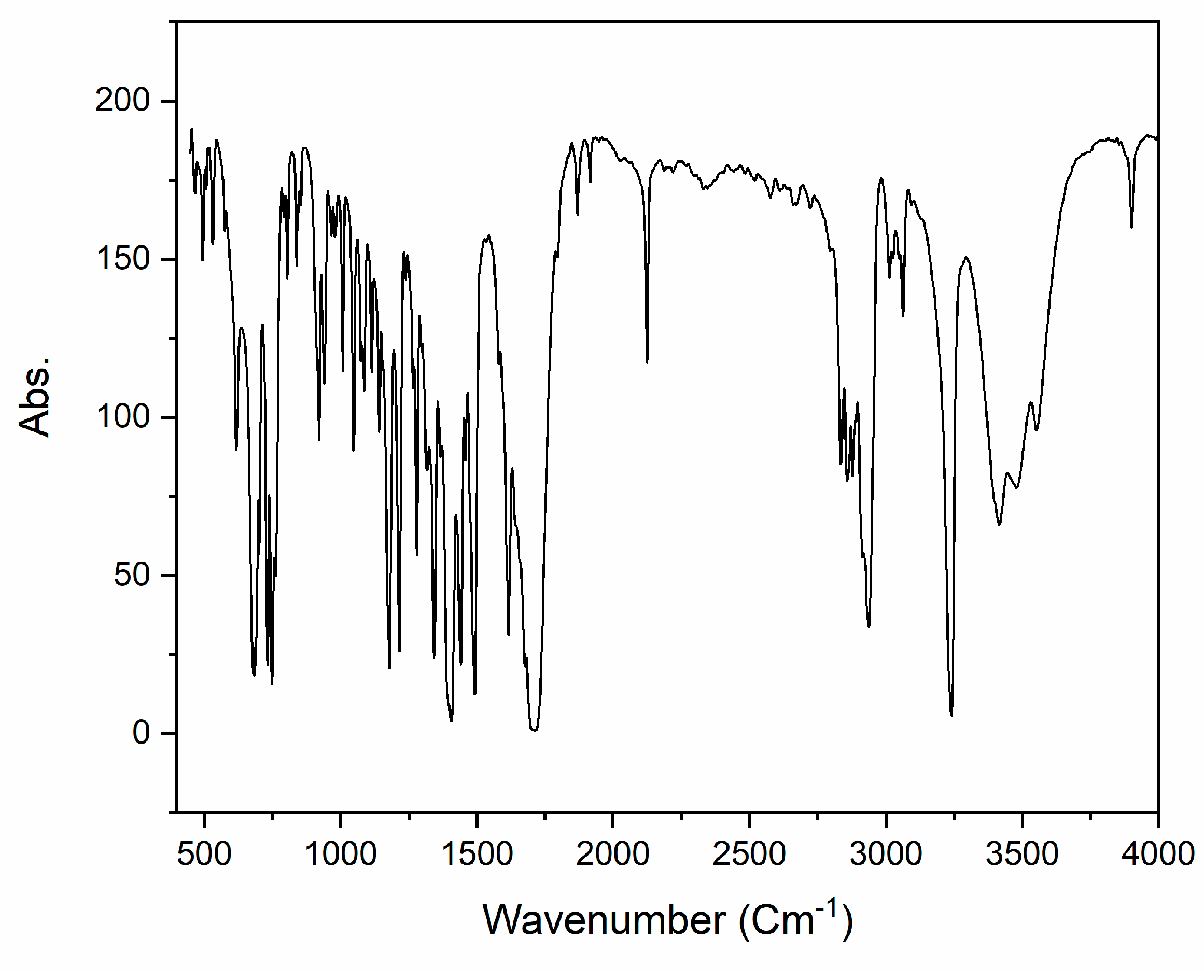
The infrared (IR) spectrum of 2 (KBr pellet, cm−1) reveals prominent characteristic peaks, as illustrated in Figure 2. Specifically, the CH2 group exhibits a notable peak at 2930 cm−1, the C−N stretch appears at 1404 cm−1, while other distinctive groups, such as the C=O stretch and the C≡C triple bond, are observed at 1710 cm−1 and 2125 cm−1, respectively. For a more comprehensive understanding, the theoretical IR spectrum was computed using the B3LYP/6-311++G(d,p) level in the gas phase and is presented in Figure 2. The most important experimental and calculated harmonic stretching frequencies are listed in Table 1. Comparing the values, it is evident that the theoretical FT-IR, derived from the same basis-set calculations, generally aligns well with the experimental data, as summarized in Table 1.
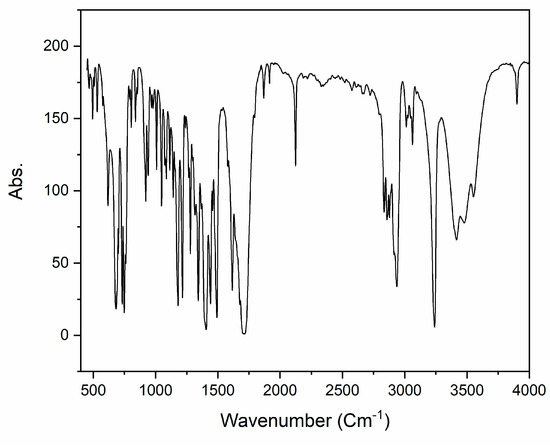
Figure 2.
Experimental IR spectrum of compound 2.

Table 1.
Vibrational study of compound 2 (cm−1).
2.3. X-ray Diffraction and Geometry Optimization
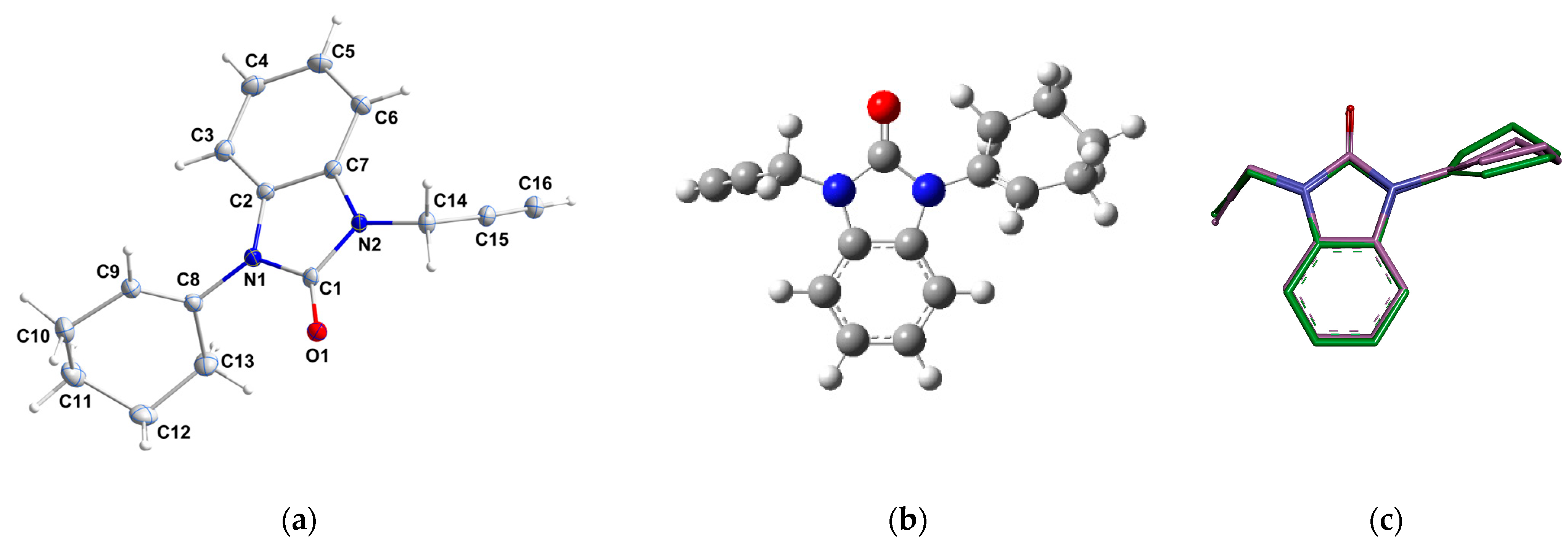
The molecular structure of 2 is depicted in Figure 3a. The benzimidazole unit (N1/C1/N2/C2-C7) is almost planar, with maximum deviations of −0.012 (1) Å for atom N1. The cyclohexenyl ring is disordered over two conformations and both it and the NCH2C≡CH group are rotated out of the mean benzimidazole plane. The benzimidazole plane makes dihedral angles of 119.26° and 111.72° with the base of the cyclohexenyl ring (N1-C8-C9) and the approximately planar (N2-C14-C15) propynyl chain, respectively. The dihedral angle between the cyclohexenyl ring and the NCH2C≡CH group is 58.99°. The major conformation of the cyclohexenyl ring has a half-chair conformation with puckering parameters of Q = 0.496(2)Å, θ = 128.1(2), and φ = 23.4 (4)° [27].
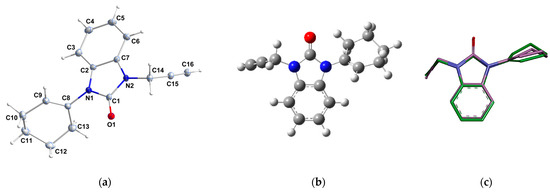
Figure 3.
(a) ORTEP of compound 2 at a 50% probability level (only the major disorder component is shown), (b) optimized structure of compound 2, and (c) superimposition of the X-ray structure (green) and the optimized structure (magenta) on the studied molecule 2.
Table 2 provides a summary of the crystal data, the data collection process, and details of the structural refinement. The positioning of C-bound H atoms was determined through calculated positions, where C-H distances ranged from 0.93 to 0.97 Å. Subsequently, refinement was conducted using the riding-model approximation, with Uiso(H) = 1.2–1.5 Ueq of the respective parent atom, as depicted in Table 3.

Table 2.
Details of the XRD data collection and structure refinement parameters for 2.

Table 3.
Hydrogen bond geometry (Å, °).
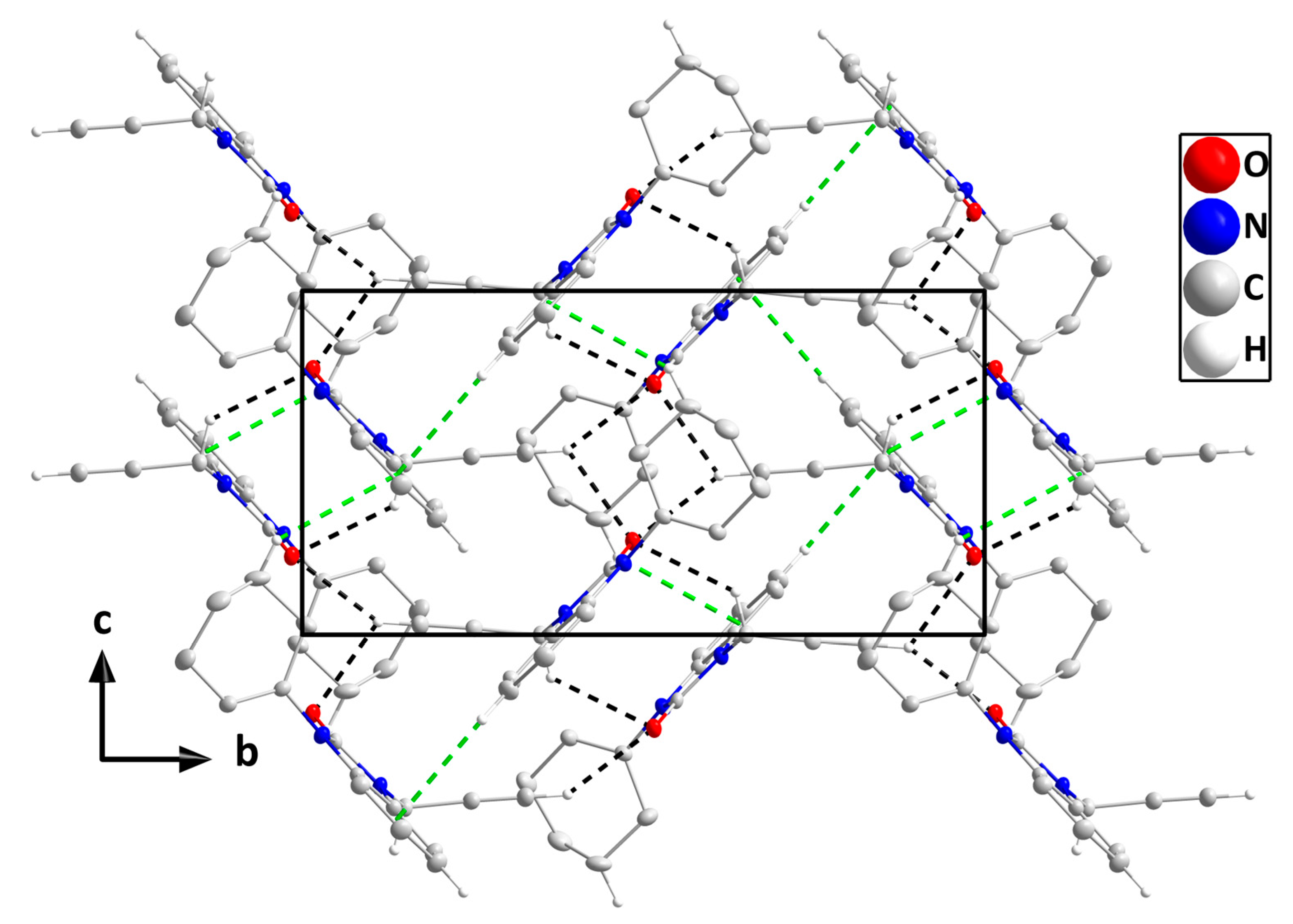
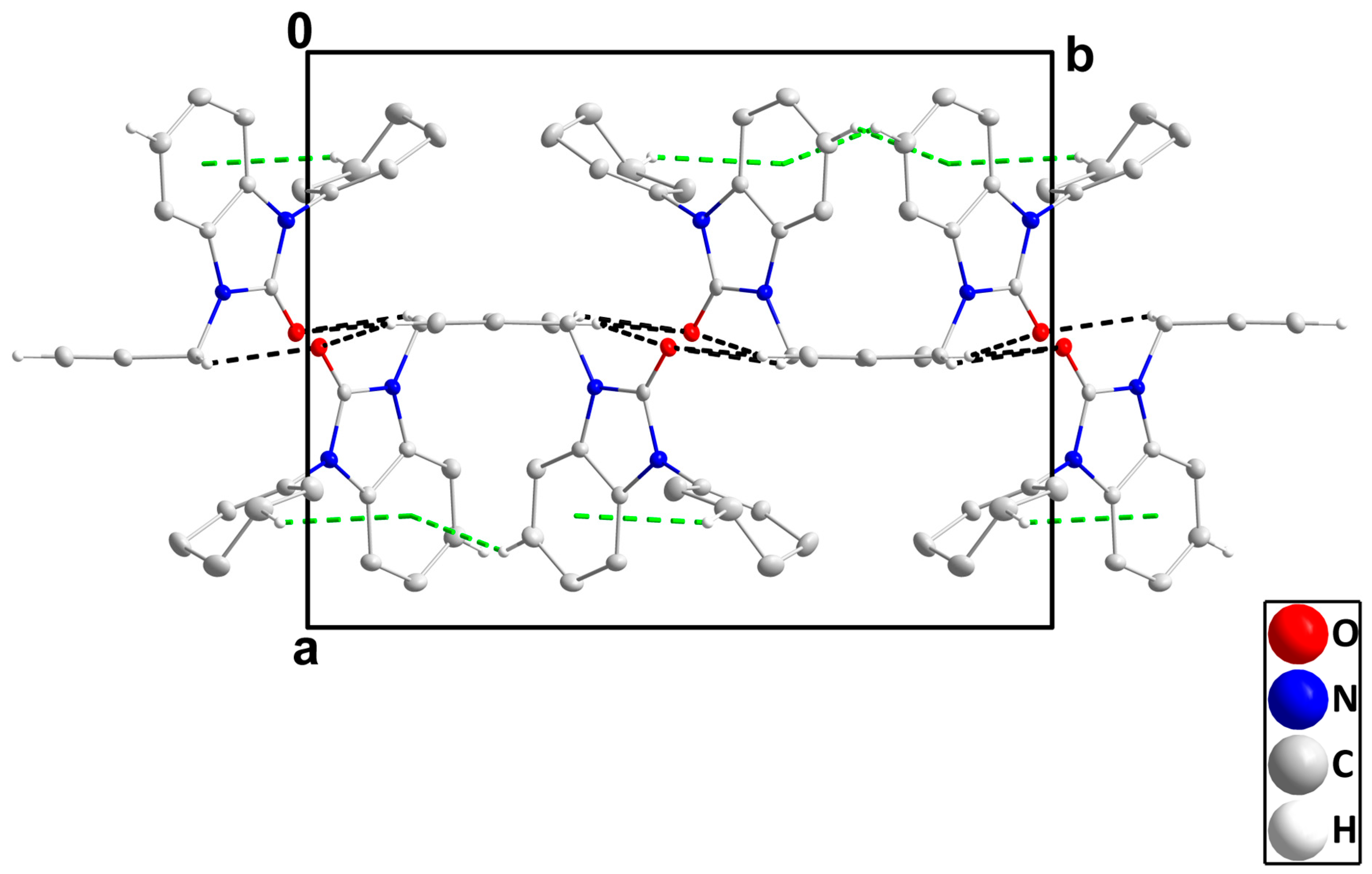
In the crystal, C14-H14B‧‧‧O1 and C16-H16‧‧‧O1 hydrogen bonds plus C5-H5‧‧‧Cg2 and C12-H12B‧‧‧Cg2 interactions (Table 3) form layers of molecules parallel to the bc plane (Figure 4), which stack along the a-axis direction with normal van der Waals contacts (Figure 5).
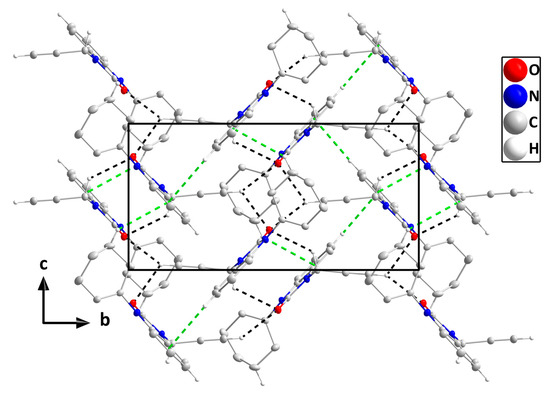
Figure 4.
A portion of one layer, observed along the a-axis orientation, illustrates C-H···O hydrogen bonds and C-H···π(ring) interactions denoted by black and green dashed lines, respectively. For clarity, hydrogen atoms with no interaction are omitted from the representation.
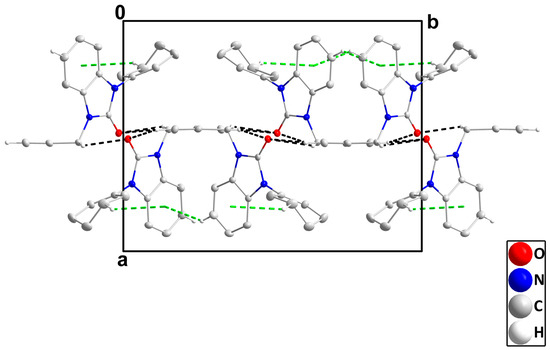
Figure 5.
Stacking of layers along the a-axis direction. Hydrogen atoms are omitted for clarity.
The measured X-ray data and the computed geometry parameters, including bond lengths and bond angles, are listed in Table 4. The ORTEP, optimized geometry, and superimposition of the X-ray structure alongside the optimized structure are shown in Figure 2. After aligning the optimized ideal molecular geometry obtained through DFT calculations with the final geometry of 2 derived from the X-ray data, the determination of the root mean square deviation (RMSD) value becomes feasible through superposition, as shown in Figure 3c. This process allows a quantitative assessment of the structural congruence between the experimentally determined X-ray structure and the theoretically optimized molecular geometry. From the alignment (Figure 3c), it is clear the optimized geometry is superimposed upon the experimental one with an overlay similarity of 0.98%, which means that the puckering potential is consistent at the B3LYP level, except for the cyclohexenyl ring that showed a small deviation. This deviation was confirmed by an RMSD value of 0.18. According to this, the DFT calculation yields a meaningful geometry of the benzimidazole-2-one (2), and the findings of the obtained experimental data from the X-ray correctly described the calculated geometric parameters. The findings revealed minimal heterogeneity between experimental and computational methods. From these results presented in Table 4 and depicted in Figure 2, the selected theoretical method and the basis set (i.e., B3LYP/6-311++G(d,p)) could be suitable for the DFT calculation study of (2).

Table 4.
Selected geometrical parameters for the title compound 2.
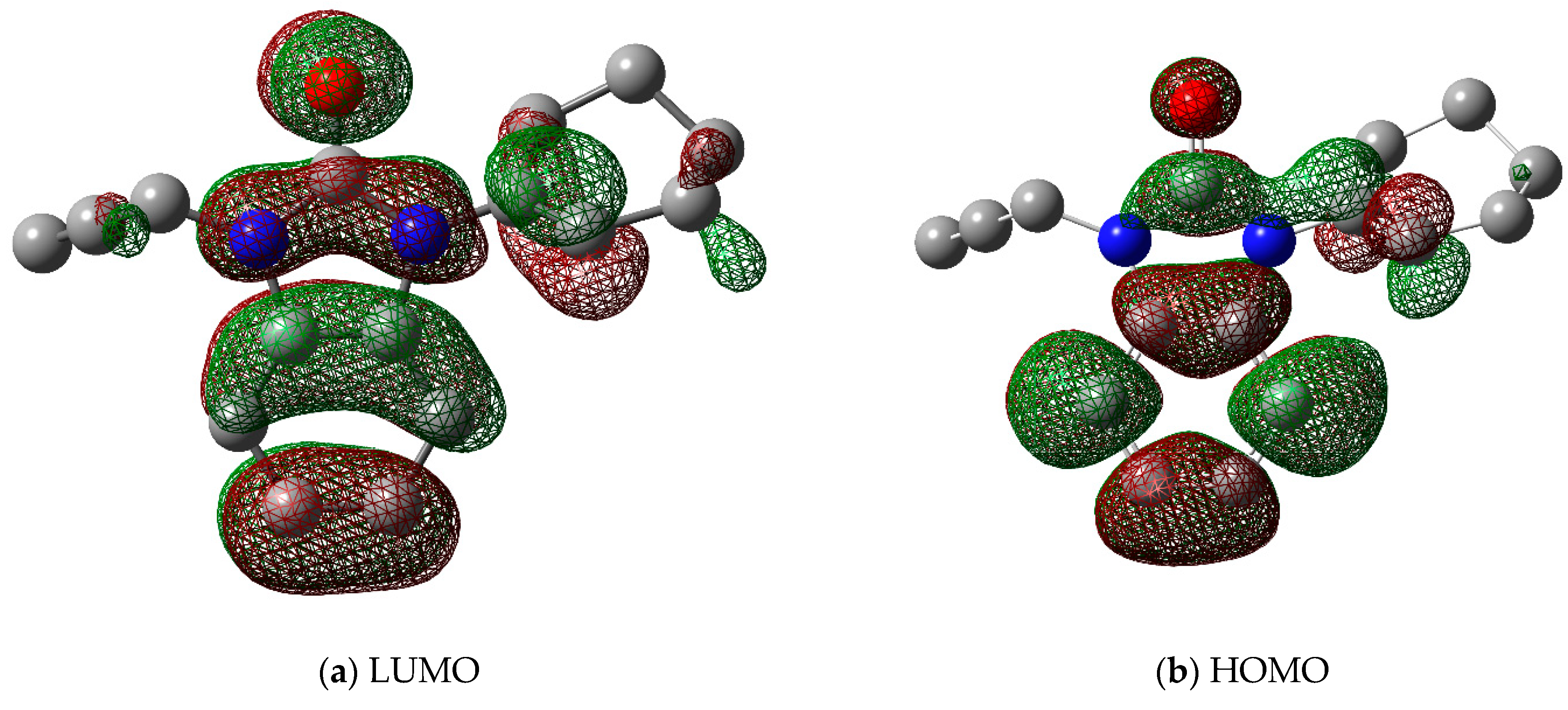
A molecule with a large gap (∆E) (i.e., the highest occupied molecular orbital (HOMO) minus the lowest unoccupied molecular orbital (LUMO)) has generally a high kinetic stability and a low chemical reactivity (Figure 6) [28]. In terms of the energetic stability of a molecule, it is unfavorable to add electrons to a high-lying HOMO and to remove electrons from a low-lying LUMO. Table 5 shows the calculated molecular reactivity parameters for the title molecule, including electronegativity (χ), ionization potential (IP), chemical potential (μ), chemical hardness (η), chemical softness (S), and electron affinity index (EA).
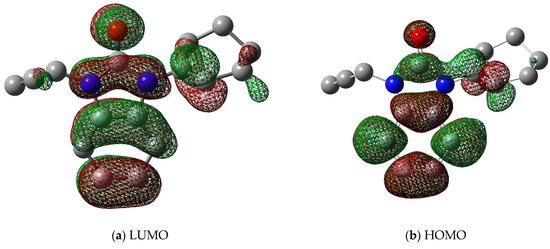
Figure 6.
Frontier molecular orbitals (a) LUMO and (b) HOMO of the title crystal 2.

Table 5.
Calculated chemical reactivity parameters for 2.
2.4. Hirshfeld Surface Analysis
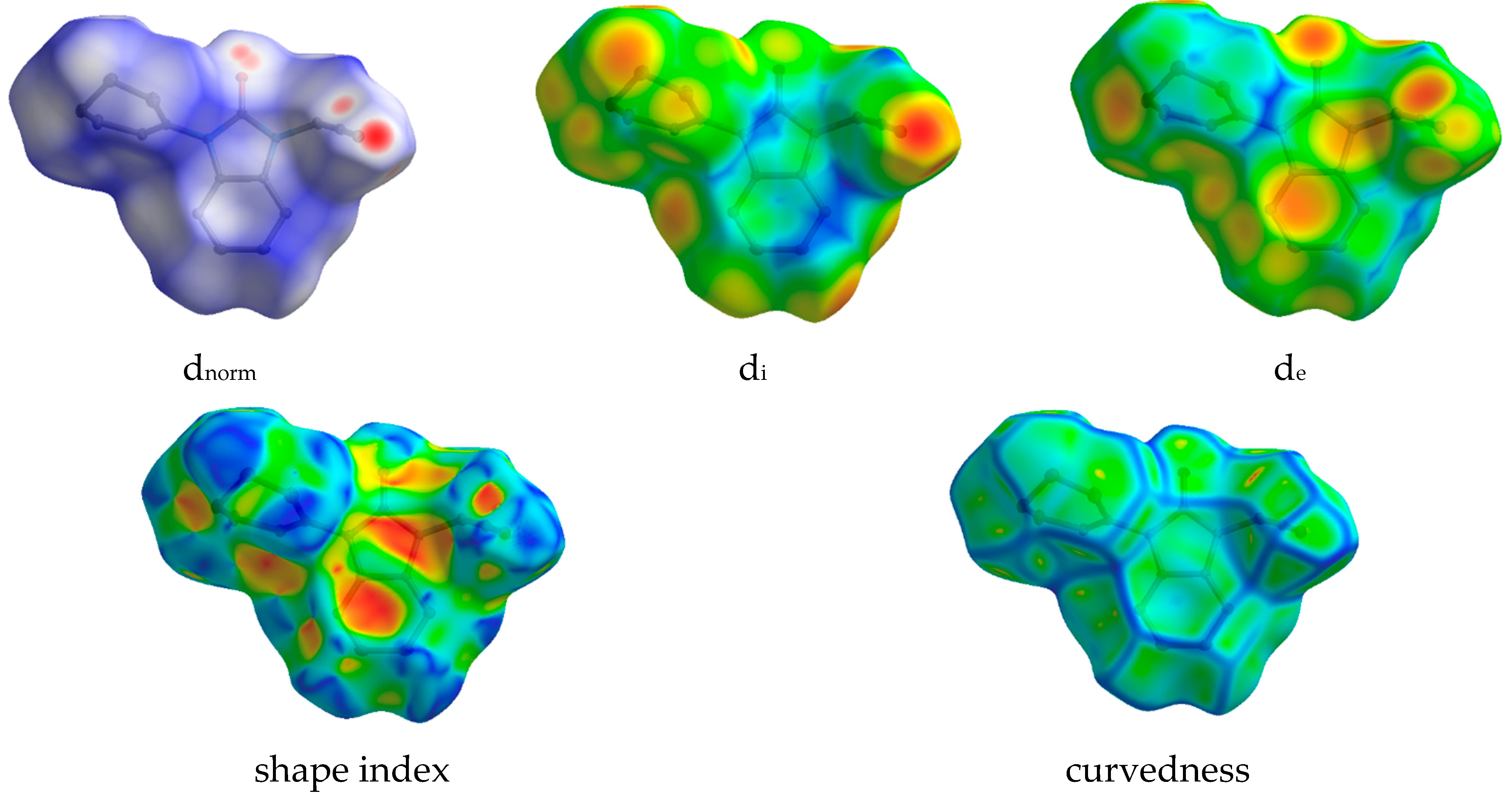
In view of visualizing the different intermolecular contacts in the crystal packing of compound 2, an analysis using the Hirshfeld surface (HS) was performed. This method facilitates the identification of regions of interaction between pairs of atoms, shedding light on the contributions of these contacts. The analysis specifically highlights atoms with the potential to engage in hydrogen bonds and π-stacking interactions, providing valuable information on the nature and strength of intermolecular forces [29,30,31]. This approach improves our understanding of the spatial distribution and importance of specific atomic interactions within the crystal lattice. The dnorm (normalized contact distance) plays a crucial role in identifying the key sites for intermolecular contacts. In the context of the HS represented in dnorm (Figure 7), the white surface signifies the points which contact the van der Waals radii at intervals equidistant from their cumulative total. The distances of the van der Waals radius that are shorter (near touch) or longer (different contact) are represented by red and blue colors [30].
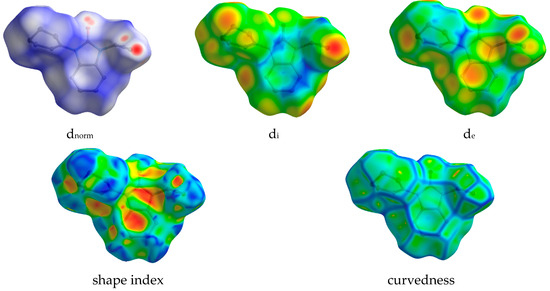
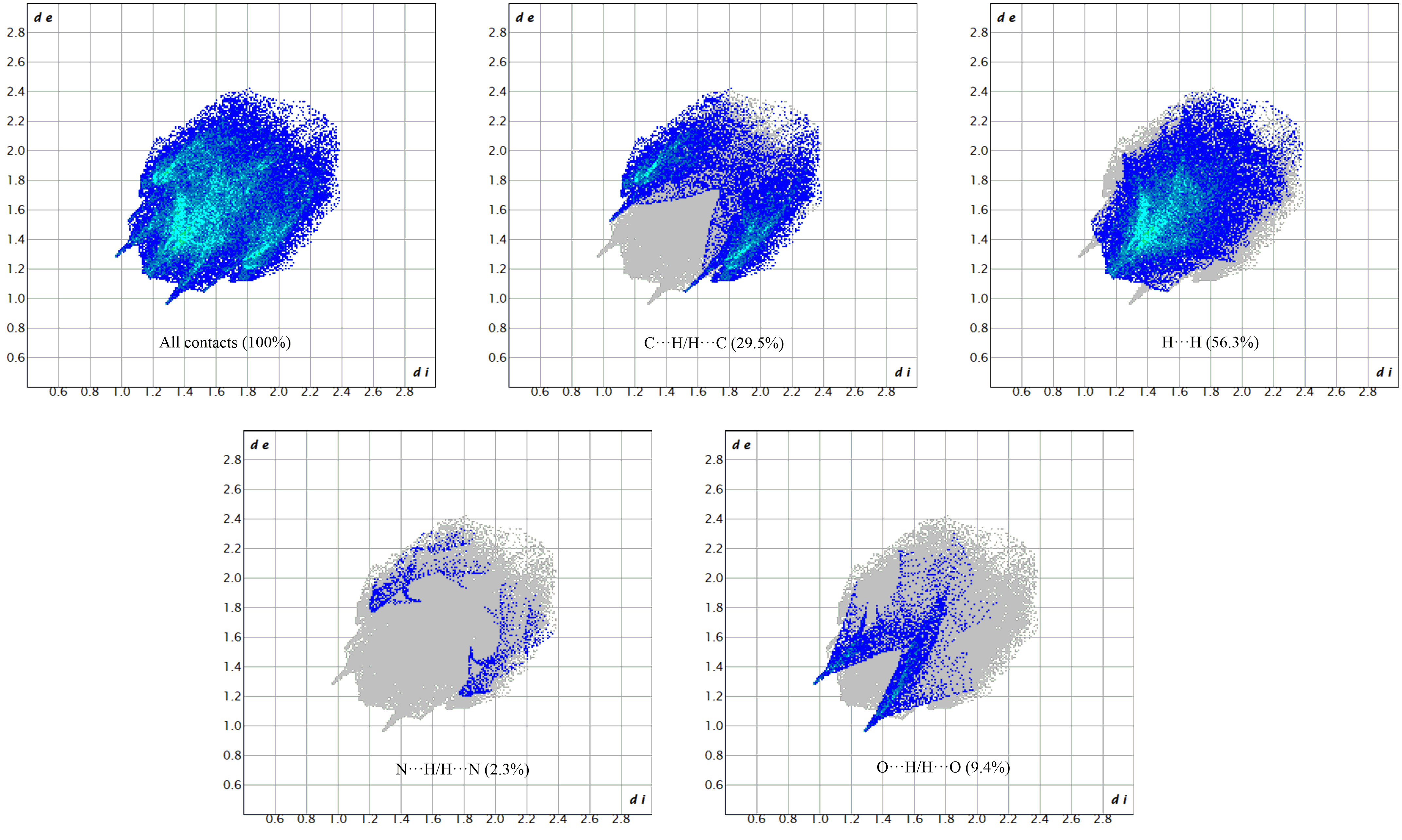
Figure 7.
Hirshfeld surface mapped with dnorm, di, de, shape index, and curvedness for compound (2).
The dnorm, di, de, curvedness, and shape index are −0.2600 to 1.1859, 0.9684 to 2.4213, 0.9696 to 2.4592 Å, −4 to 4 and −1 to 1, respectively. The dnorm, di, de, shape index, and curvedness contour maps on a molecule HS are depicted in Figure 7. The analysis of π-stacking interactions is further aided by the HS shape index and curvedness plots, which are computed based on local curvatures. The presence of blue and green colors illustrates the packed stacking of a compound throughout the curvature. The 2D fingerprint plots quantified the contribution of each type of interaction to the HS. Red spots on the surface designate intermediate interactions taking into account the hydrogen bonds and the interatomic contacts between the molecules. As can be seen on the dnorm surface (Figure 7), the C-H‧‧‧O intermolecular hydrogen bonds and C-H‧‧‧Cg over the surface were indicated by the adjacent red– blue spots.
Dominant interactions within the HS are corroborated by the fingerprint diagrams, as illustrated in Figure 8, where one molecule acts as an acceptor (di > de) while the other functions as a donor (de > di). The 2D fingerprint diagrams, depicted in Figure 8, highlighted the prevalence of specific intermolecular contacts, with H‧‧‧H and C‧‧‧H interactions being the most prevalent in 2, constituting 56.3% and 25.5% of the total area, respectively. Other dominant forces are O‧‧‧H/H‧‧‧O (9.4%) and N‧‧‧H/H‧‧‧N (2.3%) contacts for (2). The presence of π-π-stacking-type interactions in the shape index is corroborated by the flat green surface, delimited by a blue hump close to the red hollows, as well as a blue outline on the curvedness surface. The 2D fingerprint plots and the molecular HS regions are useful in clarifying the contributions of the different inter-contacts that participate in the stabilization of the crystal structure. These contributions indicate that the arrangement of 2 is predominantly influenced by the robust electrostatic interactions that drive the crystal-packing force in 2.
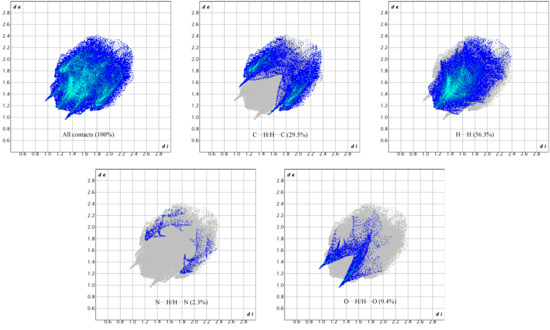
Figure 8.
Two-dimensional fingerprint plots for 2.
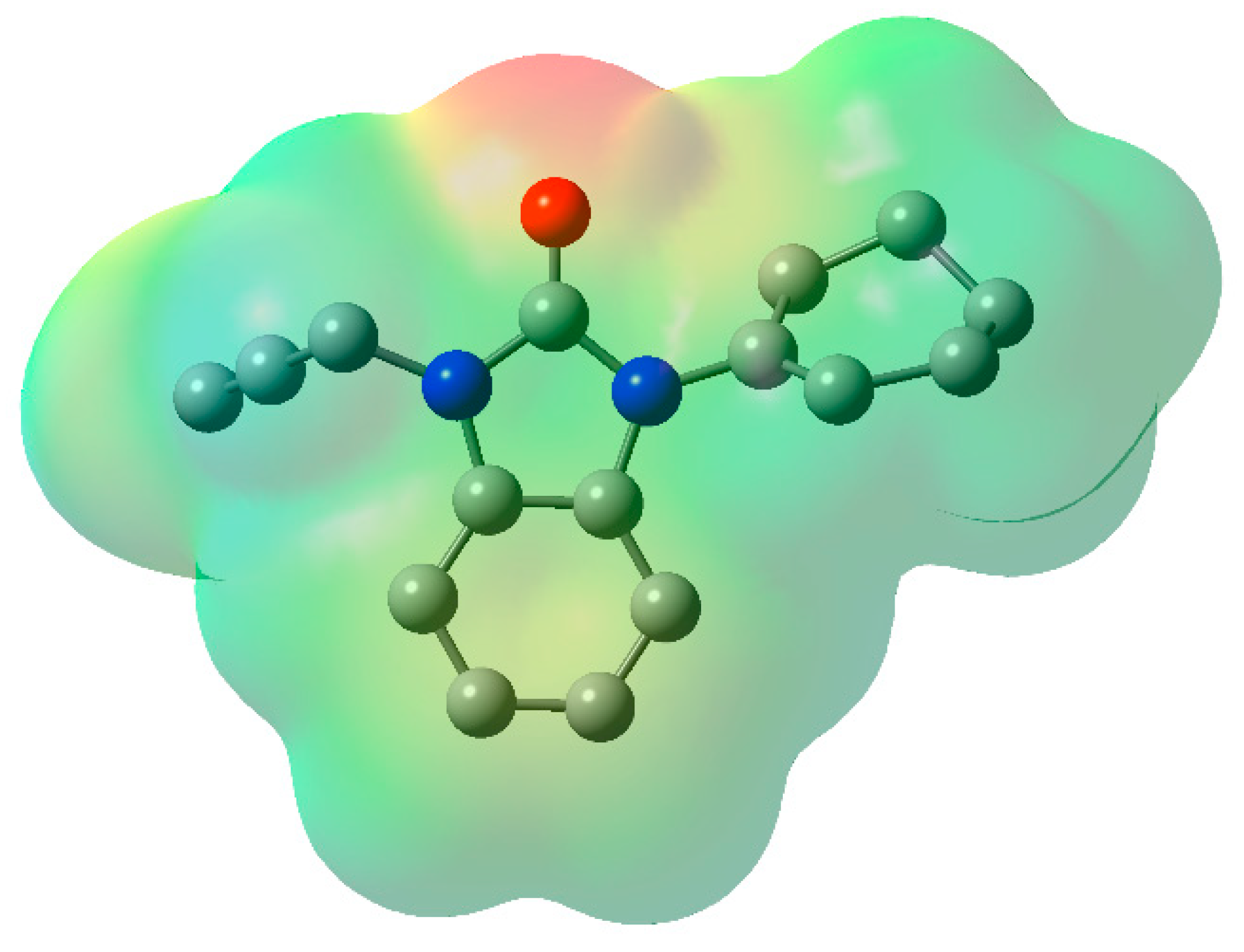
2.5. Molecular Electrostatic Potential (MEP)
The MEP is associated with the electronic density and serves as a valuable descriptor for identifying regions pertinent to nucleophilic and electrophilic reactions. It also aids in understanding chemical reactivity and hydrogen bonding interactions [32]. The various electrostatic potential values are visually represented with distinct colors: green represents zero potential regions, red represents the most negative electrostatic potential surface, and blue represents the most positive electrostatic potential areas. Within the MEP, the positive electrostatic potential (depicted by the blue areas) corresponds to the push exerted by nuclei, while the negative electrostatic potential (represented by yellow and red) is associated with the electrostatic charge of a proton and the overall electron density within the molecule. The negative electrostatic potential (shown by the yellow and red regions) is largely located on the nitrogen and oxygen atoms, making them the most reactive places for an electrophilic assault, as seen in Figure 9. In contrast, hydrogen atoms have a positive electrostatic potential (indicated by the blue patches), indicating that they are the most reactive places for a nucleophilic attack.
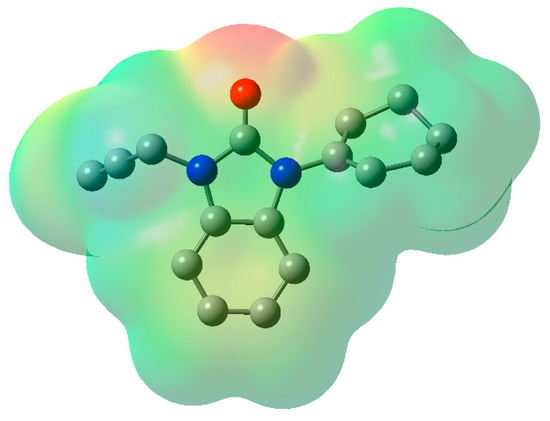
Figure 9.
Molecular electrostatic potential map of 2, Hydrogen atoms were omitted for clarity.
2.6. Pharmacological Analysis
To determine the bioactivity of benzimidazolone (2), the physicochemical and pharmacokinetic properties were examined using the SwissADME web tool, and the results are shown in Table 6. Compound (2) meets the topological polar surface area (TPSA) criteria specified by the Egan rule (TPSA ≤ 132 Å2) and the Veber rule (TPSA ≤ 140 Å2) [33,34]. The LogP values aid in determining the transport and binding behavior of medicinal molecules inside the body [35]. The SwissADME tool predicts LogP values using five different models, and the average of the five predicted LogP values represents the lipophilicity of our produced molecule. Molecule (2) has a high absorption rate in the gastrointestinal (GI) tract. The negative log Kp value indicates that title chemical (2) is less skin-permeant [36].

Table 6.
Drug-like properties of 2 estimated through the use of SwissADME online tool.
The toxicity of molecule (2) was calculated using the GUSAR online tool and is illustrated in Table 7. According to these results, title molecule (2) belongs to the toxic (Class 4) group in the oral mode and the slightly harmful (Class 4) group in the intraperitoneal (IP) and subcutaneous (SC) modes.

Table 7.
Estimated acute toxicity prediction for molecule (2) using the GUSAR server.
3. Experimental and Computational Methods
3.1. General
The determination of melting points was carried out using an open capillary Buchi 510 instrument. The spectroscopic analysis, involving 13C NMR (75 MHz) and 1H NMR (300 MHz), was conducted on a Bruker spectrometer. The recorded spectra present numerical values of chemical shifts in parts per million (ppm), relative to tetramethylsilane (TMS) set as the reference at 0.00 ppm. This methodology ensures precision and accuracy in assessing both the physical and spectroscopic characteristics of the compounds under investigation. A Triple TOFTM 5600 LC/MS-MS System (AB SCIEX) was utilized to acquire high resolution mass spectral data. The ion spray voltage, in the 5500 ionization mode, was utilized in the mass spectra. To perform column chromatography, E-Merck silica gel 60-F254 was used. To monitor the progression of the reaction, thin-layer chromatography (TLC) was employed, utilizing silica gel 60-F254. The UV-light method was employed for spot detection at a wavelength of 254 nm. Standard purification procedures were applied to purify reagents and solvents before their use in the experiment, ensuring the reliability and accuracy of the analytical results. A EuroEA Elemental Analyser was utilized to measure the elemental analysis of 2.
3.2. Synthesis of Alkylated Benzimidazolones (2)
A mixture of 1-(cyclohex-1-en-1-yl)-1,3-dihydro-2H-benzimidazol-2-one (1) (4.7 mmol), benzyltriethylammonium chloride (2.3 mmol), and potassium carbonate (14.69 mmol) in a mixture of N,N-dimethylformamide and acetonitrile (15 mL, 3:2 v/v) was heated at 90 °C for 30 min. After cooling, propargyl bromide (4.8 mmol) was added. The mixture was stirred at room temperature for 6 h. The reaction mixture was diluted with 30 mL of distilled water and then extracted three times with 30 mL of dichloromethane. Anhydrous sodium sulfate was used to dry the mixed organic layers. The solvent was evaporated under vacuum, and the residue was purified using column chromatography on silica gel using a hexane/ethyl acetate mixture (8/2) as eluent. This procedure made it possible to obtain a pure and isolated product with a yield of 85% [22].
Characterization of 1-(Cyclohex-1-en-1-yl)-3-(prop-2-yn-1-yl)-1,3-dihydro-2H-benzimidazol-2-one (2)
White solid. Yield: 85%, 0.95 g, m.p. 118–120 °C (ethyl acetate). IR (KBr, ν (cm−1), 2125 (-C≡CH), 1710 (2C=O). 1H NMR (CDCl3): δppm: 1.63, 1.73, 2.16, 2.40 (4m, 8H, 4CH2-cyclohex-1-enyl), 2.15 (t, J = 3 Hz, 1H, HC≡C), 4.56 (d, J = 3 Hz, 2H, N-CH2-C), 5.82 (m, 1H, HC=C-, H- cyclohex-1-enyl), 6.89–7.14 (m, 4H, H-Ar). 13C NMR (CDCl3): δppm: 21.63, 22.56, 24.71, 26.75 (4C, 4CH2, C- cyclohex-1-enyl), 30.37 (1C, N-CH2-C), 72.67 (1C, HC≡C), 127.41 (4C, =CH-, C-Ar), 128.56, 129.67, 132.15 (4C, =C-, C-quaternary), 154.60 (1C, C=O). HRMS of [M+H]+ m/z 253.1335, calcd for C16H17N2O, found: 253.1296. Anal. Calcd for C16H16N2O: C, 76.16; H, 6.39; N, 11.10. Found: C, 76.56; H, 6.27; N, 11.22.
3.3. Crystal Structure Determination and Refinement
The benzimidazolone (2) was selected and X-ray intensity data were collected at 296 K on a Bruker APEX-II QUAZAR CCD graphite-monochromated Mo-Kα radiation (λ = 0.71073 Å). A complete data set was processed using Bruker SAINT. The structure solution was obtained via direct methods using SHELXT [37] and refined through a full matrix, least-squares on F2 using SHELXL [37]. The cyclohexenyl ring is disordered over two sites in a 0.844(3)/0.156(3) ratio. The two moieties were resolved and refined with some restraints that their geometries be comparable. The crystallographic characteristics, details of X-ray data collection and structure refinement parameters for 1-(cyclohexenyl)-3-(prop-2-ynyl)-1,3-dihydro-2H-benzimidazol-2-one (2) are listed in Table 2. The H atoms bound to carbon were placed in calculated positions with C-H = 0.93–0.97 Å and refined in the riding-model approximation with Uiso (H) = 1.5 Ueq of the parent atom. The residual electron density in the final difference Fourier map was between −0.23 < Dq < 0.75. The geometry of the title crystal molecule 2 was resolved and refined using WinGX [38], SHELXL, and Mercury [39].
3.4. DFT Calculations
The molecular structure was optimized and the harmonic vibrational frequencies of compound 2 were computed using the same DFT/B3LYP on a 6-311G++(d,p) basis set using the Gaussian program 09W without any restrictions on geometry. The obtained vibrational frequencies of the optimized compound using the B3LYP/6-311++G(d,p) in the gas phase were scaled by 0.9627 [40].
Hirshfeld surface (HS) analysis was carried out to determine the type of intermolecular contacts. The 2D fingerprint plots and molecular surface regions for 2 were created to offer information about the percentage contribution as well as the major and minor intermolecular contributors. The HS was mapped using dnorm, the shape index, curvature, and 2D fingerprint plots generated using Crystal Explorer 17 [41]. The primary goal of quantum chemistry lies in elucidating the molecular geometry, chemical reactivity, energy, and the intricacies of chemical bonds. DFT calculations were employed in this study to gain insights into the electrophilic and nucleophilic structures. The current investigation consists of structural and chemical properties and spectroscopic details, as well as both local indicators (molecular electrostatic potential (MEP)) and global descriptors like softness and hardness parameters. These assessments were conducted using the DFT/B3LYP/6-311++G(d,p) method, providing a comprehensive understanding of the molecular behavior and reactivity.
3.5. Drug-Likeness Prediction
The drug-like physicochemical and pharmacokinetic properties of the synthesized compound were estimated using the SwissADME online server [42]. The acute toxicity of the title molecule 2 was estimated using the GUSAR (General Unrestricted Structure-Activity Relationships) web tool by calculating the LD50 values for oral, intravenous (IV), intraperitoneal (IP), and subcutaneous (SC) administration methods [43].
4. Conclusions
Benzimidazolone (2) was prepared via the liquid-solid phase-transfer alkylation method from benzimidazolone (1). The structure of the alkylated benzimidazolone was characterized via 1H-NMR, 13C-NMR, HRMS, and X-ray analysis. This research aimed to explore the various hydrogen-bonding modes and π-π interactions in the solid. The Hirshfeld surface analysis assisted in identifying and understanding these intermolecular interactions. The 2D fingerprint plots revealed that H‧‧‧H and C‧‧‧H/H‧‧‧C intermolecular interactions are the most important contributors to these interactions.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cryst13121661/s1. 1H-NMR, 13C-NMR and HRMS spectra of 2 can be found in the Supplementary Materials.
Author Contributions
Conceptualization, M.A. and I.H.; methodology, M.L., D.C. and A.B.; software, M.A., M.A.L. and I.H.; validation, M.A.L., M.M.A., D.C. and A.B.; formal analysis, M.A., H.M., S.F. and M.F.; investigation, M.A., I.H. and M.M.A.; data curation, M.A. and I.H.; writing original draft preparation, M.A. and I.H.; visualization, M.A.L., I.H.; M.M.A. and A.B.; supervision, D.C. and A.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Researchers Supporting Project number (RSPD2023R628), King Saud University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The crystallographic data for the structure reported in this article have been deposited at the Cambridge Crystallographic Data Center, under deposition number CCDC 2176903. A copy of the data can be obtained free of charge from https://www.ccdc.cam.ac.uk/structures/ (accessed on 10 October 2023). All other relevant data generated and analyzed during this study, including experimental, spectroscopic, crystallographic, and computational data, are included in this article and its Supplementary Information.
Acknowledgments
The authors would like to thank Joel T. Mague at the X-ray Crystallography Laboratory (School of Science and Engineering, Tulane University, USA) for his assistance with the final refinement of the molecule.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, S.-L.; Damu, G.L.V.; Zhang, L.; Geng, R.-X.; Zhou, C.-H. Synthesis and Biological Evaluation of Novel Benzimidazole Derivatives and Their Binding Behavior with Bovine Serum Albumin. Eur. J. Med. Chem. 2012, 55, 164–175. [Google Scholar] [CrossRef]
- Alpan, A.S.; Parlar, S.; Carlino, L.; Tarikogullari, A.H.; Alptüzün, V.; Güneş, H.S. Synthesis, Biological Activity and Molecular Modeling Studies on 1H-Benzimidazole Derivatives as Acetylcholinesterase Inhibitors. Bioorg. Med. Chem. 2013, 21, 4928–4937. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Luis, F.; Hernández-Campos, A.; Castillo, R.; Navarrete-Vázquez, G.; Soria-Arteche, O.; Hernández-Hernández, M.; Yépez-Mulia, L. Synthesis and Biological Activity of 2-(Trifluoromethyl)-1H-Benzimidazole Derivatives against Some Protozoa and Trichinella Spiralis. Eur. J. Med. Chem. 2010, 45, 3135–3141. [Google Scholar] [CrossRef] [PubMed]
- Mulugeta, E.; Samuel, Y. Synthesis of Benzimidazole-Sulfonyl Derivatives and Their Biological Activities. Biochem. Res. Int. 2022, 2022, 7255299. [Google Scholar] [CrossRef]
- Refaat, H.M. Synthesis and Anticancer Activity of Some Novel 2-Substituted Benzimidazole Derivatives. Eur. J. Med. Chem. 2010, 45, 2949–2956. [Google Scholar] [CrossRef] [PubMed]
- Chandrika, N.T.; Shrestha, S.K.; Ngo, H.X.; Garneau-Tsodikova, S. Synthesis and Investigation of Novel Benzimidazole Derivatives as Antifungal Agents. Bioorg. Med. Chem. 2016, 24, 3680–3686. [Google Scholar] [CrossRef] [PubMed]
- Starčević, K.; Kralj, M.; Ester, K.; Sabol, I.; Grce, M.; Pavelić, K.; Karminski-Zamola, G. Synthesis, Antiviral and Antitumor Activity of 2-Substituted-5-Amidino-Benzimidazoles. Bioorg. Med. Chem. 2007, 15, 4419–4426. [Google Scholar] [CrossRef] [PubMed]
- Tonelli, M.; Simone, M.; Tasso, B.; Novelli, F.; Boido, V.; Sparatore, F.; Paglietti, G.; Pricl, S.; Giliberti, G.; Blois, S.; et al. Antiviral Activity of Benzimidazole Derivatives. II. Antiviral Activity of 2-Phenylbenzimidazole Derivatives. Bioorg. Med. Chem. 2010, 18, 2937–2953. [Google Scholar] [CrossRef]
- Patagar, D.N.; Batakurki, S.R.; Kusanur, R.; Patra, S.M.; Saravanakumar, S.; Ghate, M. Synthesis, Antioxidant and Anti-Diabetic Potential of Novel Benzimidazole Substituted Coumarin-3-Carboxamides. J. Mol. Struct. 2023, 1274, 134589. [Google Scholar] [CrossRef]
- Acar Çevik, U.; Celik, I.; Paşayeva, L.; Fatullayev, H.; Bostancı, H.E.; Özkay, Y.; Kaplancıklı, Z.A. New Benzimidazole-oxadiazole Derivatives: Synthesis, A-glucosidase, A-amylase Activity, and Molecular Modeling Studies as Potential Antidiabetic Agents. Arch. Der Pharm. 2023, 356, 2200663. [Google Scholar] [CrossRef]
- Singh, N.; Pandurangan, A.; Rana, K.; Anand, P.; Ahamad, A.; Tiwari, A.K. Benzimidazole: A Short Review of Their Antimicrobial Activities. Int. Curr. Pharm. J. 2012, 1, 110–118. [Google Scholar] [CrossRef]
- El-Gohary, N.S.; Shaaban, M.I. Synthesis and Biological Evaluation of a New Series of Benzimidazole Derivatives as Antimicrobial, Antiquorum-Sensing and Antitumor Agents. Eur. J. Med. Chem. 2017, 131, 255–262. [Google Scholar] [CrossRef]
- Chen, G.; Liu, Z.; Zhang, Y.; Shan, X.; Jiang, L.; Zhao, Y.; He, W.; Feng, Z.; Yang, S.; Liang, G. Synthesis and Anti-Inflammatory Evaluation of Novel Benzimidazole and Imidazopyridine Derivatives. ACS Med. Chem. Lett. 2013, 4, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.K.; Ali, M.A.; Wei, A.C.; Choon, T.S.; Osman, H.; Parang, K.; Shirazi, A.N. Synthesis and Evaluation of Novel Benzimidazole Derivatives as Sirtuin Inhibitors with Antitumor Activities. Bioorg. Med. Chem. 2014, 22, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Obot, I.B.; Obi-Egbedi, N.O. Theoretical Study of Benzimidazole and Its Derivatives and Their Potential Activity as Corrosion Inhibitors. Corros. Sci. 2010, 52, 657–660. [Google Scholar] [CrossRef]
- Adardour, M.; Lasri, M.; Ait Lahcen, M.; Maatallah, M.; Idouhli, R.; Alanazi, M.M.; Lahmidi, S.; Abouelfida, A.; Mague, J.T.; Baouid, A. Exploring the Efficacy of Benzimidazolone Derivative as Corrosion Inhibitors for Copper in a 3.5 Wt.% NaCl Solution: A Comprehensive Experimental and Theoretical Investigation. Molecules 2023, 28, 6948. [Google Scholar] [CrossRef]
- Rossi, A.; Hunger, A.; Kebrle, J.; Hoffmann, K. Benzimidazol-Derivate und verwandte Heterocyclen V. Die Kondensation von o-Phenylendiamin mit aliphatischen und alicyclischen β-Ketoestern. Helv. Chim. Acta 1960, 43, 1298–1313. [Google Scholar] [CrossRef]
- Adardour, M.; Zaballos-García, E.; Loughzail, M.; Dahaoui, S.; Baouid, A. Synthesis, Characterization and X-Ray Structure of Heterocyclic Systems Prepared via 1,3-Dipolar Cycloaddition of Nitrile Oxides with Benzimidazolone. J. Mol. Struct. 2018, 1165, 153–161. [Google Scholar] [CrossRef]
- Nath, J.; Paul, R.; Ghosh, S.K.; Paul, J.; Singha, B.; Debnath, N. Drug Repurposing and Relabeling for Cancer Therapy: Emerging Benzimidazole Antihelminthics with Potent Anticancer Effects. Life Sci. 2020, 258, 118189. [Google Scholar] [CrossRef]
- Elhenaw, A.; Al-Wasidi, A.; Refat, M.; Naglah, A. Different Potential Biological Activities of Benzimidazole Derivatives. Egypt. J. Chem. 2021, 64, 3–5. [Google Scholar] [CrossRef]
- Hernández-López, H.; Tejada-Rodríguez, C.J.; Leyva-Ramos, S. A Panoramic Review of Benzimidazole Derivatives and Their Potential BiologicalActivity. Mini Rev. Med. Chem. 2022, 22, 1268–1280. [Google Scholar] [CrossRef] [PubMed]
- Adardour, M.; Boutafda, A.; Hdoufane, I.; Aghraz, A.; Hafidi, M.; Zaballos-García, E.; Cherqaoui, D.; Baouid, A. Efficient and Simple Synthesis of Novel 1,2,3-Triazolyl-Linked Benzimidazolone, Molecular Docking and Evaluation of Their Antimicrobial Activity. Synth. Commun. 2020, 50, 3490–3506. [Google Scholar] [CrossRef]
- Saber, A.; Sebbar, N.K.; Sert, Y.; Alzaqri, N.; Hökelek, T.; El Ghayati, L.; Talbaoui, A.; Mague, J.T.; Baba, Y.F.; Urrutigoîty, M.; et al. Syntheses of N-Substituted Benzimidazolone Derivatives: DFT Calculations, Hirshfeld Surface Analysis, Molecular Docking Studies and Antibacterial Activities. J. Mol. Struct. 2020, 1200, 127174. [Google Scholar] [CrossRef]
- Veena, K.; Raghu, M.S.; Yogesh Kumar, K.; Pradeep Kumar, C.B.; Alharti, F.A.; Prashanth, M.K.; Jeon, B.-H. Design and Synthesis of Novel Benzimidazole Linked Thiazole Derivatives as Promising Inhibitors of Drug-Resistant Tuberculosis. J. Mol. Struct. 2022, 1269, 133822. [Google Scholar] [CrossRef]
- Saber, A.; Anouar, E.H.; Sebbar, G.; Ibrahimi, B.E.; Srhir, M.; Hökelek, T.; Mague, J.T.; Ghayati, L.E.; Sebbar, N.K.; Essassi, E.M. New 1,2,3-Triazole Containing Benzimidazolone Derivatives: Syntheses, Crystal Structures, Spectroscopic Characterizations, Hirshfeld Surface Analyses, DFT Calculations, Anti-Corrosion Property Anticipation, and Antibacterial Activities. J. Mol. Struct. 2021, 1242, 130719. [Google Scholar] [CrossRef]
- Othman, D.I.A.; Hamdi, A.; Tawfik, S.S.; Elgazar, A.A.; Mostafa, A.S. Identification of New Benzimidazole-Triazole Hybrids as Anticancer Agents: Multi-Target Recognition, in Vitro and in Silico Studies. J. Enzym. Inhib. Med. Chem. 2023, 38, 2166037. [Google Scholar] [CrossRef] [PubMed]
- Cremer, D.; Pople, J.A. General Definition of Ring Puckering Coordinates. J. Am. Chem. Soc. 1975, 97, 1354–1358. [Google Scholar] [CrossRef]
- Yang, P.; Zhu, Z.; Chen, M.; Zhou, X.; Chen, W. Microwave-Assisted Synthesis of Polyamine-Functionalized Carbon Dots from Xylan and Their Use for the Detection of Tannic Acid. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 213, 301–308. [Google Scholar] [CrossRef]
- Hirshfeld, F.L. Bonded-Atom Fragments for Describing Molecular Charge Densities. Theoret. Chim. Acta 1977, 44, 129–138. [Google Scholar] [CrossRef]
- Spackman, M.A.; McKinnon, J.J.; Jayatilaka, D. Electrostatic Potentials Mapped on Hirshfeld Surfaces Provide Direct Insight into Intermolecular Interactions in Crystals. CrystEngComm 2008, 10, 377–388. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld Surface Analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Louroubi, A.; Nayad, A.; Hasnaoui, A.; Hdoufane, I.; Idouhli, R.; Saadi, M.; El Ammari, L.; Abdessalam, A.; Berraho, M.; El Firdoussi, L.; et al. Synthesis, Structural Characterization, Theoretical Studies and Corrosion Inhibition of a New Pyrrole Derivative:1-(1-Benzyl-4-(4-Chlorophenyl)-2,5-Dimethyl-1H-Pyrrol-3-Yl)Ethanone. J. Mol. Struct. 2019, 1189, 240–248. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef] [PubMed]
- Rutkowska, E.; Pajak, K.; Jóźwiak, K. Lipophilicity--Methods of Determination and Its Role in Medicinal Chemistry. Acta Pol. Pharm. 2013, 70, 3–18. [Google Scholar] [PubMed]
- Chen, C.-P.; Chen, C.-C.; Huang, C.-W.; Chang, Y.-C. Evaluating Molecular Properties Involved in Transport of Small Molecules in Stratum Corneum: A Quantitative Structure-Activity Relationship for Skin Permeability. Molecules 2018, 23, 911. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated Space-Group and Crystal-Structure Determination. Acta Crystallogr. Sect. A. Found. Crystallogr. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Farrugia, L. WinGX and ORTEP for Windows: An Update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Macrae, C.F.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Shields, G.P.; Taylor, R.; Towler, M.; Van De Streek, J. Mercury: Visualization and Analysis of Crystal Structures. J. Appl. Crystallogr. 2006, 39, 453–457. [Google Scholar] [CrossRef]
- Andersson, M.P.; Uvdal, P. New Scale Factors for Harmonic Vibrational Frequencies Using the B3LYP Density Functional Method with the Triple-ζ Basis Set 6-311+G(d,p). J. Phys. Chem. A 2005, 109, 2937–2941. [Google Scholar] [CrossRef]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Cryst. 2021, 54, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Lagunin, A.; Zakharov, A.; Filimonov, D.; Poroikov, V. QSAR Modelling of Rat Acute Toxicity on the Basis of PASS Prediction. Mol. Inf. 2011, 30, 241–250. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).