Synthesis, Crystal Structure and Supramolecular Features of Novel 2,4-Diaminopyrimidine Salts
Abstract
1. Introduction
2. Materials and Methods
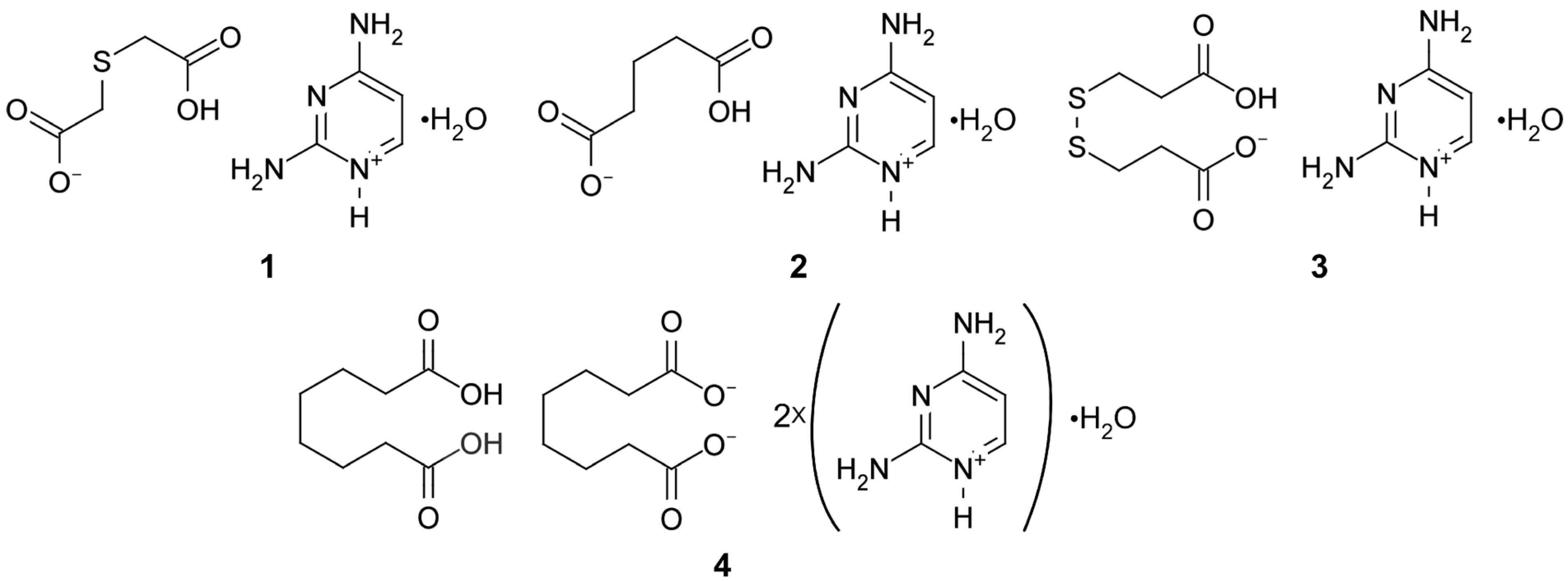
2.1. Synthesis of Compounds 1–4
2.2. Single-Crystal X-ray Diffraction
2.3. Computational Details
3. Results and Discussion
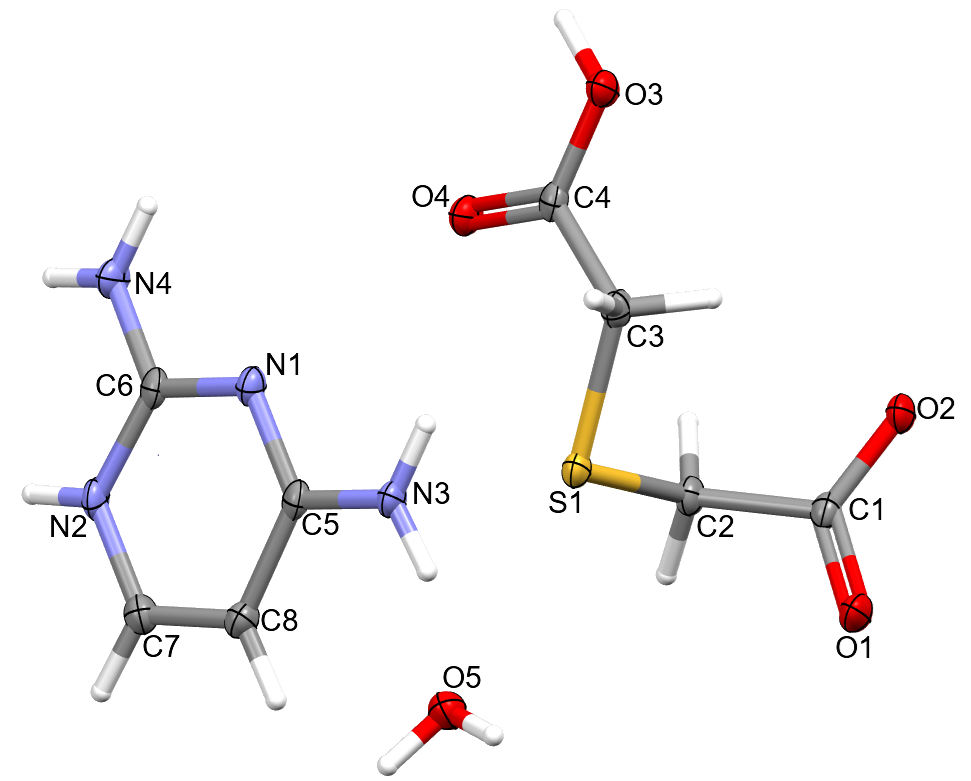
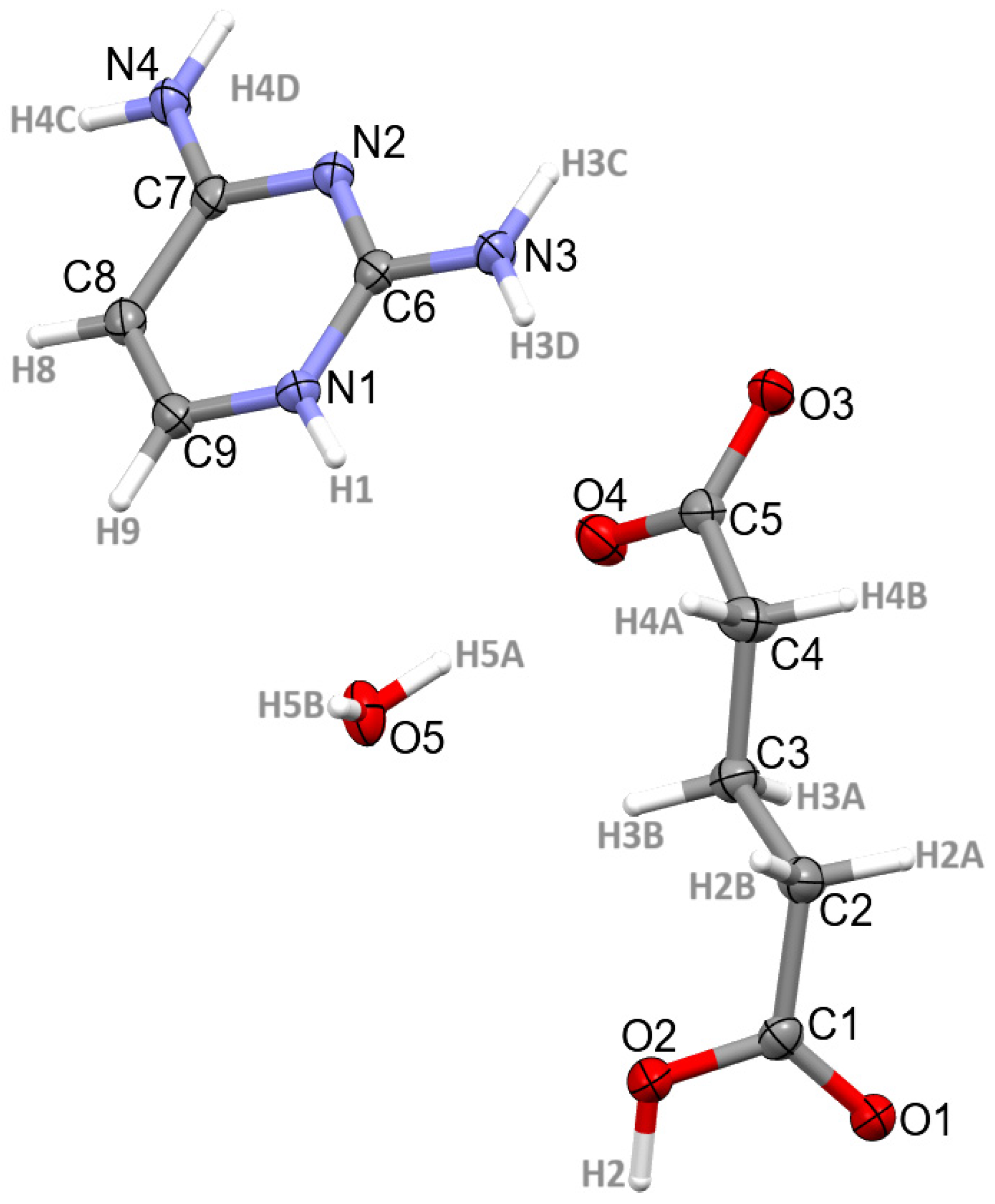
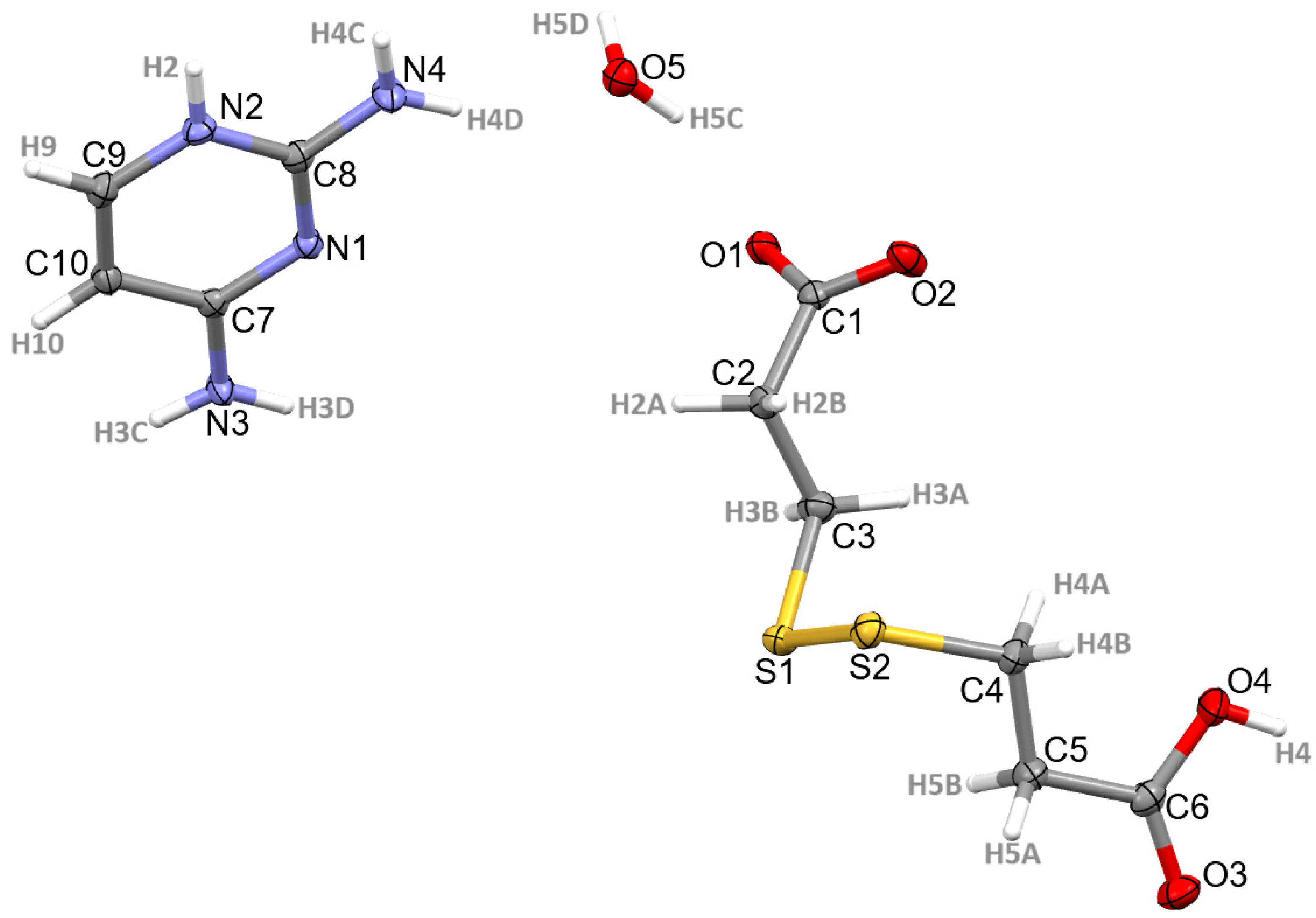
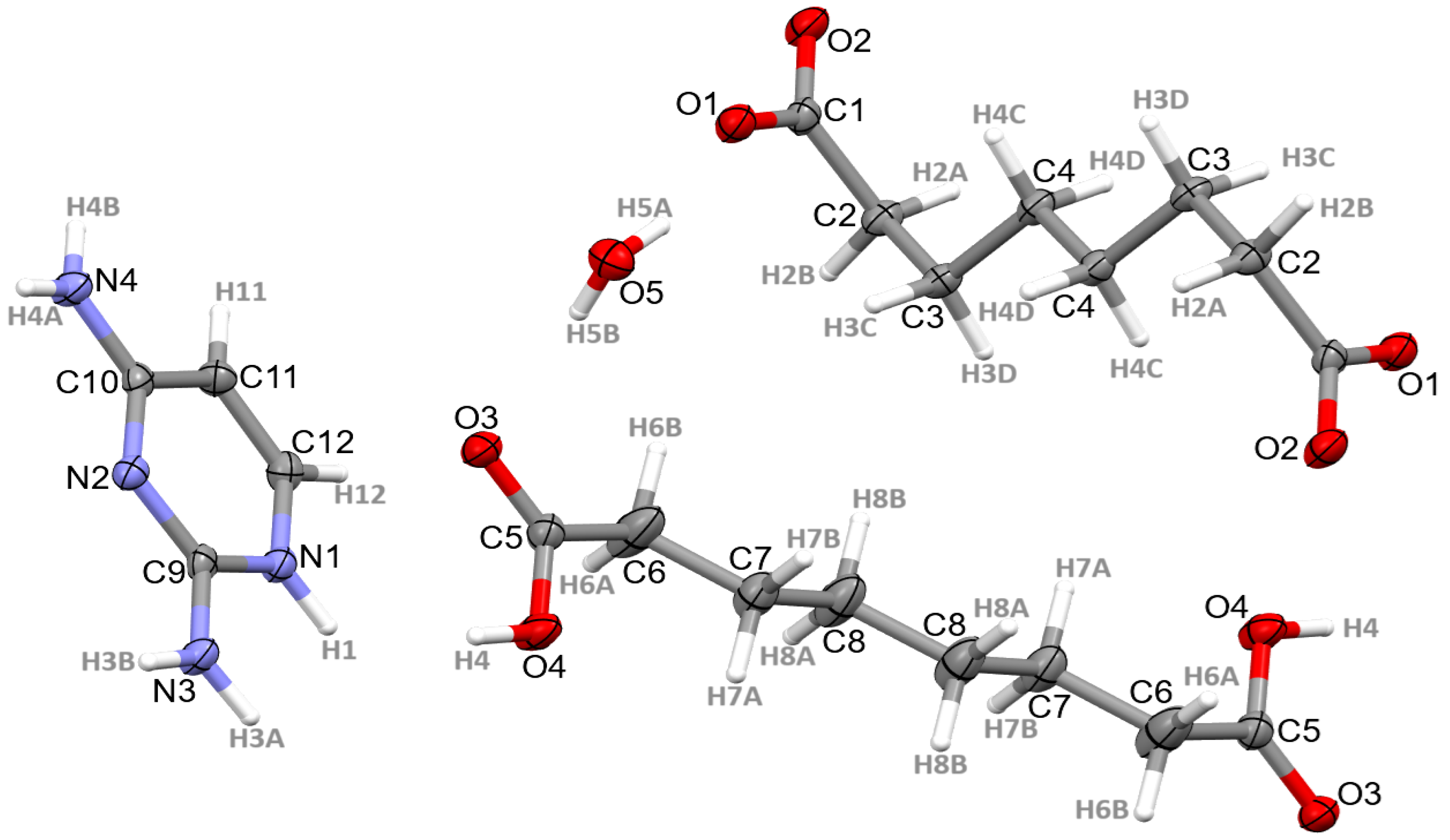
3.1. Structural Commentary
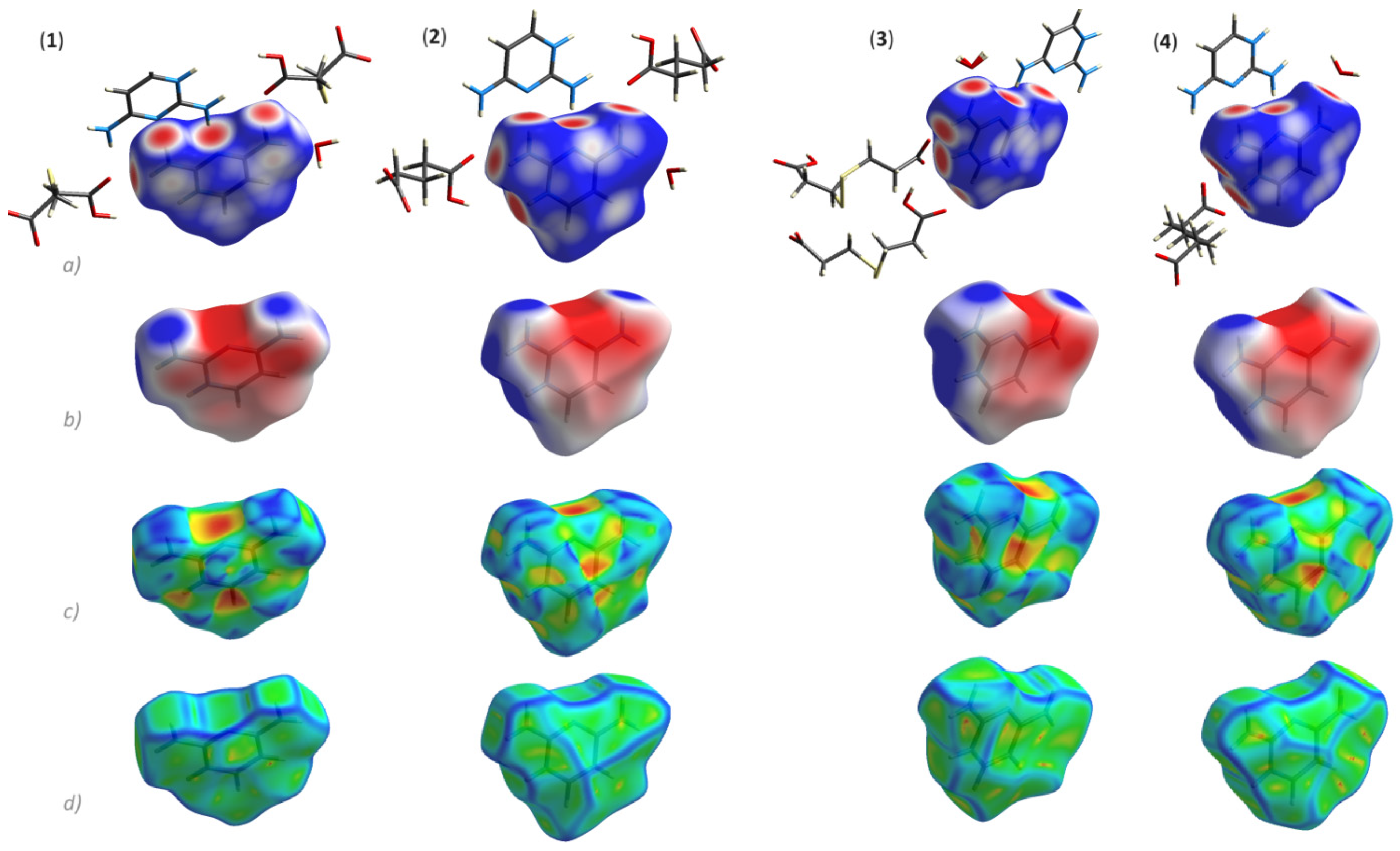
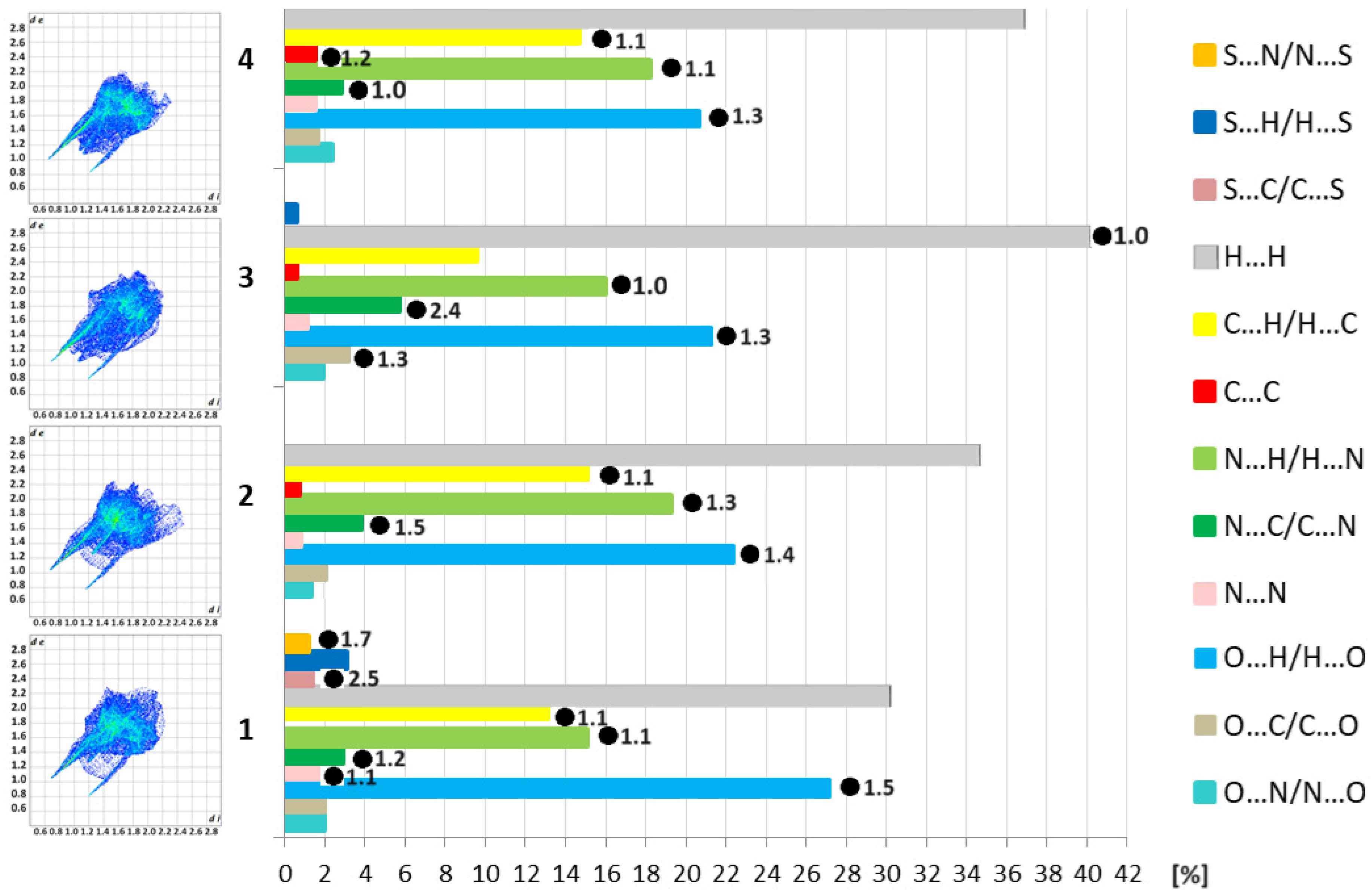
3.2. Supramolecular Features of 1–4
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ashique, S.; Sandhu, N.K.; Haque, S.N.; Koley, K. A Systemic Review on Topical Marketed Formulations, Natural Products, and Oral Supplements to Prevent Androgenic Alopecia: A Review. Nat. Prod. Bioprospect. 2020, 10, 345–365. [Google Scholar] [CrossRef]
- Wróbel, A.; Ariszewska, K.; Maliszewski, D.; Drozdowska, D. Trimethoprim and other nonclassical antifolates an excellent template for searching modifications of dihydrofolate reductase enzyme inhibitors. J. Antibiot. 2020, 73, 5–27. [Google Scholar] [CrossRef]
- Raimondi, M.V.; Randazzo, O.; La Franca, M.; Barone, G.; Vignoni, E.; Rossi, D.; Collina, S. DHFR Inhibitors: Reading the Past for Discovering Novel Anticancer Agents. Molecules 2019, 24, 1140. [Google Scholar] [CrossRef]
- Ai, M.; Wang, C.; Tang, Z.; Liu, K.; Sun, X.; Ma, T.; Li, Y.; Ma, X.; Chen, L. Design and synthesis of diphenylpyrimidine derivatives (DPPYs) as potential dual EGFR T790M and FAK inhibitors against a diverse range of cancer cell lines. Bioorg. Chem. 2020, 94, 103408. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, R.-G.; Zhang, H.; Wang, Y.-C.; Yang, D.; Zhao, Y.-L.; Yan, G.-Y.; Xu, G.-B.; Guan, H.-Y.; Zhou, Y.-H.; et al. Design, synthesis, and biological evaluation of 2,4-diamino pyrimidine derivatives as potent FAK inhibitors with anti-cancer and anti-angiogenesis activities. Eur. J. Med. Chem. 2021, 222, 113573. [Google Scholar] [CrossRef]
- Matulkova, I.; Mathauserova, J.; Cisarova, I.; Nemec, I.; Fabry, J. The study of crystal structures and vibrational spectra of inorganic salts of 2,4-diaminopyrimidine. J. Mol. Struct. 2016, 1103, 82–93. [Google Scholar] [CrossRef]
- Bertolasi, V.; Pretto, L.; Gilli, P.; Ferretti, V.; Gilli, G. Hydrogen-bonded supramolecular structures in co-crystals of β- or ζ-diketone enols with 2,6-diaminopyridine or 2,4-diaminopyrimidine. New J. Chem. 2002, 26, 1559–1566. [Google Scholar] [CrossRef]
- Draguta, S.; Fonari, M.S.; Masunov, A.E.; Zazueta, J.; Sullivan, S.; Antipin, M.Y.; Timofeeva, T.V. New acentric materials constructed from aminopyridines and 4-nitrophenol. CrystEngComm 2013, 15, 4700–4710. [Google Scholar] [CrossRef]
- Hutzler, W.M.; Egert, E.; Bolte, M. One barbiturate and two solvated thiobarbiturates containing the triply hydrogen-bonded ADA/DAD synthon, plus one ansolvate and three solvates of their coformer 2,4-diaminopyrimidine. Acta Cryst. C 2016, 72, 705–715. [Google Scholar] [CrossRef]
- Hutzler, W.M.; Egert, E.; Bolte, M. 6-Propyl-2-thiouracil versus 6-methoxymethyl-2-thiouracil: Enhancing the hydrogen-bonded synthon motif by replacement of a methylene group with an O atom. Acta Cryst. C 2016, 72, 634–646. [Google Scholar] [CrossRef]
- Hall, V.M.; Thornton, A.; Miehls, E.K.; Bertke, J.A.; Swift, J.A. Uric Acid Crystallization Interrupted with Competing Binding Agents. Cryst. Growth Des. 2019, 19, 7363–7371. [Google Scholar] [CrossRef]
- Bis, J.A.; Zaworotko, M.J. The 2-Aminopyridinium-carboxylate Supramolecular Heterosynthon: A Robust Motif for Generation of Multiple-Component Crystals. Cryst. Growth Des. 2005, 5, 1169–1179. [Google Scholar] [CrossRef]
- Mirzaei, M.; Sadeghi, F.; Molčanov, K.; Zaręba, J.K.; Gomila, R.M.; Frontera, A. Recurrent Supramolecular Motifs in a Series of Acid–Base AdductsBased on Pyridine-2,5-Dicarboxylic Acid N-Oxide and Organic Bases: Inter- and Intramolecular Hydrogen Bonding. Cryst. Growth Des. 2020, 20, 1738–1751. [Google Scholar] [CrossRef]
- Ebenezer, S.; Muthiah, P.T. Design of Co-crystals/Salts of Aminopyrimidines and Carboxylic Acids through Recurrently Occurring Synthons. Cryst. Growth Des. 2012, 12, 3766–3785. [Google Scholar] [CrossRef]
- Capelletti da Silva, C.; de Lima Cirqueira, M.; Martins, F.T. Lamivudine salts with 1,2-dicarboxylic acids: A new and a rare synthon with double pairing motif fine-tuning their solubility. CrystEngComm 2013, 15, 6311–6317. [Google Scholar] [CrossRef]
- Garg, U.; Azim, Y.; Kar, A.; Pradeep, C.P. Cocrystals/salt of 1-naphthaleneacetic acid and utilizing Hirshfeld surface calculations for acid–aminopyrimidine synthons. CrystEngComm 2020, 22, 2978–2989. [Google Scholar] [CrossRef]
- Janczak, J. Structure, vibrational characterization and DFT calculations of 1-(diaminomethylene)thiouron-1-ium 2,3-pyridinedicarboxylate. Struct. Chem. 2023. [Google Scholar] [CrossRef]
- Clarke, H.D.; Arora, K.K.; Bass, H.; Kavuru, P.; Ong, T.T.; Pujari, T.; Wojtas, L.; Zawarotko, M.J. Structure–Stability Relationships in Cocrystal Hydrates: Does the Promiscuity of Water Make Crystalline Hydrates the Nemesis of Crystal Engineering? Cryst. Growth Des. 2019, 10, 2152–2167. [Google Scholar] [CrossRef]
- Surov, A.O.; Vasilev, N.A.; Churakov, A.V.; Parashchuk, O.D.; Artobolevskii, S.V.; Alatortsev, O.A.; Makhrov, D.E.; Vener, M.V. Two Faces of Water in the Formation and Stabilization of Multicomponent Crystals of Zwitterionic Drug-Like Compounds. Symmetry 2021, 13, 425. [Google Scholar] [CrossRef]
- Bolla, G.; Nangia, A. Novel pharmaceutical salts of albendazole. CrystEngComm 2018, 20, 6394–6405. [Google Scholar] [CrossRef]
- Hu, L.; Staples, R.J.; Shreeve, J.M. Energetic compounds based on a new fused triazolo[4,5-d]pyridazine ring: Nitroimino lights up energetic performance. Chem. Eng. J. 2021, 420, 129839. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Cryst. C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Cryst. 2020, 53, 226–235. [Google Scholar] [CrossRef]
- Spek, A.L. Structure Validation in chemical crystallography. Acta Cryst. D 2009, 65, 148–155. [Google Scholar] [CrossRef]
- Turner, M.J.; McKinnon, J.J.; Wol, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer, version 3.1; The University of Western Australia: Perth, Australia, 2017.
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Jayatilaka, D.; Grimwood, D.J. Tonto: A Fortran Based Object-Oriented System for Quantum Chemistry and Crystallography. In Computational Science—ICCS 2003; Sloot, P.M.A., Abramson, D., Bogdanov, A.V., Gorbachev, Y.E., Dongarra, J.J., Zomaya, A.Y., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2003; Volume 2660, pp. 142–151. [Google Scholar] [CrossRef]
- Jelsch, C.; Ejsmont, K.; Huder, L. The enrichment ratio of atomic contacts in crystals, an indicator derived from the Hirshfeld surface analysis. IUCrJ 2014, 1, 119–128. [Google Scholar] [CrossRef]
- Bernstein, J.; Davis, R.E.; Shimoni, L.; Chang, N.L. Patterns in Hydrogen Bonding: Functionality and Graph Set Analysis in Crystals. Angew. Chem. Int. Ed. 1995, 34, 1555–1573. [Google Scholar] [CrossRef]
- Etter, M.C. Encoding and decoding hydrogen-bond patterns of organic compounds. Acc. Chem. Res. 1990, 23, 120–126. [Google Scholar] [CrossRef]
- Nishio, M. CH/interactions hydrogen bonds in crystals. CrystEngComm 2004, 6, 130–158. [Google Scholar] [CrossRef]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Cryst. 2021, 54, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 37, 3814–3816. [Google Scholar] [CrossRef] [PubMed]
- Hathwar, V.R.; Sist, M.; Jørgensen, M.R.V.; Mamakhel, A.H.; Wang, X.; Hoffmann, C.M.; Sugimoto, K.; Overgaard, J.; Iversen, B.B. Quantitative analysis of intermolecular interactions in orthorhombic rubrene. IUCrJ 2015, 2, 563–574. [Google Scholar] [CrossRef]
- Lee, S.M.; Lo, K.M.; Tan, S.L.; Tiekink, E.R.T. (Tris{2-[(5-chloro-2-oxido benzyl idene-κO)amino-κN]eth yl}amine-κN) ytterbium(III): Crystal structure and Hirshfeld surface analysis. Acta Crystallogr. E 2016, 72, 1390–1395. [Google Scholar] [CrossRef] [PubMed]


| Compound | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Chemical formula | C8H14N4O5S | C9H16N4O5 | C20H34N8O9S4 | C24H44N8O10 |
| Formula weight | 278.29 | 260.26 | 658.79 | 604.67 |
| λ (Cu Kα) (Å) | 1.54184 | 1.54184 | 1.54184 | 1.54184 |
| Crystal system | triclinic | monoclinic | monoclinic | Triclinic |
| Space group | P21/c | C2/c | ||
| a (Å) | 4.78960(16) | 4.85263(11) | 31.6649(8) | 5.3664(4) |
| b (Å) | 10.5175(3) | 26.0403(5) | 5.10031(11) | 9.2521(5) |
| c (Å) | 12.6651(4) | 9.8003(2) | 18.3024(4) | 15.2231(9) |
| α (°) | 107.397(3) | 90 | 90 | 93.347(5) |
| β (°) | 95.472(3) | 96.4945(19) | 93.141(2) | 94.615(6) |
| γ (°) | 102.363(3) | 90 | 90 | 102.566(6) |
| Volume (Å) | 585.94(3) | 1230.46(5) | 2951.41(12) | 733.10(9) |
| Z | 2 | 4 | 4 | 1 |
| Z′ | 1 | 1 | 0.5 | 0.5 |
| Dcalc (g·cm−3) | 1.577 | 1.405 | 1.483 | 1.370 |
| μ (mm−1) | 2.700 | 0.986 | 3.499 | 0.902 |
| F (000) | 292 | 552 | 1384 | 324 |
| Crystal size (mm) | 0.28 × 0.15 × 0.04 | 0.20 × 0.12 × 0.03 | 0.20 × 0.14 × 0.05 | 0.18 × 0.10 × 0.04 |
| θ range (°) | 3.712–70.035 | 3.394–70.311 | 2.795–69.226 | 2.921–70.572 |
| Reflections collected | 15217 | 13019 | 17911 | 5322 |
| Unique reflections | 2178 | 2308 | 2738 | 2740 |
| Reflections I > 2σ(I) | 1992 | 2064 | 2648 | 2192 |
| Rint | 0.0323 | 0.0253 | 0.0246 | 0.0247 |
| Restraints/parameters | 1/193 | 0/195 | 0/214 | 0/222 |
| Goodness-of-fit | 1.035 | 1.050 | 1.059 | 1.035 |
| R1, wR2 (I > 2σ(I)) | 0.0305, 0.0347 | 0.0344, 0.0391 | 0.0234, 0.0241 | 0.0488, 0.0630 |
| R1, wR2 (all data) | 0.0770, 0.0810 | 0.0885, 0.0932 | 0.0633, 0.0639 | 0.1260, 0.1373 |
| Max. peak/hole (e·Å−3) | 0.369/−0.346 | 0.260/−0.219 | 0.277/−0.200 | 0.661/−0.440 |
| K.P.I. (%) * | 74.7 | 69.9 | 70.4 | 71.3 |
| D-H…A | d(D-H) | d(H…A) | d(D…A) | <(DHA) | Symmetry Code |
|---|---|---|---|---|---|
| 1 | |||||
| N2-H2∙∙∙O2 | 0.87(2) | 1.87(2) | 2.7375(18) | 172.2(18) | x, −1 + y, z |
| O3-H3∙∙∙O2 | 0.96(2) | 1.57(2) | 2.5218(15) | 175.2(18) | 1 − x, 1 − y, −z |
| N3-H3A∙∙∙O4 | 0.87(2) | 2.21(2) | 3.0757(18) | 170.2(19) | −1 + x, y, z |
| N3-H3B∙∙∙O5 | 0.86(2) | 2.11(2) | 2.9540(18) | 169.9(18) | −1 + x, y, z |
| N4-H4A∙∙∙O4 | 0.85(2) | 2.141(19) | 2.8473(19) | 140.0(16) | 1 − x, −y, −z |
| N4-H4B∙∙∙N1 | 0.85(2) | 2.17(2) | 3.018(2) | 174.4(19) | −x, −y, −z |
| O5-H5A∙∙∙O1 | 0.85 | 1.97 | 2.8111(18) | 173 | 1 − x, 1 − y, 1 − z |
| O5-H5B∙∙∙O5 | 0.85 | 2.16 | 2.991(2) | 164 | 2 − x, 1 − y, 1 − z |
| O5-H5C∙∙∙O5 | 0.85 | 2.18 | 3.012(2) | 166 | 1 − x, 1 − y, 1 − z |
| C2-H2B∙∙∙O3 | 0.99 | 2.41 | 3.1713(19) | 133 | 1 − x, 1 − y, −z |
| C3-H3C∙∙∙O4 | 0.99 | 2.31 | 3.2956(19) | 175 | −1 + x, y, z |
| 2 | |||||
| N1-H1∙∙∙O3 | 0.93(2) | 1.80(2) | 2.7207(15) | 171.6(17) | −1 + x, y, z |
| N1-H1∙∙∙O4 | 0.93(2) | 2.51(2) | 3.1815(14) | 129.0(15) | −1 + x, y, z |
| O2-H2∙∙∙O3 | 0.97(2) | 1.60(2) | 2.5647(14) | 171(2) | −1 + x, ½ − y, −1/2 + z |
| N3-H3C∙∙∙N2 | 0.893(19) | 2.078(18) | 2.9679(17) | 174.5(17) | 2 − x, 1 − y, 1 − z |
| N3-H3D∙∙∙O1 | 0.877(18) | 2.103(17) | 2.8148(16) | 137.8(16) | x, ½ − y, ½ + z |
| N4-H4C∙∙∙O5 | 0.898(18) | 1.925(18) | 2.8214(15) | 175.3(18) | 1 − x, 1 − y, −z |
| N4-H4D∙∙∙O1 | 0.909(19) | 2.060(19) | 2.9605(14) | 170.7(16) | 2 − x, ½ + y, ½ − z |
| O5-H5A∙∙∙O4 | 0.87(3) | 1.96(3) | 2.8160(14) | 171(2) | |
| O5-H5B∙∙∙O4 | 0.86(2) | 2.04(2) | 2.8973(14) | 172(2) | −1 + x, y, z |
| C4-H4B∙∙∙O2 | 0.99 | 2.51 | 3.2596(17) | 132 | 1 + x, ½ − y, ½ + z |
| 3 | |||||
| N2-H2∙∙∙O2 | 0.84 | 1.92 | 2.7553(13) | 172 | 1 − x, 1 + y, ½ − z |
| N3-H3C∙∙∙O1 | 0.85 | 2.14 | 1.9730(12) | 168 | 1 − x, 1 − y, 1 − z |
| N3-H3D∙∙∙N1 | 0.87 | 2.17 | 3.0322(14) | 175 | 1 − x, 1 − y, 1 − z |
| O4-H4∙∙∙O2 | 0.83 | 1.78 | 2.6053(14) | 174 | 3/2 − x, −1/2 + y, ½ − z |
| N4-H4C∙∙∙O1 | 0.84 | 2.08 | 2.9087(12) | 170 | 1 − x, 1 + y, ½ − z |
| O5-H5C∙∙∙O1 | 0.87 | 1.87 | 2.7334(10) | 174 | 1 − x, y, ½ − z |
| O5-H5D∙∙∙O1 | 0.87 | 1.93 | 2.7334(10) | 153 | |
| *C5-H5B∙∙∙S1 | 0.99 | 2.85 | 3.3970(14) | 115 | |
| C9-H9∙∙∙O3 | 0.95 | 2.31 | 3.1211(15) | 143 | −1/2 + x, 3/3 + y, z |
| 4 | |||||
| N1-H1∙∙∙O2 | 0.92(3) | 1.77(3) | 2.682(2) | 174(2) | x, −1 + y, z |
| N3-H3A∙∙∙O1 | 0.92(3) | 1.90(3) | 2.814(2) | 173(2) | x, −1 + y, z |
| N3-H3B∙∙∙N2 | 0.89(3) | 2.19(3) | 3.080(3) | 174(3) | −x, −y, −z |
| O4-H4∙∙∙O2 | 0.98(3) | 1.54(3) | 2.518(2) | 175(3) | −1 + x, −1 + y, z |
| N4-H4A∙∙∙O5 | 0.87(3) | 2.08(3) | 2.854(3) | 149(2) | −x, 1 − y, −z |
| N4-H4B∙∙∙O3 | 0.88(3) | 2.17(3) | 3.049(2) | 177(3) | 1 − x, 1 − y, −z |
| O5-H5A∙∙∙O1 | 0.90(3) | 1.81(3) | 2.685(2) | 164(3) | |
| O5-H5B∙∙∙O3 | 0.78(3) | 2.14(3) | 2.921(2) | 177(3) | |
| C2-H2B∙∙∙O5 | 0.99 | 2.58 | 3.524(3) | 158 | 1 + x, y, z |
| C12-H12∙∙∙O3 | 0.95 | 2.54 | 3.437(3) | 157 | 1 + x, y, z |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bojarska, J.; Łyczko, K.; Mieczkowski, A. Synthesis, Crystal Structure and Supramolecular Features of Novel 2,4-Diaminopyrimidine Salts. Crystals 2024, 14, 133. https://doi.org/10.3390/cryst14020133
Bojarska J, Łyczko K, Mieczkowski A. Synthesis, Crystal Structure and Supramolecular Features of Novel 2,4-Diaminopyrimidine Salts. Crystals. 2024; 14(2):133. https://doi.org/10.3390/cryst14020133
Chicago/Turabian StyleBojarska, Joanna, Krzysztof Łyczko, and Adam Mieczkowski. 2024. "Synthesis, Crystal Structure and Supramolecular Features of Novel 2,4-Diaminopyrimidine Salts" Crystals 14, no. 2: 133. https://doi.org/10.3390/cryst14020133
APA StyleBojarska, J., Łyczko, K., & Mieczkowski, A. (2024). Synthesis, Crystal Structure and Supramolecular Features of Novel 2,4-Diaminopyrimidine Salts. Crystals, 14(2), 133. https://doi.org/10.3390/cryst14020133







