FDIP—A Fast Diffraction Image Processing Library for X-ray Crystallography Experiments
Abstract
:1. Introduction
2. Materials and Methods
2.1. Handling a Bad Pixel Mask
2.2. Algorithm Descriptions
- Radial background subtraction—a set of functions used to calculate radial background for further subtraction on the pattern-by-pattern basis.
- —an algorithm to mask the radial streaks at the diffraction patterns.
- —an algorithm to mask the sharp rings at the diffraction patterns. It also works with just a set of strong Bragg peaks at a single radius (not just uninterrupted rings).
- —an algorithm to find Bragg peaks in diffraction patterns, that is, using local background estimation to determine the peaks’ location.
- —a GUI that can be used for the optimisation of parameters for all the algorithms mentioned above.
2.2.1. Radial Background Subtraction
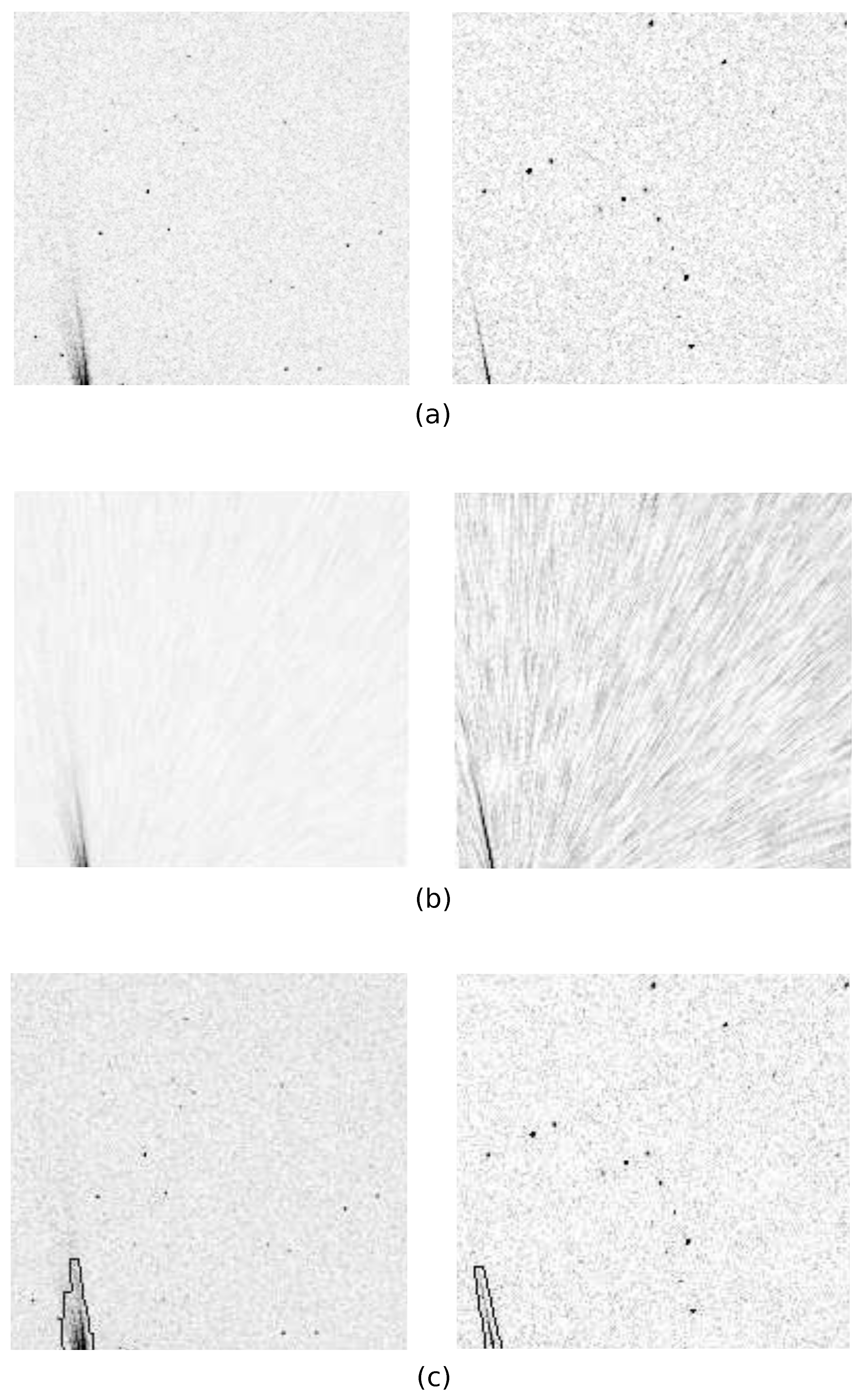
2.2.2. Streak Masking
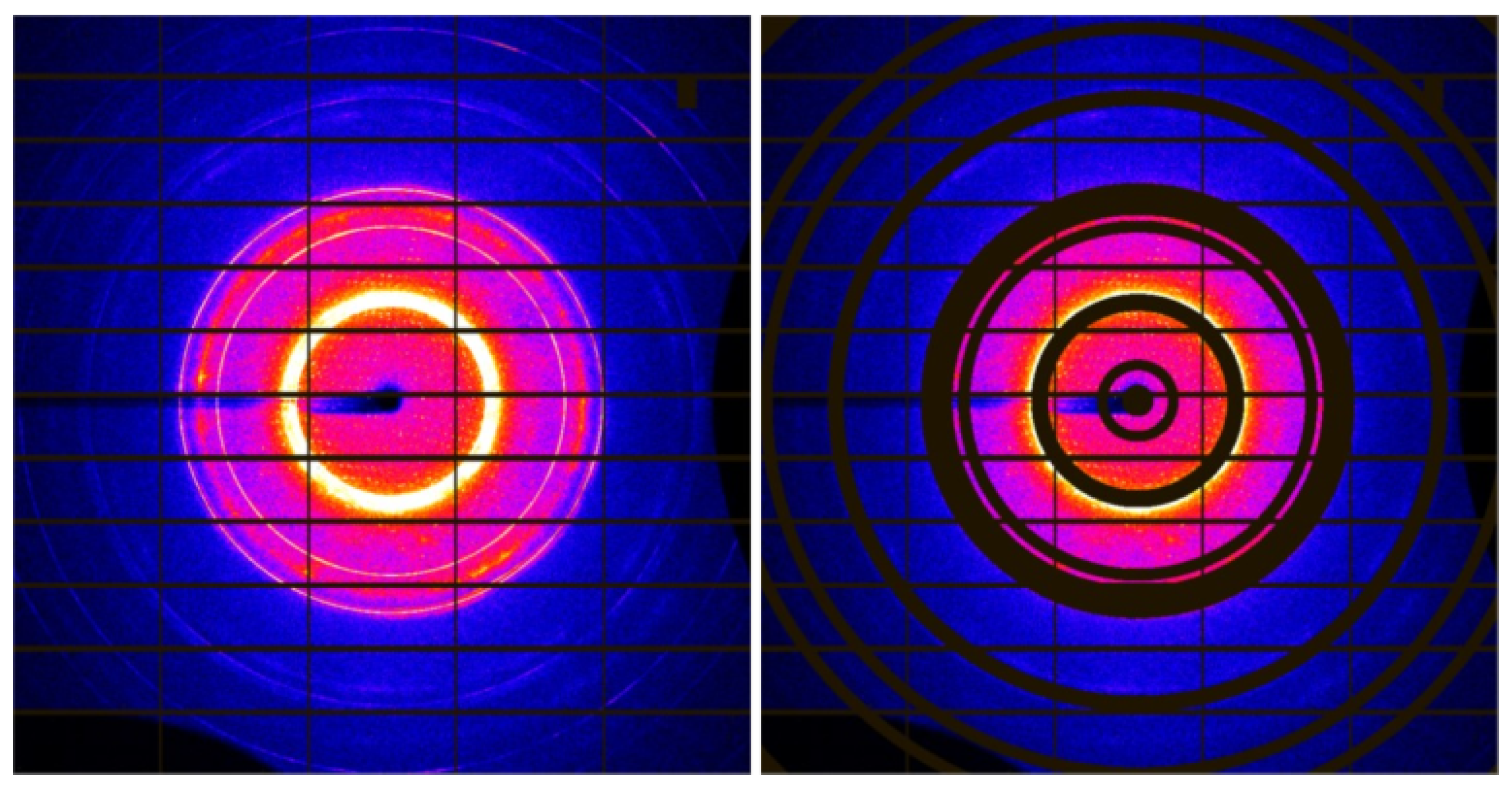
2.2.3. Ring Masking
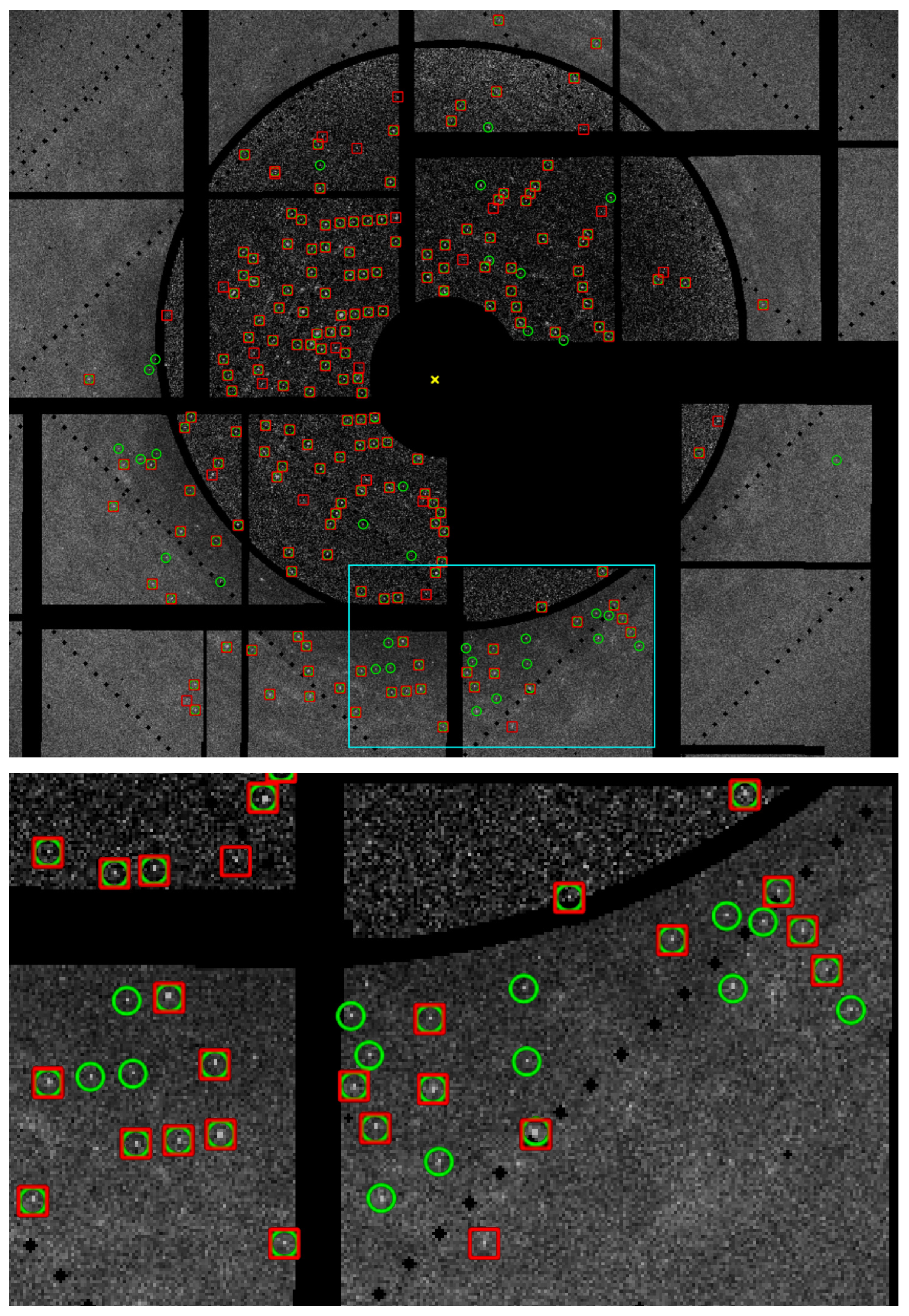
2.2.4. Peak Finding
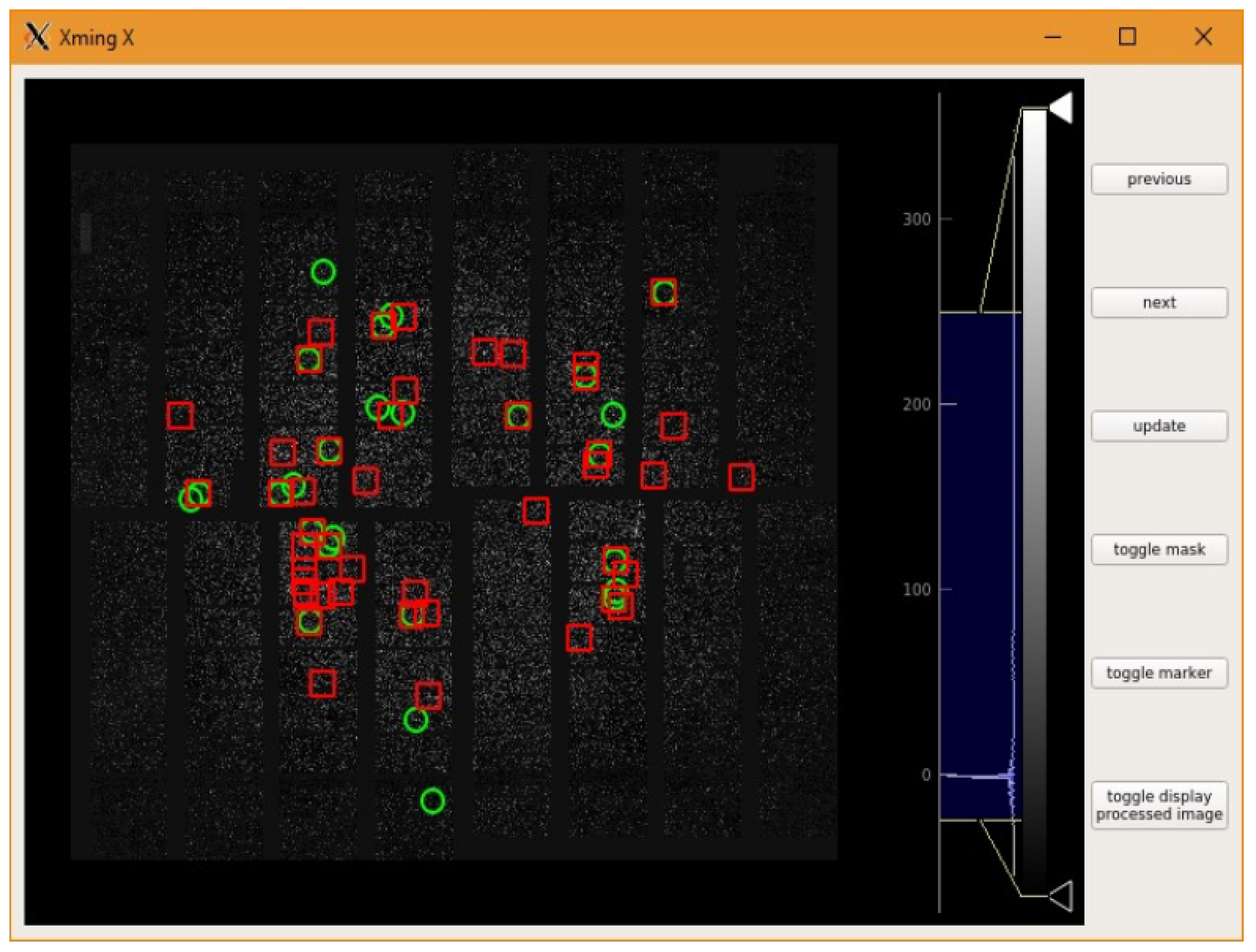
2.3. Parameter Tweaker
- ;
- ;
- ;
- ;
3. Results
3.1. Execution Time Measurement
3.2. Radial Background Subtraction
3.3. Streak Masking
3.4. Ring Masking
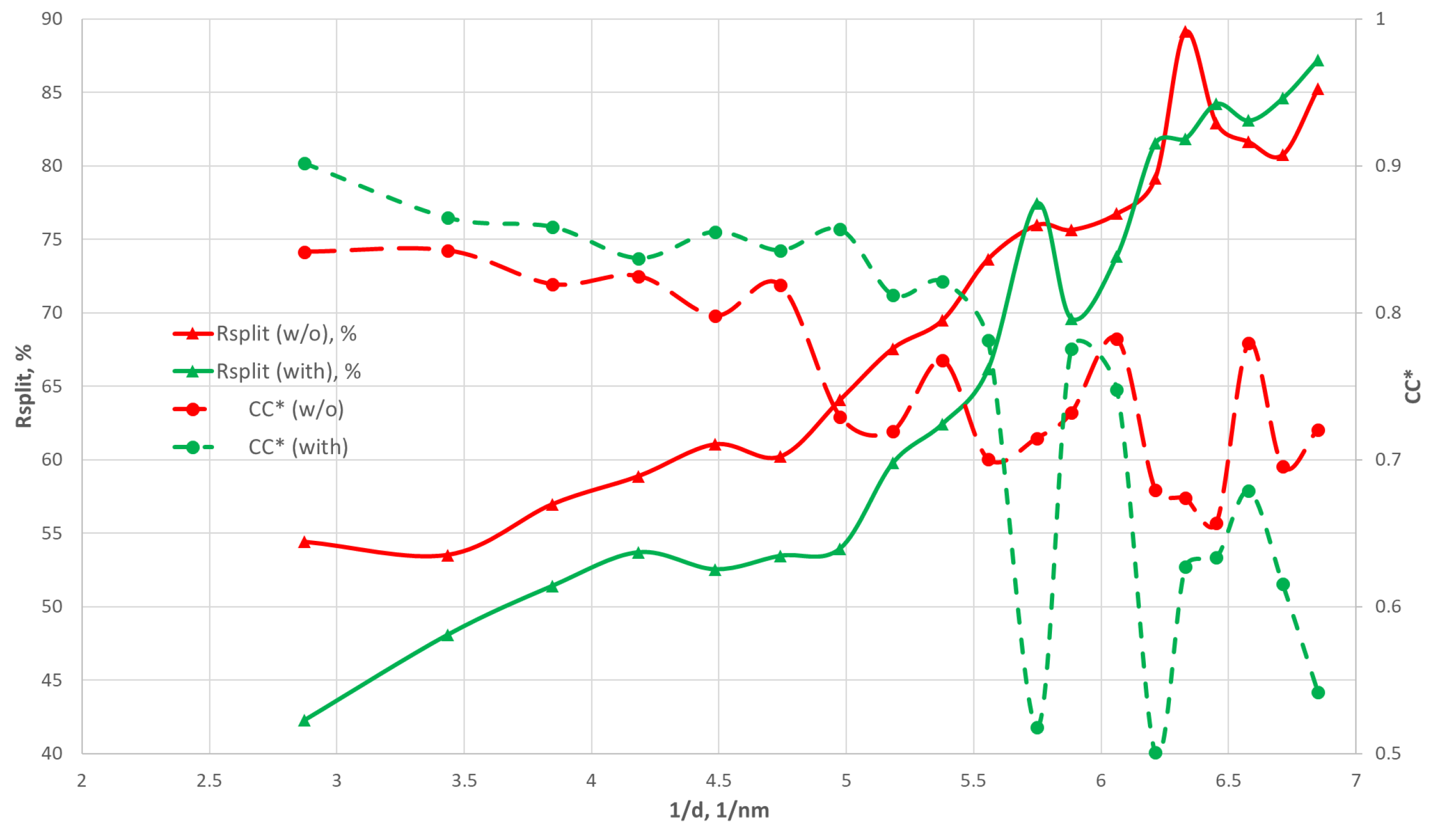
3.5. Peak Finding
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SX | Serial crystallography |
| GUI | Graphical user interface |
| OnDA | Online data analysis |
| FDIP | Fast diffraction image processing library |
References
- Chapman, H.N.; Fromme, P.; Barty, A.; White, T.A.; Kirian, R.A.; Aquila, A.; Hunter, M.S.; Schulz, J.; DePonte, D.P.; Weierstall, U.; et al. Femtosecond X-ray protein nanocrystallography. Nature 2011, 470, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Boutet, S.; Lomb, L.; Williams, G.J.; Barends, T.R.; Aquila, A.; Doak, R.B.; Weierstall, U.; DePonte, D.P.; Steinbrener, J.; Shoeman, R.L.; et al. High-resolution protein structure determination by serial femtosecond crystallography. Science 2012, 337, 362–364. [Google Scholar] [CrossRef]
- Barends, T.R.; Foucar, L.; Ardevol, A.; Nass, K.; Aquila, A.; Botha, S.; Doak, R.B.; Falahati, K.; Hartmann, E.; Hilpert, M.; et al. Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation. Science 2015, 350, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Pande, K.; Hutchison, C.D.; Groenhof, G.; Aquila, A.; Robinson, J.S.; Tenboer, J.; Basu, S.; Boutet, S.; DePonte, D.P.; Liang, M.; et al. Femtosecond structural dynamics drives the trans/cis isomerization in photoactive yellow protein. Science 2016, 352, 725–729. [Google Scholar] [CrossRef]
- Stagno, J.R.; Knoska, J.; Chapman, H.N.; Wang, Y.X. Mix-and-Inject Serial Femtosecond Crystallography to Capture RNA Riboswitch Intermediates. In RNA Structure and Dynamics; Springer: Berlin/Heidelberg, Germany, 2022; pp. 243–249. [Google Scholar]
- Barends, T.R.; Stauch, B.; Cherezov, V.; Schlichting, I. Serial femtosecond crystallography. Nat. Rev. Methods Prim. 2022, 2, 59. [Google Scholar] [CrossRef]
- Tolstikova, A.; Levantino, M.; Yefanov, O.; Hennicke, V.; Fischer, P.; Meyer, J.; Mozzanica, A.; Redford, S.; Crosas, E.; Opara, N.L.; et al. 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector. IUCrJ 2019, 6, 927–937. [Google Scholar] [CrossRef]
- Yefanov, O.; Oberthür, D.; Bean, R.; Wiedorn, M.O.; Knoska, J.; Pena, G.; Awel, S.; Gumprecht, L.; Domaracky, M.; Sarrou, I.; et al. Evaluation of serial crystallographic structure determination within megahertz pulse trains. Struct. Dyn. 2019, 6, 064702. [Google Scholar] [CrossRef] [PubMed]
- Mariani, V.; Morgan, A.; Yoon, C.H.; Lane, T.J.; White, T.A.; O’Grady, C.; Kuhn, M.; Aplin, S.; Koglin, J.; Barty, A.; et al. OnDA: Online data analysis and feedback for serial X-ray imaging. J. Appl. Crystallogr. 2016, 49, 1073–1080. [Google Scholar] [CrossRef]
- Barty, A.; Kirian, R.A.; Maia, F.R.; Hantke, M.; Yoon, C.H.; White, T.A.; Chapman, H. Cheetah: Software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Crystallogr. 2014, 47, 1118–1131. [Google Scholar] [CrossRef]
- Leonarski, F.; Mozzanica, A.; Brückner, M.; Lopez-Cuenca, C.; Redford, S.; Sala, L.; Babic, A.; Billich, H.; Bunk, O.; Schmitt, B.; et al. JUNGFRAU detector for brighter x-ray sources: Solutions for IT and data science challenges in macromolecular crystallography. Struct. Dyn. 2020, 7, 014305. [Google Scholar] [CrossRef]
- Grünbein, M.L.; Nass Kovacs, G. Sample delivery for serial crystallography at free-electron lasers and synchrotrons. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 178–191. [Google Scholar] [CrossRef]
- Zhao, F.Z.; Zhang, B.; Yan, E.K.; Sun, B.; Wang, Z.J.; He, J.H.; Yin, D.C. A guide to sample delivery systems for serial crystallography. FEBS J. 2019, 286, 4402–4417. [Google Scholar] [CrossRef]
- DePonte, D.; Weierstall, U.; Schmidt, K.; Warner, J.; Starodub, D.; Spence, J.; Doak, R. Gas dynamic virtual nozzle for generation of microscopic droplet streams. J. Phys. D Appl. Phys. 2008, 41, 195505. [Google Scholar] [CrossRef]
- Oberthuer, D.; Knoška, J.; Wiedorn, M.O.; Beyerlein, K.R.; Bushnell, D.A.; Kovaleva, E.G.; Heymann, M.; Gumprecht, L.; Kirian, R.A.; Barty, A.; et al. Double-flow focused liquid injector for efficient serial femtosecond crystallography. Sci. Rep. 2017, 7, 44628. [Google Scholar] [CrossRef]
- Hunter, M.S.; Segelke, B.; Messerschmidt, M.; Williams, G.J.; Zatsepin, N.A.; Barty, A.; Benner, W.H.; Carlson, D.B.; Coleman, M.; Graf, A.; et al. Fixed-target protein serial microcrystallography with an x-ray free electron laser. Sci. Rep. 2014, 4, 6026. [Google Scholar] [CrossRef]
- Maia, F.R.N.C. The Coherent X-ray Imaging Data Bank. Nat. Methods 2012, 9, 854–855. [Google Scholar] [CrossRef]
- Bernstein, H.J.; Förster, A.; Bhowmick, A.; Brewster, A.S.; Brockhauser, S.; Gelisio, L.; Hall, D.R.; Leonarski, F.; Mariani, V.; Santoni, G.; et al. Gold Standard for macromolecular crystallography diffraction data. IUCrJ 2020, 7, 784–792. [Google Scholar] [CrossRef]
- White, T.A.; Kirian, R.A.; Martin, A.V.; Aquila, A.; Nass, K.; Barty, A.; Chapman, H.N. CrystFEL: A software suite for snapshot serial crystallography. J. Appl. Crystallogr. 2012, 45, 335–341. [Google Scholar] [CrossRef]
- Yefanov, O.; Mariani, V.; Gati, C.; White, T.A.; Chapman, H.N.; Barty, A. Accurate determination of segmented X-ray detector geometry. Opt. Express 2015, 23, 28459. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, J.; Karkoulis, D. PyFAI, a versatile library for azimuthal regrouping. J. Phys. Conf. Ser. 2013, 425, 202012. [Google Scholar] [CrossRef]
- Wiedorn, M.O.; Awel, S.; Morgan, A.J.; Ayyer, K.; Gevorkov, Y.; Fleckenstein, H.; Roth, N.; Adriano, L.; Bean, R.; Beyerlein, K.R.; et al. Rapid sample delivery for megahertz serial crystallography at X-ray FELs. IUCrJ 2018, 5, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Yefanov, O.; Gati, C.; Bourenkov, G.; Kirian, R.A.; White, T.A.; Spence, J.C.H.; Chapman, H.N.; Barty, A. Mapping the continuous reciprocal space intensity distribution of X-ray serial crystallography. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130333. [Google Scholar] [CrossRef] [PubMed]
- Ayyer, K.; Yefanov, O.M.; Oberthür, D.; Roy-Chowdhury, S.; Galli, L.; Mariani, V.; Basu, S.; Coe, J.; Conrad, C.E.; Fromme, R.; et al. Macromolecular diffractive imaging using imperfect crystals. Nature 2016, 530, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Guenther, S.; Reinke, P.Y.; Fernandez-Garcia, Y.; Lieske, J.; Lane, T.J.; Ginn, H.; Koua, F.; Ehrt, C.; Ewert, W.; Oberthuer, D.; et al. Massive X-ray screening reveals two allosteric drug binding sites of SARS-CoV-2 main protease. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hart, P.; Boutet, S.; Carini, G.; Dubrovin, M.; Duda, B.; Fritz, D.; Haller, G.; Herbst, R.; Herrmann, S.; Kenney, C.; et al. The CSPAD megapixel X-ray camera at LCLS. In Proceedings of the X-ray Free-Electron Lasers: Beam Diagnostics, Beamline Instrumentation, and Applications, Proceeding of the SPIE Optica Engineering + Applications, San Diego, CA, USA, 12–16 August 2012; SPIE: Bellingham, WA, USA, 2012; Volume 8504, pp. 51–61. [Google Scholar]
- Günther, S.; Reinke, P.Y.; Fernández-García, Y.; Lieske, J.; Lane, T.J.; Ginn, H.M.; Koua, F.H.; Ehrt, C.; Ewert, W.; Oberthuer, D.; et al. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science 2021, 372, 642–646. [Google Scholar] [CrossRef]
- Karplus, P.A.; Diederichs, K. Linking crystallographic model and data quality. Science 2012, 336, 1030–1033. [Google Scholar] [CrossRef]
- Assmann, G.; Brehm, W.; Diederichs, K. Identification of rogue datasets in serial crystallography. J. Appl. Crystallogr. 2016, 49, 1021–1028. [Google Scholar] [CrossRef]
- Gasparotto, P.; Barba, L.; Stadler, H.C.; Assmann, G.; Mendonça, H.; Ashton, A.; Janousch, M.; Leonarski, F.; Béjar, B. TORO Indexer: A PyTorch-Based Indexing Algorithm for kHz Serial Crystallography. ChemRxiv 2023. [Google Scholar] [CrossRef]
- Swanson, H.E.; McMurdie, H.F.; Morris, M.C.; Evans, E.H. Standard X-ray Diffraction Powder Patterns; Number Bd. 1–10 in NBS Monograph; U.S. Department of Commerce, National Bureau of Standards: Washington, DC, USA, 1953.
- Owen, N.; Smith, P.; Martin, J.; Wright, A. X-ray diffraction at ultra-high pressures. J. Phys. Chem. Solids 1963, 24, 1519–1524. [Google Scholar] [CrossRef]
- Varma, M.; Krottenmüller, M.; Poswal, H.K.; Kuntscher, C.A. Pressure-Induced Structural Phase Transitions in the Chromium Spinel LiInCr4O8 with Breathing Pyrochlore Lattice. Crystals 2023, 13, 170. [Google Scholar] [CrossRef]




| Eiger 16M | PILATUS 6M | |
|---|---|---|
| FDIP | 0.05 s | 0.02 s |
| Cheetah | 4.96 s | 1.63 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gevorkov, Y.; Galchenkova, M.; Mariani, V.; Barty, A.; White, T.A.; Chapman, H.N.; Yefanov, O. FDIP—A Fast Diffraction Image Processing Library for X-ray Crystallography Experiments. Crystals 2024, 14, 164. https://doi.org/10.3390/cryst14020164
Gevorkov Y, Galchenkova M, Mariani V, Barty A, White TA, Chapman HN, Yefanov O. FDIP—A Fast Diffraction Image Processing Library for X-ray Crystallography Experiments. Crystals. 2024; 14(2):164. https://doi.org/10.3390/cryst14020164
Chicago/Turabian StyleGevorkov, Yaroslav, Marina Galchenkova, Valerio Mariani, Anton Barty, Thomas A. White, Henry N. Chapman, and Oleksandr Yefanov. 2024. "FDIP—A Fast Diffraction Image Processing Library for X-ray Crystallography Experiments" Crystals 14, no. 2: 164. https://doi.org/10.3390/cryst14020164
APA StyleGevorkov, Y., Galchenkova, M., Mariani, V., Barty, A., White, T. A., Chapman, H. N., & Yefanov, O. (2024). FDIP—A Fast Diffraction Image Processing Library for X-ray Crystallography Experiments. Crystals, 14(2), 164. https://doi.org/10.3390/cryst14020164





