Crystal Structure of Chaperonin GroEL from Xanthomonas oryzae pv. oryzae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Gene Cloning and Protein Expression and Purification
2.3. Crystallization and X-ray Data Collection
2.4. Structure Determination
3. Results
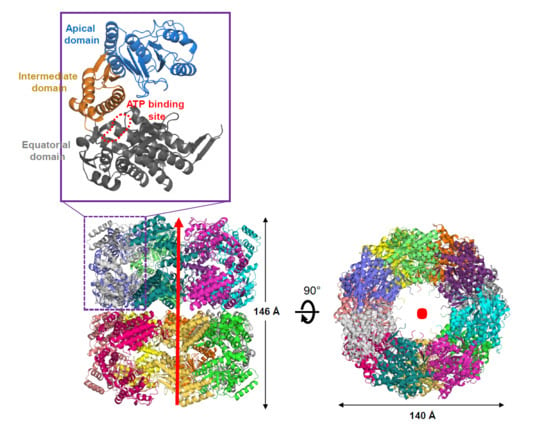
3.1. Molecular Packing of XoGroEL in Crystal
3.2. Overall Structure of XoGroEL
3.3. Conformation of XoGroEL Protomer
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Available online: http://faostat.fao.org (accessed on 1 August 2019).
- Khush, G.S. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 2005, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Verdier, V.; Vera Cruz, C.; Leach, J.E. Controlling rice bacterial blight in Africa: needs and prospects. J. Biotechnol. 2012, 159, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Alfano, J.R.; Collmer, A. Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 2004, 42, 385–414. [Google Scholar] [CrossRef] [PubMed]
- Nans, A.; Kudryashev, M.; Saibil, H.R.; Hayward, R.D. Structure of a bacterial type III secretion system in contact with a host membrane in situ. Nat. Commun. 2015, 6, 10114. [Google Scholar] [CrossRef] [PubMed]
- Portaliou, A.G.; Tsolis, K.C.; Loos, M.S.; Zorzini, V.; Economou, A. Type III secretion: Building and operating a remarkable nanomachine. Trends Biochem. Sci. 2016, 41, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Parsot, C.; Hamiaux, C.; Page, A.L. The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 2003, 6, 7–14. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Skjaerven, L.; Cuellar, J.; Martinez, A.; Valpuesta, J.M. Dynamics, flexibility, and allostery in molecular chaperonins. FEBS Lett. 2015, 589, 2522–2532. [Google Scholar] [CrossRef]
- Tran, H.T.; Pham, T.V.; Ngo, H.P.; Hong, M.K.; Kim, J.G.; Lee, S.H.; Ahn, Y.J.; Kang, L.W. Crystallization and preliminary X-ray crystallographic analysis of the XoGroEL chaperonin from Xanthomonas oryzae pv. oryzae. Acta Crystallogr. F Struct. Biol. Commun. 2014, 70, 604–607. [Google Scholar] [CrossRef]
- Park, S.Y.; Ha, S.C.; Kim, Y.G. The protein crystallography beamlines at the pohang light source II. Biodesign 2017, 5, 30–34. [Google Scholar]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997, 277, 307–326. [Google Scholar]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Fei, X.; Ye, X.; LaRonde, N.A.; Lorimer, G.H. Formation and structures of GroEL: GroES2 chaperonin footballs, the protein-folding functional form. Proc. Natl. Acad. Sci. USA 2014, 111, 12775–12780. [Google Scholar] [CrossRef] [PubMed]
- Moodie, E.E.; Richardson, T.S.; Stephens, D.A. Demystifying optimal dynamic treatment regimes. Biometrics 2007, 63, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Mansell, W. An integrative formulation-based cognitive treatment of bipolar disorders: Application and illustration. J. Clin. Psychol. 2007, 63, 447–461. [Google Scholar] [CrossRef] [PubMed]
- DeFrank, J.J.; Cheng, T.C. Purification and properties of an organophosphorus acid anhydrase from a halophilic bacterial isolate. J. Bacteriol. 1991, 173, 1938–1943. [Google Scholar] [CrossRef] [Green Version]
- Steegborn, C.; Messerschmidt, A.; Laber, B.; Streber, W.; Huber, R.; Clausen, T. The crystal structure of cystathionine gamma-synthase from nicotiana tabacum reveals its substrate and reaction specificity. J. Mol. Biol. 1999, 290, 983–996. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System; Version 1.3r1; Schrodinger: New York, NY, USA, 2010. [Google Scholar]
- Clare, D.K.; Vasishtan, D.; Stagg, S.; Quispe, J.; Farr, G.W.; Topf, M.; Horwich, A.L.; Saibil, H.R. ATP-triggered conformational changes delineate substrate-binding and -folding mechanics of the GroEL chaperonin. Cell 2012, 149, 113–123. [Google Scholar] [CrossRef]
- Bartolucci, C.; Lamba, D.; Grazulis, S.; Manakova, E.; Heumann, H. Crystal structure of wild-type chaperonin GroEL. J. Mol. Biol. 2005, 354, 940–951. [Google Scholar] [CrossRef]
- Koike-Takeshita, A.; Arakawa, T.; Taguchi, H.; Shimamura, T. Crystal structure of a symmetric football-shaped GroEL: GroES2-ATP14 complex determined at 3.8A reveals rearrangement between two GroEL rings. J. Mol. Biol. 2014, 426, 3634–3641. [Google Scholar] [CrossRef]
- Govada, L.; Chayen, N.E. Choosing the method of crystallization to obtain optimal results. Crystals 2019, 9, 106. [Google Scholar] [CrossRef]
- Blackburn, A.; Partowmah, S.H.; Brennan, H.M.; Mestizo, K.E.; Stivala, C.D.; Petreczky, J.; Perez, A.; Horn, A.; McSweeney, S.; Soares, A.S. A simple technique to improve microcrystals using gel exclusion of nucleation inducing elements. Crystals 2018, 8, 464. [Google Scholar] [CrossRef]
- Kim, S.; Cho, Y.J.; Song, E.S.; Lee, S.H.; Kim, J.G.; Kang, L.W. Time-resolved pathogenic gene expression analysis of the plant pathogen Xanthomonas oryzae pv. oryzae. BMC Genom. 2016, 17, 345. [Google Scholar] [CrossRef] [PubMed]



| Data | XoGroEL (PDB ID: 6KFV) |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.97949 |
| Resolution range (Å) | 47.2–3.2 (3.3–3.2) * |
| Space group | P212121 |
| Unit cell (Å) | 137.1 239.5 278.3 90 90 90 |
| Total reflections | 683,467 |
| Unique reflections | 136,180 (10475) |
| Multiplicity | 4.1 (3.0) |
| Completeness (%) | 91.7 (71.3) |
| Mean I/sigma (I) | 8.5 (2.1) |
| Wilson B-factor | 68.9 |
| R-merge | 18.9 (59.0) |
| Refinement | |
| Reflections used in refinement | 136,115 (10,470) |
| Reflections used for R-free | 6839 (542) |
| R-work | 0.21 (0.31) |
| R-free | 0.29 (0.37) |
| Number of non-hydrogen atoms | |
| In macromolecules | 53,872 |
| In ligands | 28 |
| In solvent | 33 |
| Protein residues | 7342 |
| RMS (bonds) | 0.015 |
| RMS (angles) | 1.87 |
| Ramachandran favored (%) | 82.8 |
| Ramachandran allowed (%) | 13.2 |
| Ramachandran outliers (%) | 4.0 |
| Rotamer outliers (%) | 16.5 |
| Average B-factor | |
| Macromolecules | 82.3 |
| Ligands | 99.3 |
| Solvent | 36.9 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, H.-T.; Lee, J.; Park, H.; Kim, J.-G.; Kim, S.; Ahn, Y.-J.; Kang, L.-W. Crystal Structure of Chaperonin GroEL from Xanthomonas oryzae pv. oryzae. Crystals 2019, 9, 399. https://doi.org/10.3390/cryst9080399
Tran H-T, Lee J, Park H, Kim J-G, Kim S, Ahn Y-J, Kang L-W. Crystal Structure of Chaperonin GroEL from Xanthomonas oryzae pv. oryzae. Crystals. 2019; 9(8):399. https://doi.org/10.3390/cryst9080399
Chicago/Turabian StyleTran, Huyen-Thi, Jongha Lee, Hyunjae Park, Jeong-Gu Kim, Seunghwan Kim, Yeh-Jin Ahn, and Lin-Woo Kang. 2019. "Crystal Structure of Chaperonin GroEL from Xanthomonas oryzae pv. oryzae" Crystals 9, no. 8: 399. https://doi.org/10.3390/cryst9080399
APA StyleTran, H. -T., Lee, J., Park, H., Kim, J. -G., Kim, S., Ahn, Y. -J., & Kang, L. -W. (2019). Crystal Structure of Chaperonin GroEL from Xanthomonas oryzae pv. oryzae. Crystals, 9(8), 399. https://doi.org/10.3390/cryst9080399