Synthesis, X-ray Crystal Structure, and Computational Characterization of Tetraphenylborate, 3-(5H-Dibenzo[a,d] cyclohepten-5-ylidene)-N, N-Dimethyl-1-propanamine
Abstract
1. Introduction
2. Methods
2.1. Experimental (Chemistry)
2.1.1. General
2.1.2. Synthesis
2.2. Theoretical Methodology
2.2.1. Single-Crystal X-ray Diffraction
2.2.2. Computational Details
3. Results and Discussion
3.1. Chemistry
3.2. Molecular Geometries and Computational Details
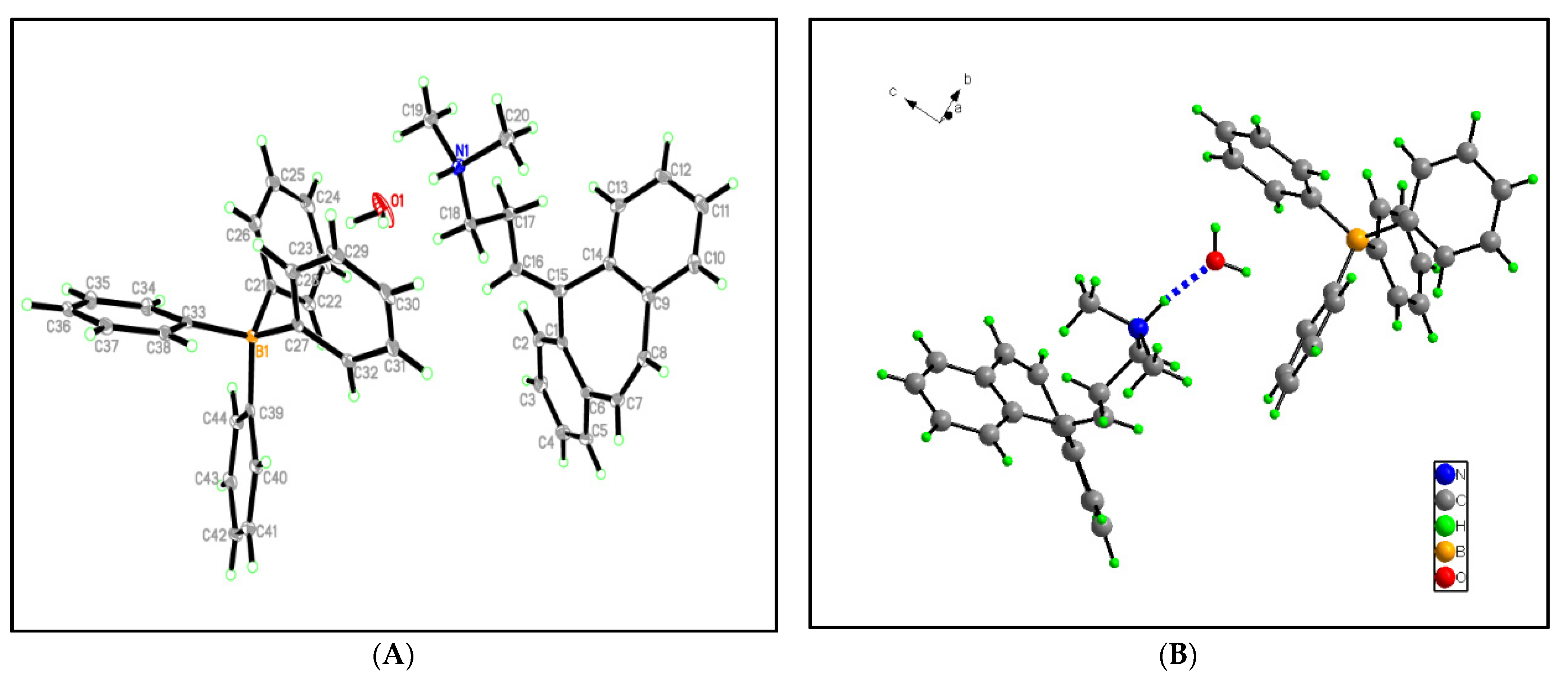
3.2.1. An Investigation through Crystallography
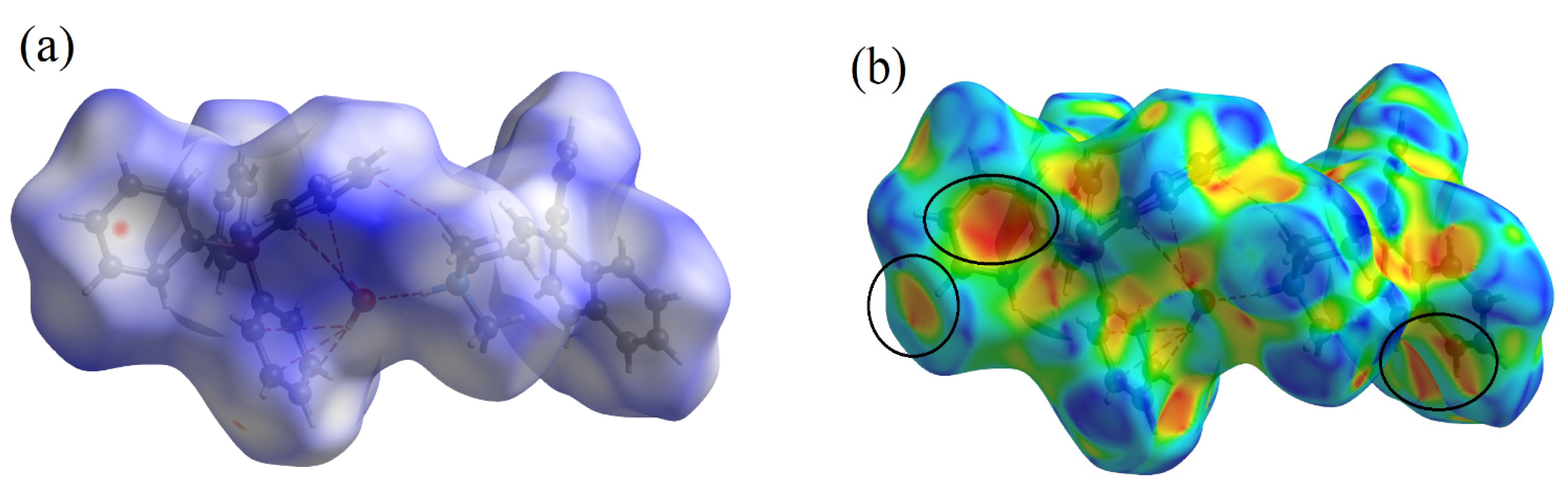
3.2.2. Quantifying Intermolecular Interactions: A Hirshfeld Surface Analysis
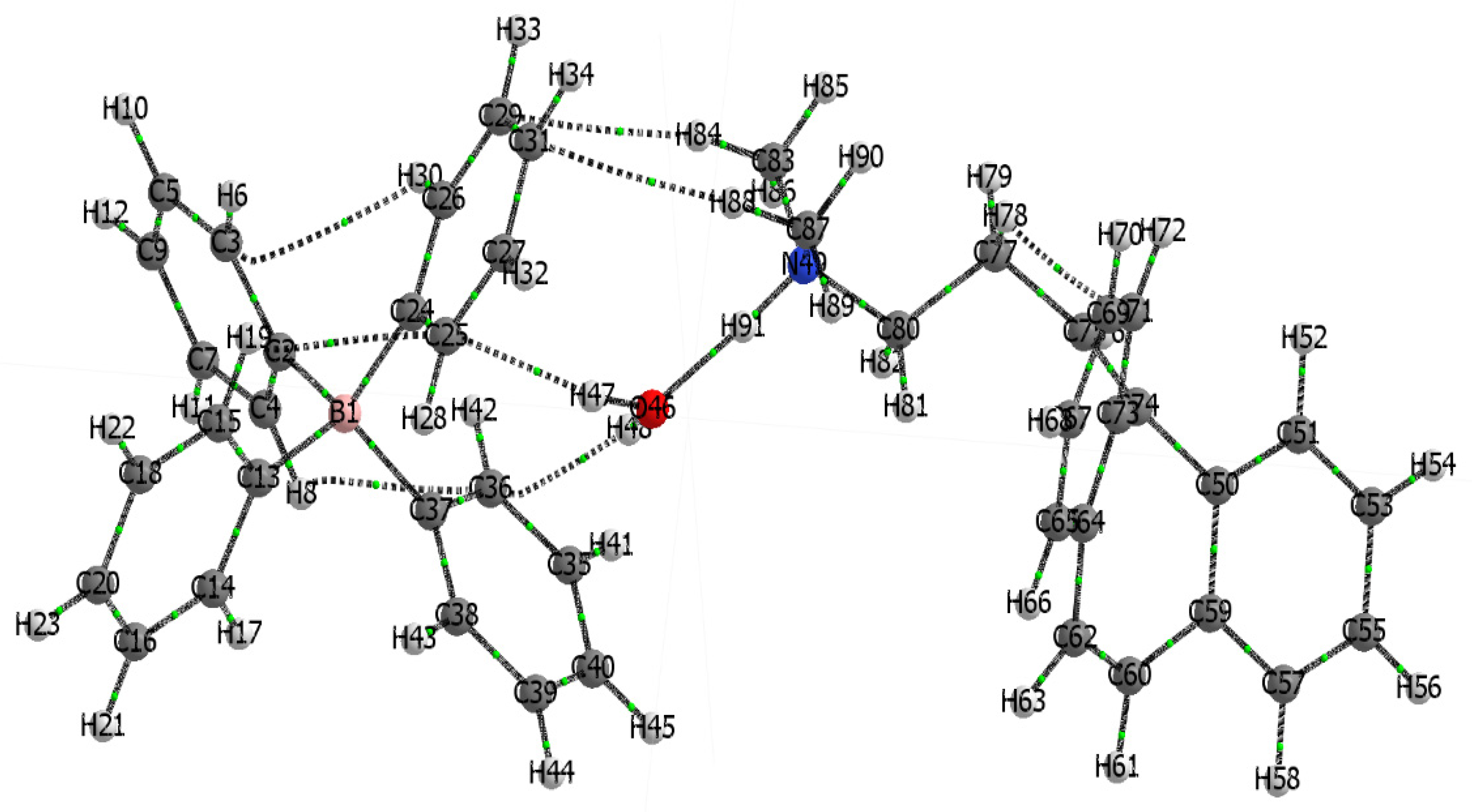
3.2.3. Quantum Theory of Atoms in Molecules (QTAIM) Analysis
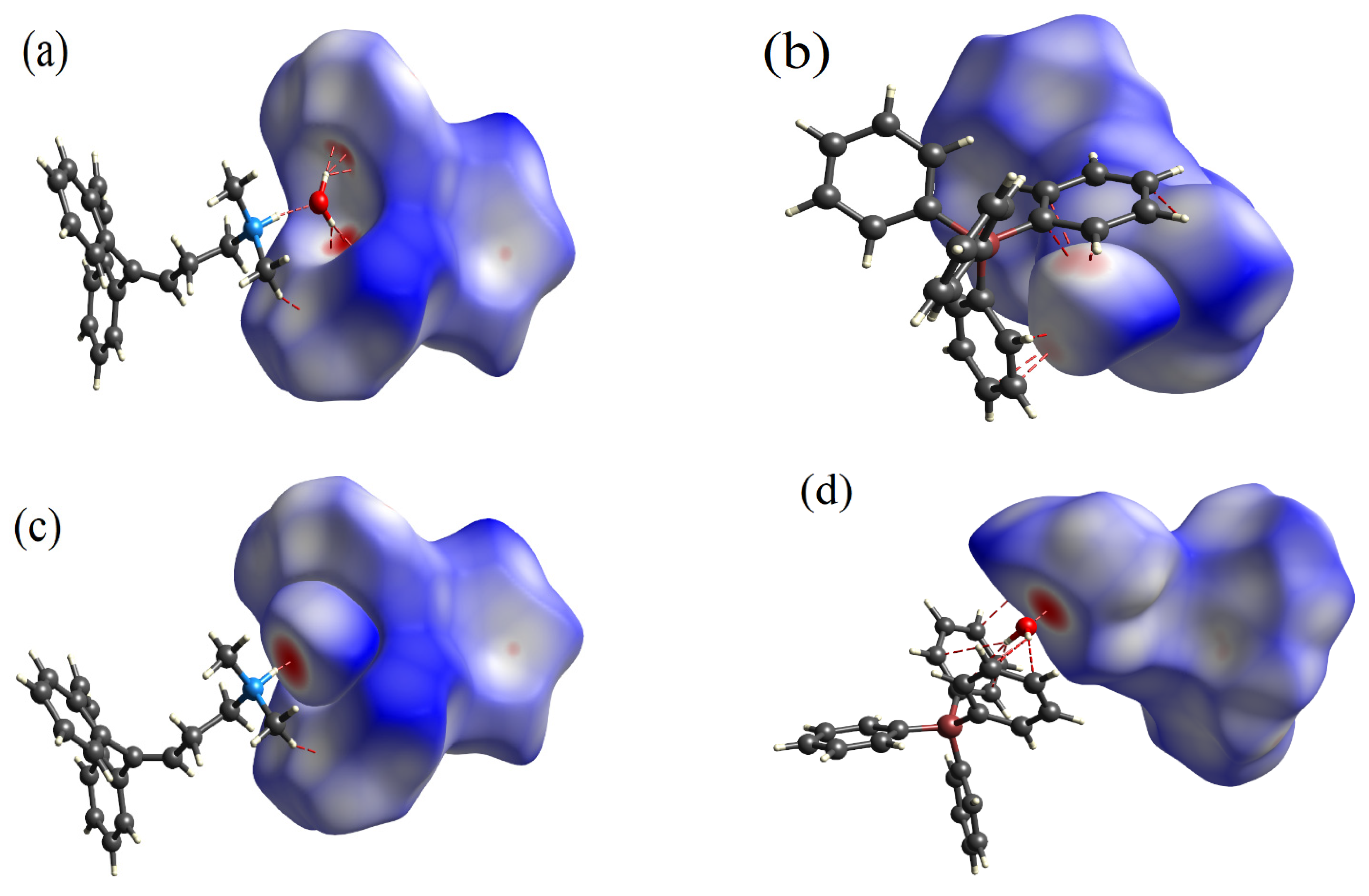
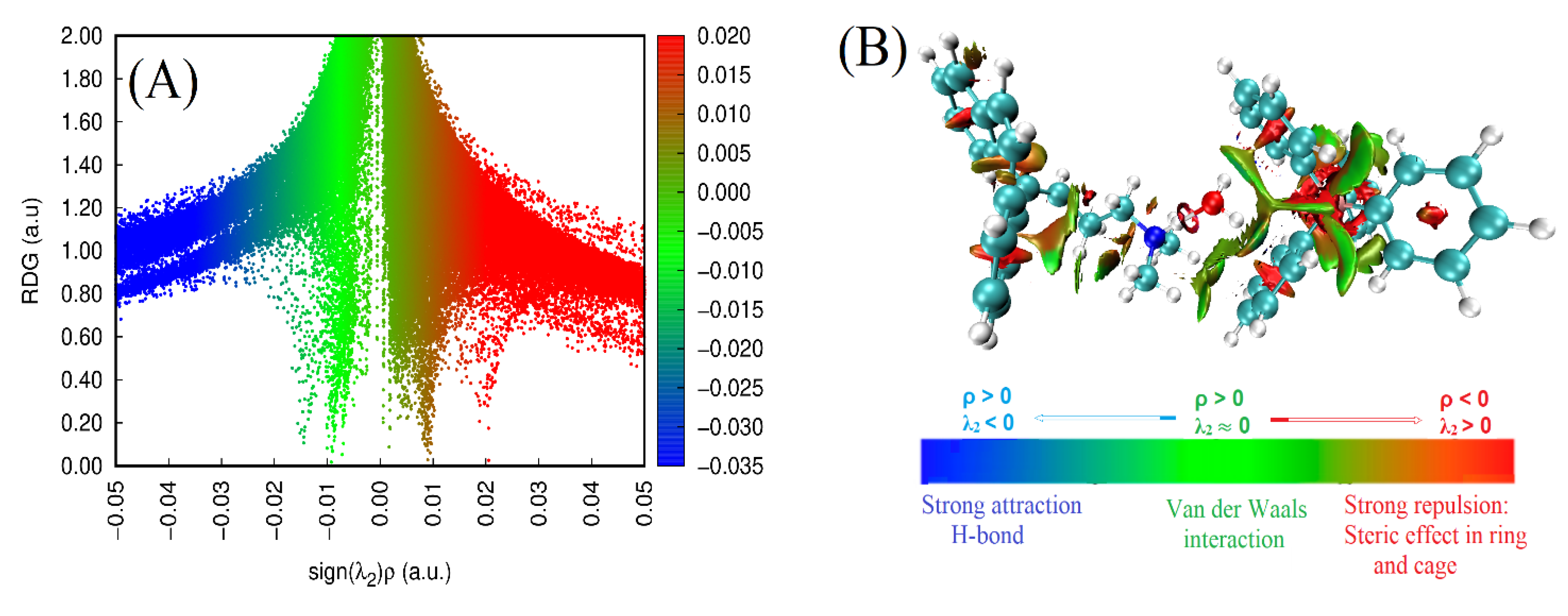
3.2.4. NCl-RDG Analysis
3.2.5. Interaction Energies (IE)
3.2.6. Chemical Reactivity Study
The Global Reactivity Descriptors
Discussion on the Analysis of Frontier Molecular Orbitals (FMOs) Based on the Given Results
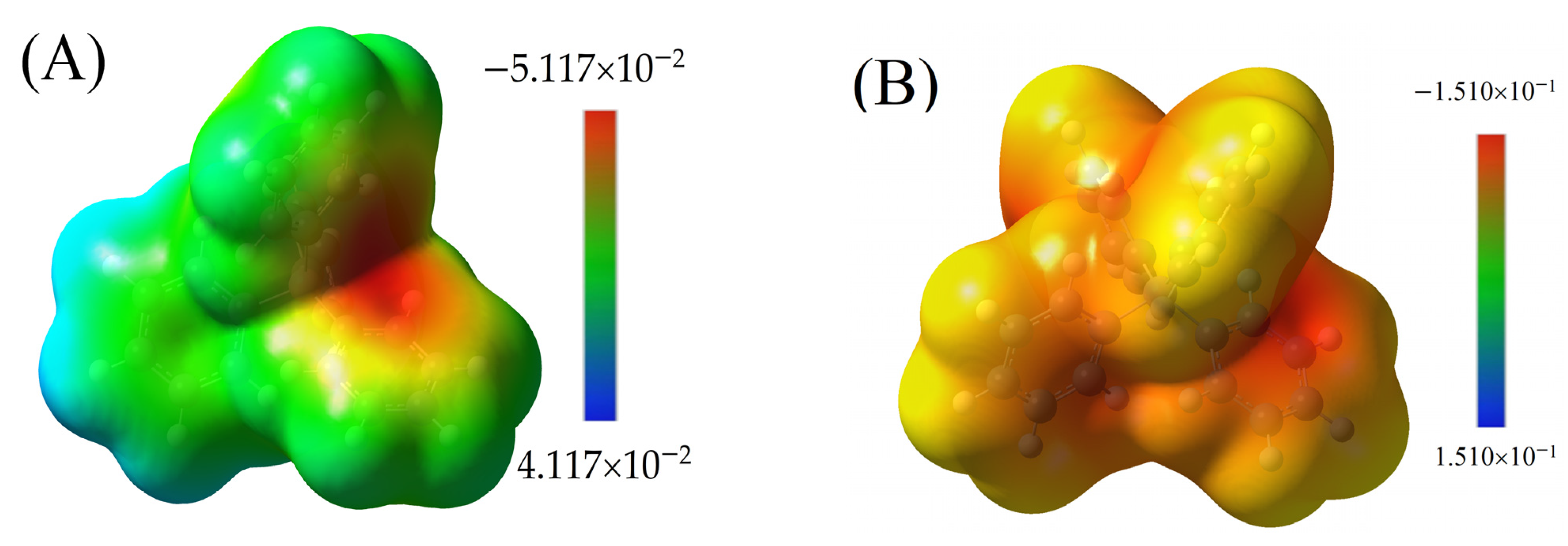
Molecular Electrostatic Nature of Interactions—MESP
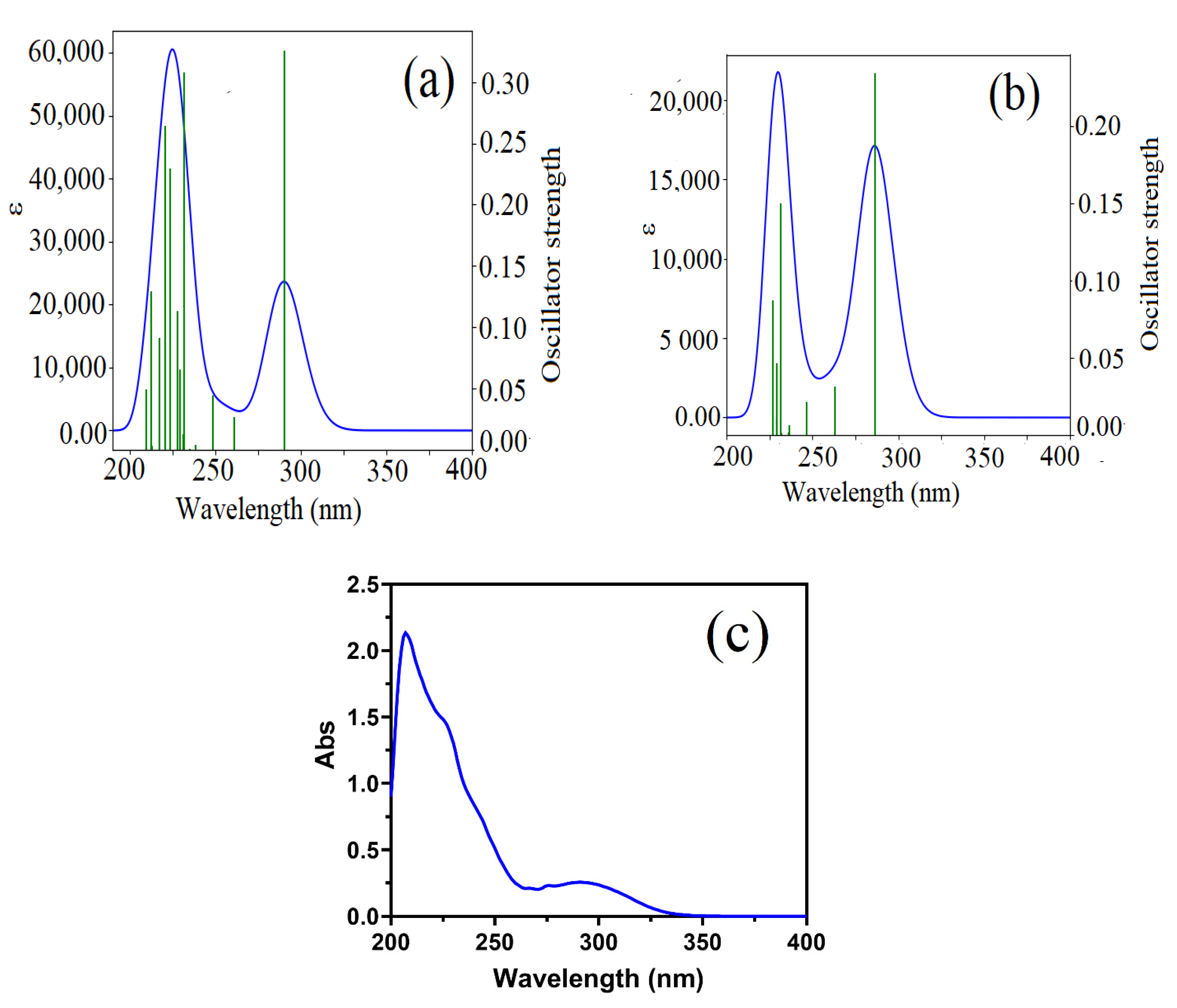
3.2.7. UV Spectral Analysis
3.2.8. NMR Spectral Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chou, R.; Peterson, K.; Helfand, M. Comparative efficacy and safety of skeletal muscle relaxants for spasticity and musculoskeletal conditions: A systematic review. J. Pain Symptom Manag. 2004, 28, 140–175. [Google Scholar] [CrossRef]
- Toth, P.E.; Urtis, J. Commonly used muscle relaxant therapies for acute low back pain: A review of carisoprodol, cyclobenzaprine hydrochloride, and metaxalone. Clin. Ther. 2004, 26, 1355–1367. [Google Scholar] [CrossRef]
- Khater, M.M.; Hassib, H.B.; Issa, Y.M.; Mohammed, S.H. Surface morphology changes of polymer membrane and carbon paste sertraline sensors. Talanta 2015, 134, 546–553. [Google Scholar] [CrossRef]
- AlRabiah, H.; Abounassif, M.A.; Al-Majed, A.; Mostafa, G.A.E. Comparative investigation of β- and γ-cyclodextrin as ionophores in potentiometric based sensors for naltrexone. Int. J. Electrochem. Sci. 2016, 11, 4930–4942. [Google Scholar] [CrossRef]
- Abdel-Raoof, A.M.; Osman, A.O.E.; El-Desouky, E.A.; Abdel-Fattah, A.; Abdul-Kareem, R.F.; Elgazzar, E. Fabrication of an (α-Mn2O3: Co)-decorated CNT highly sensitive screen printed electrode for the optimization and electrochemical determination of cyclobenzaprine hydrochloride using response surface methodology. RSC Adv. 2020, 10, 24985–24993. [Google Scholar] [CrossRef]
- Brown, T.; Lemay, E., Jr.; Bursten, B.; Murphy, C.; Langford, S.; Sagatys, D. Chemistry the Central Science: A Broad Perspective; Pearson Australia Group: Parkside, Australia, 2010. [Google Scholar]
- Giner, B.; Mergenbayeva, S.; Lomba, L.; Rafikova, K.; Dauletbakov, A.; Belyankova, Y.; Seilkhanov, T.; Zazybin, A. Synthesis and Ecotoxicological Studies of Ionic Compounds Based on Tolperisone, Diphenhydramine and Trimecaine. Chem. Sel. 2020, 5, 12823–12828. [Google Scholar] [CrossRef]
- Noda, A.; Hayamizu, K.; Watanabe, M. Pulsed-gradient spin− echo 1H and 19F NMR ionic diffusion coefficient, viscosity, and ionic conductivity of non-chloroaluminate room-temperature ionic liquids. J. Phys. Chem. B 2001, 105, 4603–4610. [Google Scholar] [CrossRef]
- Tokuda, H.; Ishii, K.; Susan, M.A.B.H.; Tsuzuki, S.; Hayamizu, K.; Watanabe, M. Physicochemical properties and structures of room-temperature ionic liquids. 3. Variation of cationic structures. J. Phys. Chem. B 2006, 110, 2833–2839. [Google Scholar] [CrossRef]
- Tokuda, H.; Tsuzuki, S.; Susan, M.A.B.H.; Hayamizu, K.; Watanabe, M. How ionic are room-temperature ionic liquids? An indicator of the physicochemical properties. J. Phys. Chem. B 2006, 110, 19593–19600. [Google Scholar] [CrossRef]
- Hunt, P.A.; Gould, I.R.; Kirchner, B. The structure of imidazolium-based ionic liquids: Insights from ion-pair interactions. Aust. J Chem. 2007, 60, 9–14. [Google Scholar] [CrossRef]
- Maiti, A.; Rogers, R.D. A correlation-based predictor for pair-association in ionic liquids. Phys. Chem. Chem. Phys. 2011, 13, 12138–12145. [Google Scholar] [CrossRef]
- Sofian, Z.M.; Harun, N.; Mahat, M.M.; Hashim, N.A.N.; Jones, S.A. Investigating how amine structure influences drug-amine ion-pair formation and uptake via the polyamine transporter in A549 lung cells. Eur. J. Pharm. Biopharm. 2021, 168, 53–61. [Google Scholar] [CrossRef]
- Wang, H.; Tian, Q.; Quan, P.; Liu, C.; Fang, L. Probing the role of ion-pair strategy in controlling dexmedetomidine penetrate through drug-in-adhesive patch: Mechanistic insights based on release and percutaneous absorption process. AAPS Pharm. Sci. Tech. 2020, 21, 4. [Google Scholar] [CrossRef]
- Al-Hamdani, Y.S.; Tkatchenko, A. Understanding non-covalent interactions in larger molecular complexes from first principles. J. Chem. Phys. 2019, 150, 010901. [Google Scholar] [CrossRef]
- Hajji, M.; Abad, N.; Habib, M.A.; Elmgirhi, S.M.H.; Guerfel, T. Computational chemistry methods for modelling non-covalent interactions and chemical reactivity—An overview. J. Indian Chem. Soc. 2021, 98, 100208. [Google Scholar] [CrossRef]
- Bakheit, A.H.; Attwa, M.W.; Kadi, A.A.; Ghabbour, H.A.; Alkahtani, H.M. Exploring the Chemical Reactivity, Molecular Docking, Molecular Dynamic Simulation and ADMET Properties of a Tetrahydrothienopyridine Derivative Using Computational Methods. Crystals 2023, 13, 1020. [Google Scholar] [CrossRef]
- Abuelizz, H.A.; Taie, H.A.; Bakheit, A.H.; Marzouk, M.; Abdellatif, M.M.; Al-Salahi, R. Biological Evaluation of 4-(1H-triazol-1-yl)benzoic Acid Hybrids as Antioxidant Agents: In Vitro Screening and DFT Study. Appl. Sci. 2021, 11, 11642. [Google Scholar] [CrossRef]
- Szatylowicz, H.; Jezierska, A.; Sadlej-Sosnowska, N. Correlations of NBO energies of individual hydrogen bonds in nucleic acid base pairs with some QTAIM parameters. Struct. Chem. 2016, 27, 367–376. [Google Scholar] [CrossRef]
- Hajji, M.; Al-Otaibi, J.S.; Belkhiria, M.; Dhifaoui, S.; Habib, M.A.; Elmgirhi, S.M.H.; Mtiraoui, H.; Bel-Hadj-Tahar, R.; Msaddek, M.; Guerfel, T. Structural and computational analyses of a 2-propanolammonium-chlorocadmate (II) assembly: Pivotal role of hydrogen bonding and H—H interactions. J. Mol. Struct. 2021, 1223, 128998. [Google Scholar] [CrossRef]
- Rezac, J.; Hobza, P. Benchmark calculations of interaction energies in noncovalent complexes and their applications. Chem. Rev. 2016, 116, 5038–5071. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. Crystal Explorer (v17); The University of Western Australia: Crawley, Australia, 2017. [Google Scholar]
- Altomare, A.; Ciriaco, F.; Cuocci, C.; Falcicchio, A.; Fanelli, F. Combined powder X-ray diffraction data and quantum-chemical calculations in EXPO2014. Powder Diffr. 2017, 32, S123–S128. [Google Scholar] [CrossRef]
- Brandenburg, K. DIAMOND; Crystal Impact GbR: Bonn, Germany, 2006. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision D.01; Gaussian Inc.: Wallingford, CT, USA, 2009; Volume 121, pp. 150–166. [Google Scholar]
- Dennington, R.; Keith, T.; Millam, J. Gauss View; Version 5; Semichem Inc.: Shawnee Mission, KS, USA, 2016; Volume 6. [Google Scholar]
- Raghavachari, K.; Trucks, G.W.; Pople, J.A.; Head-Gordon, M. A fifth-order perturbation comparison of electron correlation theories. Chem. Phys. Lett. 1989, 157, 479–483. [Google Scholar] [CrossRef]
- Bakheit, A.H.; Al-Salahi, R.; Al-Majed, A.A. Thermodynamic and Computational (DFT) Study of Non-Covalent Interaction Mechanisms of Charge Transfer Complex of Linagliptin with 2,3-Dichloro-5,6-dicyano-1,4-benzoquinone (DDQ) and Chloranilic acid (CHA). Molecules 2022, 27, 6320. [Google Scholar] [CrossRef]
- Ghabbour, H.A.; Bakheit, A.H.; Ezzeldin, E.; Mostafa, G.A.E. Synthesis Characterization and X-ray Structure of 2-(2,6-Dichlorophenylamino)-2-imidazoline Tetraphenylborate: Computational Study. Appl. Sci. 2022, 12, 3568. [Google Scholar] [CrossRef]
- Bakheit, A.H.; Ghabbour, H.A.; Hussain, H.; Al-Salahi, R.; Ali, E.A.; Mostafa, G.A. Synthesis and Computational and X-ray Structure of 2, 3, 5-Triphenyl Tetrazolium, 5-Ethyl-5-phenylbarbituric Acid Salt. Crystals 2022, 12, 1706. [Google Scholar] [CrossRef]
- Mostafa, G.A.; Bakheit, A.H.; Al-Agamy, M.H.; Al-Salahi, R.; Ali, E.A.; Alrabiah, H. Synthesis of 4-Amino-N-[2 (diethylamino)Ethyl]Benzamide Tetraphenylborate Ion-Associate Complex: Characterization, Antibacterial and Computational Study. Molecules 2023, 28, 2256. [Google Scholar] [CrossRef]
- Jeroundi, D.; Sebbar, N.K.; Hökelek, T.; Rodi, Y.K.; Mazzah, A.; Renard, C.; Chakroune, S. Crystal structure, Hirshfeld surface analysis and interaction energy and DFT studies of 10-allyl-pyrrolo [2, 1-c][1, 4] benzodiazepine-5, 11-dione. Moroc. J. Heterocycl. Chem. 2020, 9, 43–54. [Google Scholar]
- Pendás, Á.M.; Gatti, C. 3 Quantum Theory of Atoms in Molecules and the AIMAll Software. Complement. Bond. Anal. 2021, 43. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R. Chemical reactivity theory (CRT) study of small drug-like biologically active molecules. J. Biomol. Struct. Dyn. 2021, 39, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. Cryst. Eng. Comm. 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Ahmed, A.; Fatima, A.; Shakya, S.; Rahman, Q.I.; Ahmad, M.; Javed, S.; AlSalem, H.S.; Ahmad, A. Crystal structure, topology, DFT and hirshfeld surface analysis of a novel charge transfer complex (L3) of anthraquinone and 4-{[(anthracen-9-yl) meth-yl] amino}-benzoic acid (L2) exhibiting photocatalytic properties: An experimental and theoretical approach. Molecules 2022, 27, 1724. [Google Scholar] [PubMed]
- Kolandaivel, P.; Nirmala, V. Study of proper and improper hydrogen bonding using Bader’s atoms in molecules (AIM) theory and NBO analysis. J. Mol. Struct. 2004, 694, 33–38. [Google Scholar] [CrossRef]
- Rozas, I.; Alkorta, I.; Elguero, J. Behavior of ylides containing N, O, and C atoms as hydrogen bond acceptors. J. Am. Chem. Soc. 2000, 122, 11154–11161. [Google Scholar] [CrossRef]
- Fuster, G.; Schuhmacher, M.; Domingo, J.L. Human exposure to dioxins and furans: Application of the substance flow analysis to health risk assessment. Environ. Sci. Pollut. Res. 2002, 9, 241–249. [Google Scholar] [CrossRef]
- Abuelizz, H.A.; Bakheit, A.H.; Marzouk, M.; Abdellatif, M.M.; Al-Salahi, R. Reactivity of 4, 5-Dichlorophthalic Anhydride towards Thiosemicarbazide and Amines: Synthesis, Spectroscopic Analysis, and DFT Study. Molecules 2022, 27, 3550. [Google Scholar] [CrossRef]
- Haque, A.; Alenezi, K.M.; Khan, M.S.; Wong, W.Y.; Raithby, P.R. Non-covalent interactions (NCIs) in π-conjugated functional materials: Advances and perspectives. Chem. Soc. Rev. 2023, 52, 454–472. [Google Scholar] [CrossRef]
- Mostafa, G.A.; Bakheit, A.; AlMasoud, N.; AlRabiah, H. Charge Transfer Complexes of Ketotifen with 2,3-Dichloro-5,6-dicyano-p-benzoquinone and 7,7,8,8-Tetracyanoquodimethane: Spectroscopic Characterization Studies. Molecules 2021, 26, 2039. [Google Scholar] [CrossRef]
- Abuelizz, H.A.; Taie, H.A.A.; Bakheit, A.H.; Mostafa, G.A.E.; Marzouk, M.; Rashid, H.; Al-Salahi, R. Investigation of 4-Hydrazinobenzoic Acid Derivatives for Their Antioxidant Activity: In Vitro Screening and DFT Study. ACS Omega 2021, 6, 31993–32004. [Google Scholar] [CrossRef] [PubMed]
- Nemykin, V.N.; Hadt, R.G.; Belosludov, R.V.; Mizuseki, H.; Kawazoe, Y. Influence of Molecular Geometry, Exchange-Correlation Functional, and Solvent Effects in the Modeling of Vertical Excitation Energies in Phthalocyanines Using Time-Dependent Density Functional Theory (TDDFT) and Polarized Continuum Model TDDFT Methods: Can Modern Computational Chemistry Methods Explain Experimental Controversies? J. Phys. Chem. A 2007, 111, 12901–12913. [Google Scholar] [PubMed]
- Ayyappan, S.; Sundaraganesan, N.; Aroulmoji, V.; Murano, E.; Sebastian, S. Molecular structure, vibrational spectra and DFT molecular orbital calculations (TD-DFT and NMR) of the antiproliferative drug Methotrexate. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2010, 77, 264–275. [Google Scholar] [CrossRef] [PubMed]



| Empirical Formula | C44H44BNO |
|---|---|
| Formula weight | 613.61 |
| Temperature/K | 293 (2) |
| Crystal system | monoclinic |
| Space group | C2/c |
| a/Å | 41.072 (2) |
| b/Å | 9.9974 (6) |
| c/Å | 16.8148 (8) |
| α/° | 90 |
| β/° | 93.115 (2) |
| γ/° | 90 |
| Volume/Å3 | 6894.2 (6) |
| Z | 8 |
| ρcalcg/cm3 | 1.182 |
| μ/mm−1 | 0.069 |
| F(000) | 2624.0 |
| Crystal size/mm3 | 0.27 × 0.19 × 0.14 |
| Radiation | MoKα (λ = 0.71073) |
| 2Θ range for data collection/° | 4.194 to 66.446 |
| Index ranges | −63 ≤ h ≤ 62, −12 ≤ k ≤ 15, −25 ≤ l ≤ 25 |
| Reflections collected | 55,034 |
| Independent reflections | 13,176 [Rint = 0.1232, Rsigma = 0.1548] |
| Data/restraints/parameters | 13,176/0/600 |
| Goodness-of-fit on F2 | 1.014 |
| Final R indexes [I ≥ 2σ (I)] | R1 = 0.0785, wR2 = 0.1430 |
| Final R indexes [all data] | R1 = 0.1724, wR2 = 0.1711 |
| Largest diff. peak/hole/e Å−3 | 0.40/−0.28 |
| D–H···A | d(D–H)/Å | d(H–A)/Å | d(D–A)/Å | D–H–A/° |
|---|---|---|---|---|
| N1–H1···O1 | 10.99 (2) | 1.69 (2) | 2.680 (2) | 171.8 (18) |
| BCP | Atoms | ρ(r) | ∇2ρ(r) | Ellipticity | H(r) | GBL_I | G | V | BPL—GBL_I | |V(r)|/G (r) | Interaction |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 20 | C25···H47 | 0.0156 | 0.0439 | 0.3638 | 0.0007 | 4.3745 | 0.0103 | −0.0096 | 0.0248 | 0.9318 | TPB-Water |
| 29 | H19···C25 | 0.0101 | 0.039 | 0.5689 | 0.0021 | 4.9259 | 0.0076 | −0.0055 | 0.1849 | 0.72 | TPB-TPB |
| 34 | C3···H30 | 0.0092 | 0.0346 | 1.311 | 0.0019 | 5.0574 | 0.0067 | −0.0048 | 0.4009 | 0.7144 | CBP-TPB |
| 41 | C36···H48 | 0.0147 | 0.0431 | 1.0738 | 0.001 | 4.5084 | 0.0098 | −0.0088 | 0.1609 | 0.8965 | TPB-Water |
| 49 | H8···C36 | 0.0092 | 0.0355 | 1.1298 | 0.002 | 5.045 | 0.0069 | −0.0049 | 0.2378 | 0.7094 | TPB-TPB |
| 56 | O46···H91 | 0.0796 | 0.2191 | 0.0504 | −0.012 | 2.844 | 0.0668 | −0.0788 | 0.0015 | 1.1799 | TPB-Water |
| 91 | C71···H78 | 0.0097 | 0.0346 | 1.0606 | 0.0017 | 4.9338 | 0.0069 | −0.0052 | 0.0988 | 0.7537 | CBP-CBP |
| 95 | C29···H84 | 0.007 | 0.0203 | 0.2076 | 0.001 | 5.1945 | 0.0041 | −0.0031 | 0.0106 | 0.7584 | CBP-TPB |
| 100 | C31···H88 | 0.0072 | 0.0208 | 0.208 | 0.001 | 5.1949 | 0.0042 | −0.0032 | 0.0095 | 0.7605 | CBP-TPB |
| Complexes | (∆E) Raw (kcal/mol) | (∆E) Corrected (kcal/mol) | ∆EBSSE |
|---|---|---|---|
| CBP-TPB-Water | −170.59 | −146.86 | 0.0378 |
| Global Reactivity Descriptors | CBP-TPB | TPB | CBP | Unit |
|---|---|---|---|---|
| E_HOMO(N) | −4.5807 | −5.7199 | −1.0073 | eV |
| E_HOMO(N + 1) | 1.0594 | −2.4276 | 2.6896 | eV |
| E_HOMO(N − 1) | −7.5916 | −9.4675 | −8.2117 | eV |
| Vertical IP | 6.0209 | 7.5047 | 2.3711 | eV |
| Vertical EA | 0.4338 | 3.7312 | −1.4214 | eV |
| Mulliken electronegativity | 3.2273 | 5.6179 | 0.4748 | eV |
| Chemical potential | −3.2273 | −5.6179 | −0.4748 | eV |
| Hardness (=fundamental gap) | 5.587 | 3.7735 | 3.7925 | eV |
| Softness | 0.179 | 0.265 | 0.2637 | eV−1 |
| Electrophilicity index | 0.9321 | 4.182 | 0.0297 | eV |
| Nucleophilicity index | 4.5405 | 3.4013 | 8.1139 | eV |
| TPB | CBP | H2O | CBP-TPB | |
|---|---|---|---|---|
| LUMO | −1.395 | −3.821 | −0.063 | −0.079 |
| HOMO | −5.937 | −8.211 | −0.291 | −0.178 |
| (ω) | 4.1820 | 0.0297 | 0.460 | 0.9321 |
| Solvent | ETotal (a.u) | Dipole Moment | λmax | ƒ | Transition Energy (eV) | Electronic Transition | Major % Contribution |
|---|---|---|---|---|---|---|---|
| Gas phase | 34,916.5 | 21.9 | 286.4 | 0.235 | 4.27 | H–5 → L | 94 |
| 43,207.1 | 231.4 | 0.1502 | 5.35 | H–12 → L H–5 → L + 1 H–5 → L + 2 | 23 20 21 | ||
| Acetonitrile solvent | 34,456.8 | 17.6 | 290.2 | 0.326 | 4.33 | H–1 → L | 95 |
| 43,105.5 | 231.98 | 0.3081 | 5.34 | H–10 → L H–1 → L + 1 H–1 → L + 4 | 26 27 19 | ||
| 44,739.6 | 223.5 | 0.2296 | 5.547 | H → L + 6 H–12 → L + 6 | 79 3 | ||
| 45,292.1 | 220.8 | 0.2641 | 5.616 | H–6 → L H–1 → L + 5 H–14 → L | 21 45 9 |
| Atom Position Hydrogen | CBP—TPB | Atom Position Carbon | CBP—TPB | ||
|---|---|---|---|---|---|
| B3LYP/6–311G(d) | Exp | B3LYP/6–311G(d) | Exp | ||
| 91 | 8.0644 | 37, 24 | 175.8 | ||
| 42, 30 | 7.59305 | 7.43 .. 7.37 | 13, 2 | 169.94 | 164.54 |
| 28, 43, 6, 8 | 7.35675 | 7.34 .. 7.29 | 74 | 154.9247 | 163.7 |
| 19, 17, 70, 54, 68, 56 | 7.2118 | 7.20 .. 7.11 | 50 | 147.8053 | 144.48 |
| 66, 52, 41, 58, 33 | 7.05614 | 6.95 .. 6.85 | 73, 36, 26, 64 | 141.8078 | 142.1 |
| 72, 32, 44, 10, 22, 21, 11, 34, 45 | 6.8379 | 6.83 .. 6.77 | 59, 3, 14, 14, 4 | 138.4057 | |
| 23, 61, 63, 12 | 6.631 | 5.46 .. 5.35 | 15, 25 | 137.8711 | 136.59 |
| 76 | 5.04 | 3.13 .. 3.06 | 60 | 135.5883 | 136.29 |
| 82 | 1.9995 | 2.63 .. 2.57 | 62 | 135.3271 | 134.99 |
| 78, 79, 85, 84 | 1.7552 | 75 | 134.4803 | 134.18 | |
| 86 | 1.6041 | 2.54 .. 2.50 | 71, 53 | 132.628 | 132 |
| 90, 88 | 1.5217 | 2.50 .. 2.42 | 65, 69, 29, 35, 57, 67, 51 | 131.9058 | 131.59 |
| 81 | 1.4379 | 2.30 .. 2.24 | 55 | 130.2556 | 129.9 |
| 89 | 1.0723 | 7.43 .. 7.37 | 27 | 129.7315 | 129.67 |
| Atom positioncarbon | 39 | 129.6271 | 129.37 | ||
| 20 | 124.2605 | 126.32 | 18 | 128.9938 | 129.21 |
| 9 | 124.1405 | 122.67 | 5 | 128.9252 | 129.14 |
| 80 | 56.4247 | 56.88 | 16 | 128.7955 | 128.51 |
| 83 | 39.6608 | 43.01 | 7 | 128.6164 | 128.24 |
| 87 | 38.5887 | 39.16 | 40 | 126.7416 | 128.19 |
| 77 | 21.2847 | 24.75 | 31 | 126.5995 | 126.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakheit, A.H.; Al-Salahi, R.; Ghabbour, H.A.; Ali, E.A.; AlRuqi, O.S.; Mostafa, G.A.E. Synthesis, X-ray Crystal Structure, and Computational Characterization of Tetraphenylborate, 3-(5H-Dibenzo[a,d] cyclohepten-5-ylidene)-N, N-Dimethyl-1-propanamine. Crystals 2023, 13, 1088. https://doi.org/10.3390/cryst13071088
Bakheit AH, Al-Salahi R, Ghabbour HA, Ali EA, AlRuqi OS, Mostafa GAE. Synthesis, X-ray Crystal Structure, and Computational Characterization of Tetraphenylborate, 3-(5H-Dibenzo[a,d] cyclohepten-5-ylidene)-N, N-Dimethyl-1-propanamine. Crystals. 2023; 13(7):1088. https://doi.org/10.3390/cryst13071088
Chicago/Turabian StyleBakheit, Ahmed H., Rashad Al-Salahi, Hazem A. Ghabbour, Essam A. Ali, Obaid S. AlRuqi, and Gamal A. E. Mostafa. 2023. "Synthesis, X-ray Crystal Structure, and Computational Characterization of Tetraphenylborate, 3-(5H-Dibenzo[a,d] cyclohepten-5-ylidene)-N, N-Dimethyl-1-propanamine" Crystals 13, no. 7: 1088. https://doi.org/10.3390/cryst13071088
APA StyleBakheit, A. H., Al-Salahi, R., Ghabbour, H. A., Ali, E. A., AlRuqi, O. S., & Mostafa, G. A. E. (2023). Synthesis, X-ray Crystal Structure, and Computational Characterization of Tetraphenylborate, 3-(5H-Dibenzo[a,d] cyclohepten-5-ylidene)-N, N-Dimethyl-1-propanamine. Crystals, 13(7), 1088. https://doi.org/10.3390/cryst13071088








