The Use of Size Exclusion Chromatography to Monitor Protein Self-Assembly
Abstract
:1. Introduction
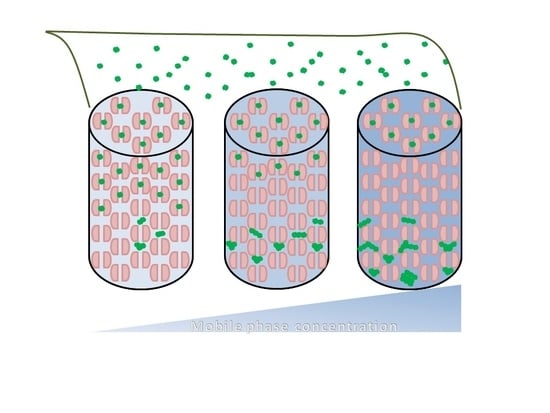
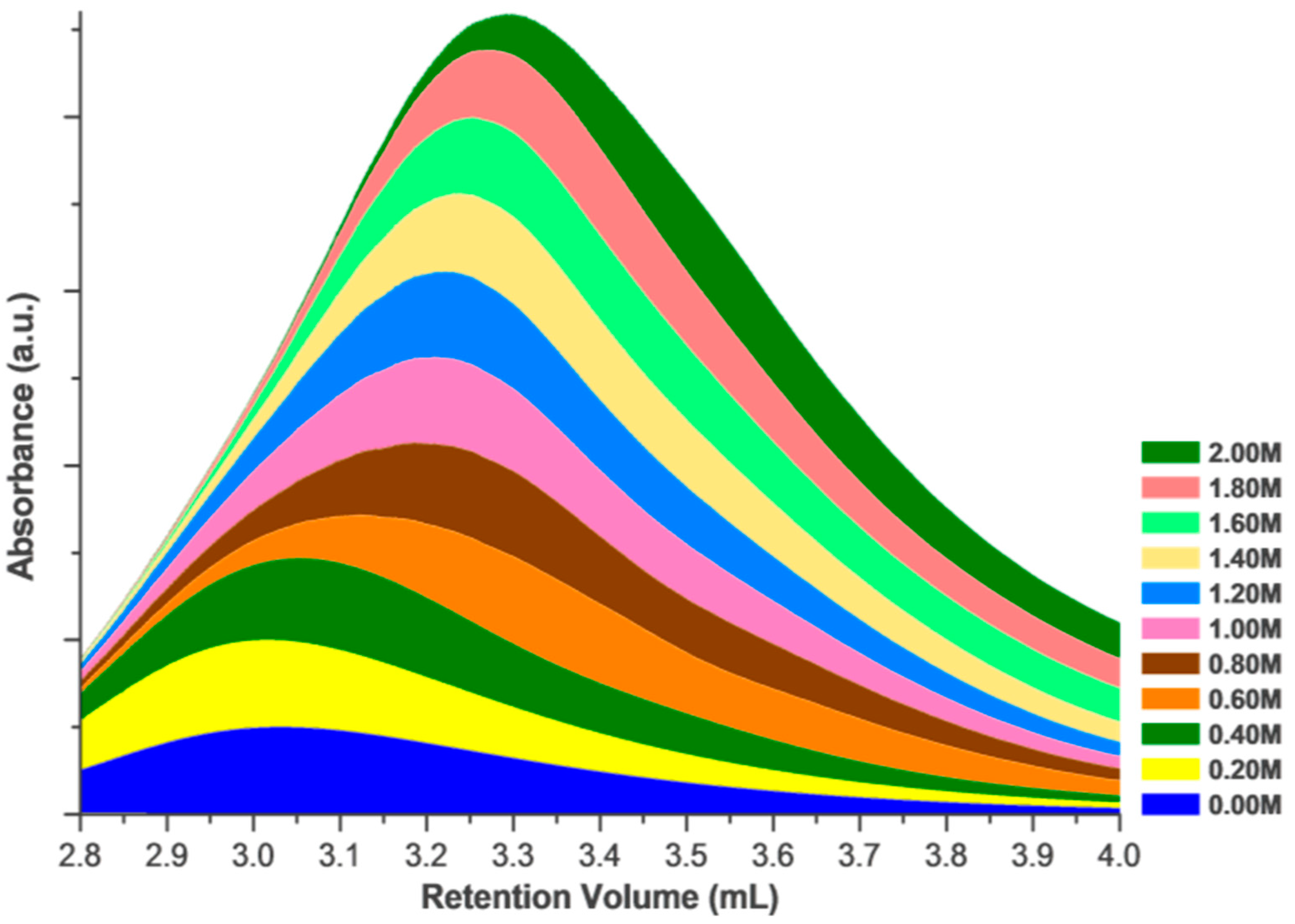
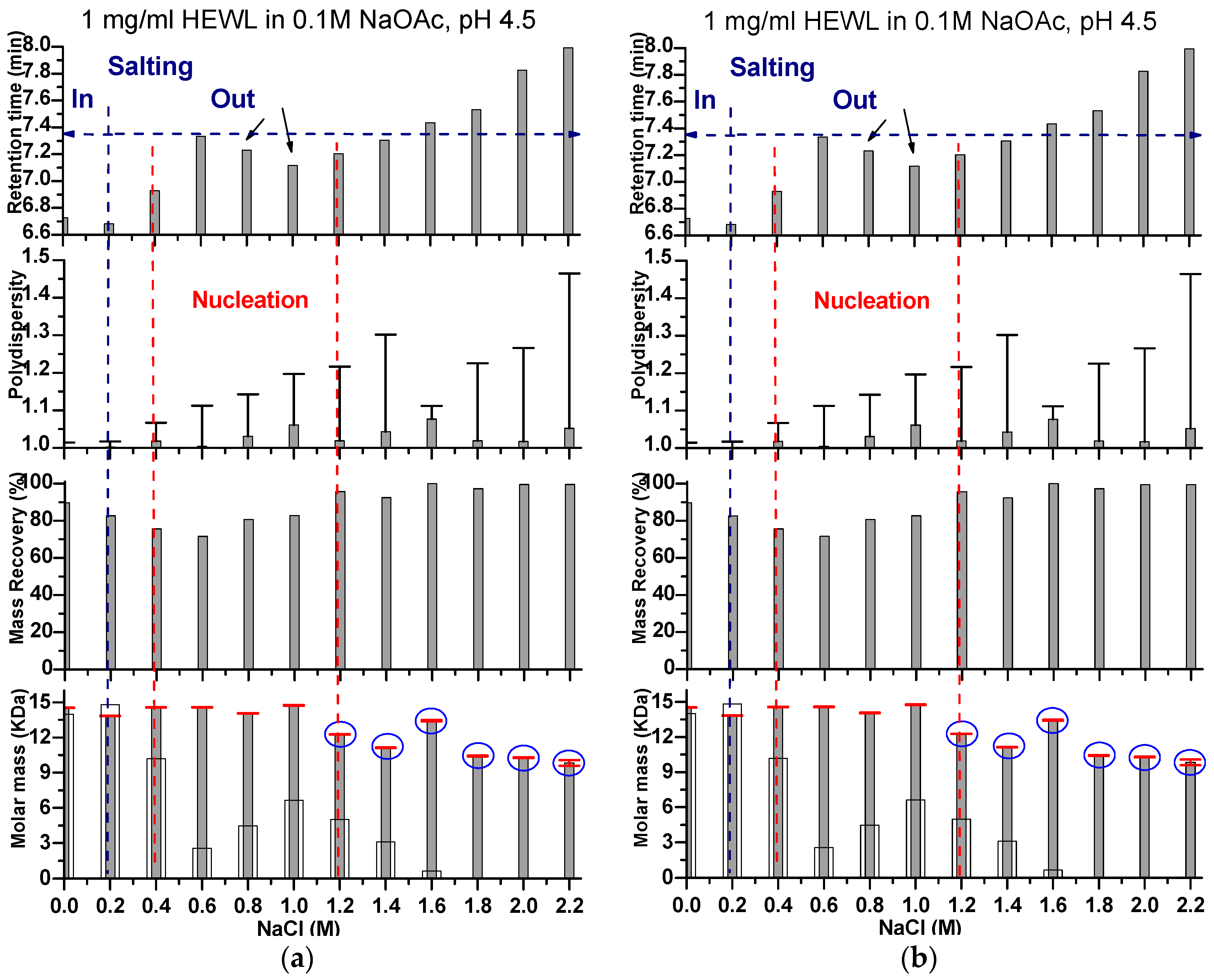
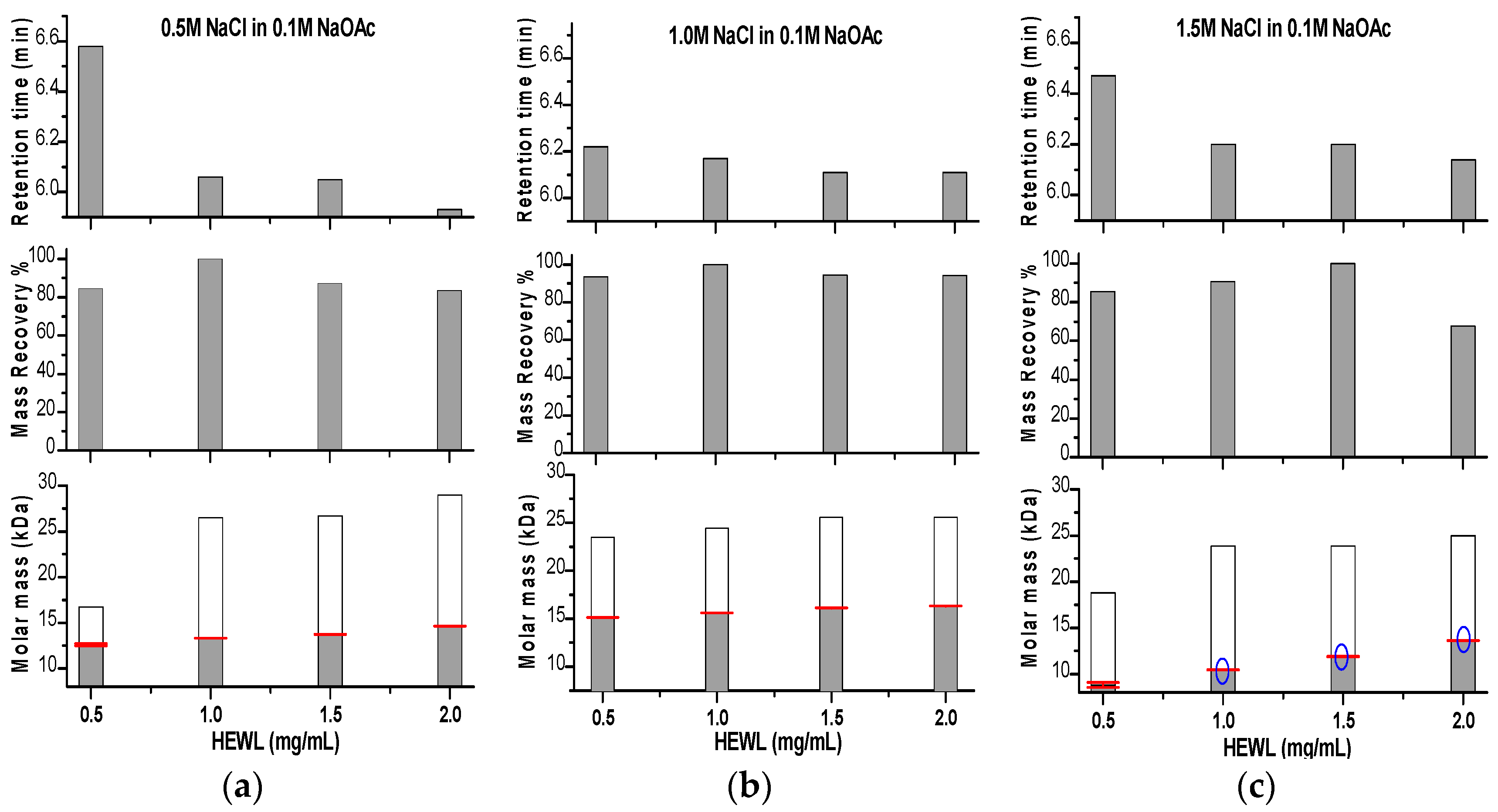
2. Results and Discussion
Effects of Ionic Strength on Elution (Salting in)
3. Discussion
4. Materials and Methods
4.1. Solution Conditions
4.2. Experimental Setups
Acknowledgments
Conflicts of Interest
References
- Hefti, F. High-performance size-exclusion chromatography: A buffer for the reliable determination of molecular weights of proteins. Anal. Biochem. 1982, 121, 378–381. [Google Scholar] [CrossRef]
- Lodish, H.; Berk, A.; Zipursky, S.L.; Matsudaira, P.; Baltimore, D.; Darnell, J. Section 3.5, Purifying, Detecting, and Characterizing Proteins. In Molecular Cell Biology; W. H. Freeman: New York, NY, USA, 2000. [Google Scholar]
- Irvine, G.B. High-performance size-exclusion chromatography of peptides. J. Biochem. Biophys. Methods 2003, 56, 233–242. [Google Scholar] [CrossRef]
- Janson, J.-C. Protein Purification: Principles, High Resolution Methods, and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Mori, S.; Barth, H.G. Size Exclusion Chromatography; Springer Science & Business Media: Berlin, Germany, 2013. [Google Scholar]
- Sigma-Aldrich1. Molecular Weight Markers for Gel Filtration Chromatography. Available online: http://www.sigmaaldrich.com/life-science/proteomics/protein-chromatography/mw-markers.html (accessed on 6 September 2017).
- Sigma-Aldrich2. Gel Filtration Chromatography. Available online: http://www.sigmaaldrich.com/life-science/proteomics/protein-chromatography/gel-filtration-chromatography.html (accessed on 6 September 2017).
- Grubisic, Z.; Rempp, P.; Benoit, H. A universal calibration for gel permeation chromatography. J. Polym. Sci. B Polym. Lett. 1967, 5, 753–759. [Google Scholar] [CrossRef]
- Ricker, R.; Sandoval, L. Fast, reproducible size-exclusion chromatography of biological macromolecules. J. Chromatogr. A 1996, 743, 43–50. [Google Scholar] [CrossRef]
- Kopaciewicz, W.; Regnier, F. Nonideal size-exclusion chromatography of proteins: Effects of pH at low ionic strength. Anal. Biochem. 1982, 126, 8–16. [Google Scholar] [CrossRef]
- Kamberi, M.; Chung, P.; DeVas, R.; Li, L.; Li, Z.; Ma, X.; Fields, S.; Riley, C.M. Analysis of non-covalent aggregation of synthetic hPTH (1–34) by size-exclusion chromatography and the importance of suppression of non-specific interactions for a precise quantitation. J. Chromatogr. B 2004, 810, 151–155. [Google Scholar] [CrossRef]
- Roumeliotis, P.; Unger, K. Assessment and optimization of system parameters in size exclusion separation of proteins on diol-modified silica columns. J. Chromatogr. A 1981, 218, 535–546. [Google Scholar] [CrossRef]
- Fekete, S.; Beck, A.; Veuthey, J.-L.; Guillarme, D. Theory and practice of size exclusion chromatography for the analysis of protein aggregates. J. Pharm. Biomed. Anal. 2014, 101, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.J.; Peterson, E.C.; Henry, R.; Hoffman, N.E. The L18 domain of light-harvesting chlorophyll proteins binds to chloroplast signal recognition particle. J. Biol. Chem. 2000, 275, 13187–13190. [Google Scholar] [CrossRef] [PubMed]
- Groves, M.R.; Mant, A.; Kuhn, A.; Koch, J.; Dübel, S.; Robinson, C.; Sinning, I. Functional characterization of recombinant chloroplast signal recognition particle. J. Biol. Chem. 2001, 276, 27778–27786. [Google Scholar] [CrossRef] [PubMed]
- Attri, A.K.; Minton, A.P. New methods for measuring macromolecular interactions in solution via static light scattering: Basic methodology and application to non-associating and self-associating proteins. Anal. Biochem. 2005, 337, 103–110. [Google Scholar] [CrossRef] [PubMed]
- George, A.; Wilson, W.W. Predicting protein crystallization from a dilute solution property. Acta Crystallogr. D 1994, 50, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.W. Monitoring crystallization experiments using dynamic light scattering: Assaying and monitoring protein crystallization in solution. Methods 1990, 1, 110–117. [Google Scholar] [CrossRef]
- Neal, B.L.; Asthagiri, D.; Velev, O.D.; Lenhoff, A.M.; Kaler, E.W. Why is the osmotic second virial coefficient related to protein crystallization? J. Cryst. Growth 1999, 196, 377–387. [Google Scholar] [CrossRef]
- Wilson, W.W. Light scattering as a diagnostic for protein crystal growth—A practical approach. J. Struct. Biol. 2003, 142, 56–65. [Google Scholar] [CrossRef]
- Sleutel, M.; Lutsko, J.; Van Driessche, A.E.S.; Durán-Olivencia, M.A.; Maes, D. Observing classical nucleation theory at work by monitoring phase transitions with molecular precision. Nat. Commun. 2014, 5, 5598. [Google Scholar] [CrossRef] [PubMed]
- Nettleship, J.E.; Brown, J.; Groves, M.R.; Geerlof, A. Methods for protein characterization by mass spectrometry, thermal shift (ThermoFluor) assay, and multiangle or static light scattering. In Structural Proteomics: High-throughput Methods; Springer: Berlin, Germany, 2008; Volume 426, pp. 299–318. [Google Scholar]
- Ye, H. Simultaneous determination of protein aggregation, degradation, and absolute molecular weight by size exclusion chromatography–multiangle laser light scattering. Anal. Biochem. 2006, 356, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Mogridge, J. Using light scattering to determine the stoichiometry of protein complexes. In Protein-Protein Interactions: Methods and Applications; Springer: Berlin, Germany, 2004; Volume 1278, pp. 113–118. [Google Scholar]
- Sahin, E.; Roberts, C.J. Size-exclusion chromatography with multi-angle light scattering for elucidating protein aggregation mechanisms. In Therapeutic Proteins: Methods and Protocols; Springer: Berlin, Germany, 2012; pp. 403–423. [Google Scholar]
- Ahrer, K.; Buchacher, A.; Iberer, G.; Josic, D.; Jungbauer, A. Analysis of aggregates of human immunoglobulin G using size-exclusion chromatography, static and dynamic light scattering. J. Chromatogr. A 2003, 1009, 89–96. [Google Scholar] [CrossRef]
- Mathew, E.; Mirza, A.; Menhart, N. Liquid-chromatography-coupled SAXS for accurate sizing of aggregating proteins. J. Synchr. Rad. 2004, 11, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Inoko, Y. Size-exclusion chromatography combined with small-angle X-ray scattering optics. J. Chromatogr. A 2009, 1216, 7461–7465. [Google Scholar] [CrossRef] [PubMed]
- David, G.; Pérez, J. Combined sampler robot and high-performance liquid chromatography: A fully automated system for biological small-angle X-ray scattering experiments at the Synchrotron SOLEIL SWING beamline. J. Appl. Crystallogr. 2009, 42, 892–900. [Google Scholar] [CrossRef]
- Graewert, M.A.; Franke, D.; Jeffries, C.M.; Blanchet, C.E.; Ruskule, D.; Kuhle, K.; Flieger, A.; Schäfer, B.; Tartsch, B.; Meijers, R.; et al. Automated pipeline for purification, biophysical and X-ray analysis of biomacromolecular solutions. Sci. Rep. 2015, 5, 10734. [Google Scholar] [CrossRef] [PubMed]
- Hutin, S.; Brennich, M.; Maillot, B.; Round, A. Online ion-exchange chromatography for small-angle X-ray scattering. Acta Crystallogr. D 2016, 72, 1090–1099. [Google Scholar] [CrossRef] [PubMed]
- Stradner, A.; Sedgwick, H.; Cardinaux, F.; Poon, W.C.K.; Egelhaaf, S.U.; Schurtenberger, P. Equilibrium cluster formation in concentrated protein solutions and colloids. Nature 2004, 432, 492–495. [Google Scholar] [CrossRef] [PubMed]
- Leberman, R. The Hofmeister series and ionic strength. FEBS Lett. 1991, 284, 293–294. [Google Scholar] [CrossRef]
- Arakawa, T.; Timasheff, S.N. Mechanism of protein salting in and salting out by divalent cation salts: Balance between hydration and salt binding. Biochemistry 1984, 23, 5912–5923. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.; Walker, J. Principles and Techniques of Biochemistry and Molecular Biology; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Ozbek, H.; JoAo, F.; Phillips, S.L. Viscosity of Aqueous Sodium Chloride Solutions from 0–150 °C. No. LBL-5931; Ernest Orlando Lawrence Berkeley National Laboratory: Berkeley, CA, USA, 1977.
- Dumetz, A.C.; Snellinger-O’Brien, A.M.; Kaler, E.W.; Lenhoff, A.M. Patterns of protein–protein interactions in salt solutions and implications for protein crystallization. Protein Sci. 2007, 16, 1867–1877. [Google Scholar] [CrossRef] [PubMed]
- Ewing, F.; Forsythe, E.; Pusey, M. Orthorhombic lysozyme solubility. Acta Crystallogr. D 1994, 50, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Ataka, M.; Asai, M. Systematic studies on the crystallization of lysozyme: Determination and use of phase diagrams. J. Cryst. Growth 1988, 90, 86–93. [Google Scholar] [CrossRef]
- Ataka, M.; Tanaka, S. The growth of large single crystals of lysozyme. Biopolymers 1986, 25, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Adawy, A.; Corbeek, W.; de Ronde, E.; van Enckevort, W.J.; de Grip, W.J.; Vlieg, E. A practical kit for micro-scale application of the ceiling crystallisation method. CrystEngComm 2015, 17, 2602–2605. [Google Scholar] [CrossRef]
- Asherie, N. Protein crystallization and phase diagrams. Methods 2004, 34, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.B.; Twigg, P.J.; Baird, J.K.; Meehan, E.J. The solubility of hen egg-white lysozyme. J. Cryst. Growth 1988, 90, 94–104. [Google Scholar] [CrossRef]
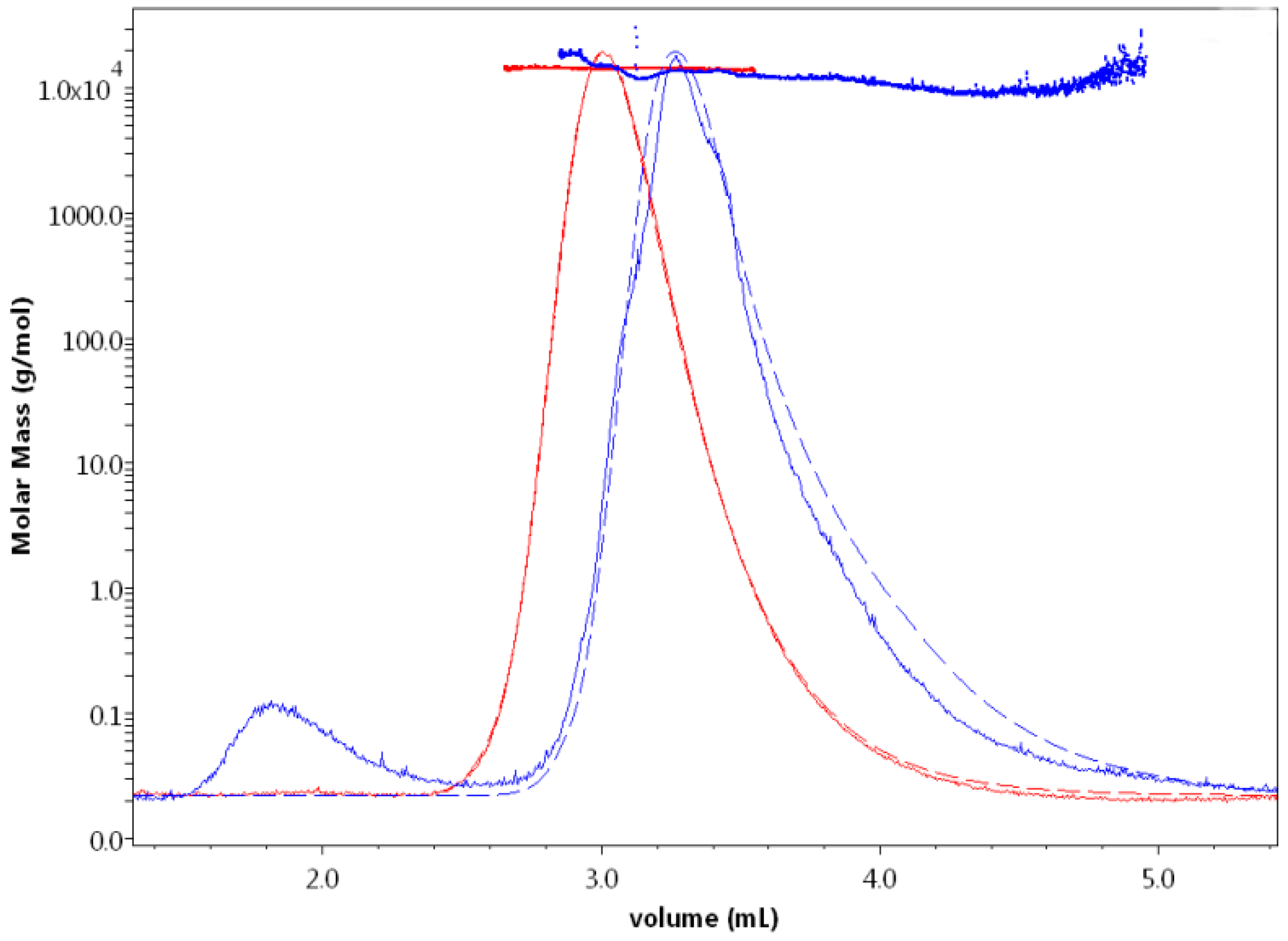
| (NH4)2SO4 (M) | Retention Volume (mL) | Measured Molar Mass (kDa) | Polydispersity |
|---|---|---|---|
| 0.0 | 2.73 | 28.688 (±0.846) | 1.002 (0.042) |
| 0.5 | 2.55 | 37.571 (±6.305) | 1.042 (0.248) |
| 1.0 | 2.91 | 25.673 (±2.765) | 1.033 (0.143) |
| 1.5 | 2.73 | 85.202 (±7.919) | 1.061 (0.177) |
| 2.96 | 26.516 (±7.972) | 1.014 (0.387) | |
| 2.0 | 1.21 | 1232.341 (±88.439) | 1.019 (0.099) |
| 1.46 | 1119.304 (±28.018) | 1.018 (0.035) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adawy, A.; Groves, M.R. The Use of Size Exclusion Chromatography to Monitor Protein Self-Assembly. Crystals 2017, 7, 331. https://doi.org/10.3390/cryst7110331
Adawy A, Groves MR. The Use of Size Exclusion Chromatography to Monitor Protein Self-Assembly. Crystals. 2017; 7(11):331. https://doi.org/10.3390/cryst7110331
Chicago/Turabian StyleAdawy, Alaa, and Matthew R. Groves. 2017. "The Use of Size Exclusion Chromatography to Monitor Protein Self-Assembly" Crystals 7, no. 11: 331. https://doi.org/10.3390/cryst7110331
APA StyleAdawy, A., & Groves, M. R. (2017). The Use of Size Exclusion Chromatography to Monitor Protein Self-Assembly. Crystals, 7(11), 331. https://doi.org/10.3390/cryst7110331