Hydrogen-Deuterium Exchange Profiles of Polyubiquitin Fibrils
Abstract
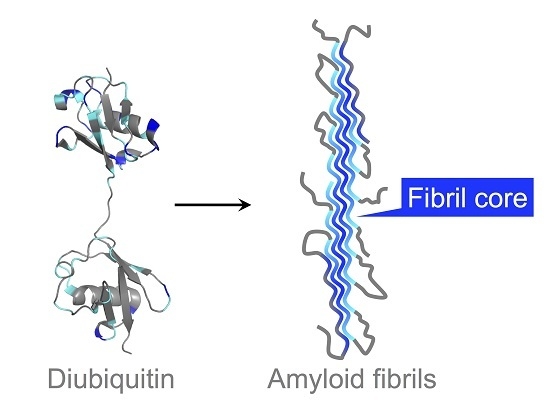
:1. Introduction
2. Materials and Methods
2.1. Protein Sample Preparation
2.2. Hydrogen-Deuterium Exchange Experiments
2.3. NMR Spectroscopy
2.4. Real-Time NMR Analysis
3. Results
3.1. Backbone Assignment of Unfolded Ubiquitin in DMSO
3.2. Solvent-Protected Core of M1-Linked Diubiquitin Fibrils
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Mori, H.; Kondo, J.; Ihara, Y. Ubiquitin is a component of paired helical filaments in Alzheimer’s disease. Science 1987, 235, 1641–1644. [Google Scholar] [CrossRef] [PubMed]
- Gallo, J.M.; Anderton, B.H. Brain diseases. Ubiquitous variations in nerves. Nature 1989, 337, 687–688. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, D.; Walinda, E.; Fukada, H.; Sou, Y.S.; Kageyama, S.; Hoshino, M.; Fujii, T.; Tsuchiya, H.; Saeki, Y.; Arita, K.; et al. The unexpected role of polyubiquitin chains in the formation of fibrillar aggregates. Nat. Commun. 2015, 6, 6116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kageyama, S.; Sou, Y.S.; Uemura, T.; Kametaka, S.; Saito, T.; Ishimura, R.; Kouno, T.; Bedford, L.; Mayer, R.J.; Lee, M.S.; et al. Proteasome dysfunction activates autophagy and the Keap1-Nrf2 pathway. J. Biol. Chem. 2014, 289, 24944–24955. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, D.; Shirakawa, M. The evolving world of ubiquitin: Transformed polyubiquitin chains. Biomol. Concepts 2016, 7, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Kheterpal, I.; Zhou, S.; Cook, K.D.; Wetzel, R. Aβ amyloid fibrils possess a core structure highly resistant to hydrogen exchange. Proc. Natl. Acad. Sci. USA 2000, 97, 13597–13601. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, D.; Walinda, E.; Fukada, H.; Sugase, K.; Shirakawa, M. Ubiquitylation directly induces fold destabilization of proteins. Sci. Rep. 2016, 6, 39453. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, D.; Isogai, S.; Tenno, T.; Tochio, H.; Shirakawa, M.; Ariyoshi, M. Purification, crystallization and preliminary crystallographic studies of Lys48-linked polyubiquitin chains. Acta. Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 834–837. [Google Scholar] [CrossRef] [PubMed]
- Hwang, T.L.; Shaka, A.J. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. Ser. A 1995, 112, 275–279. [Google Scholar] [CrossRef]
- Wittekind, M.; Mueller, L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J. Magn. Reson. Ser. B 1993, 101, 201–205. [Google Scholar] [CrossRef]
- Grzesiek, S.; Bax, A. Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J. Biomol. NMR 1993, 3, 185–204. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, Y.; Kulminskaya, N.V.; Mulder, F.A. Easy and unambiguous sequential assignments of intrinsically disordered proteins by correlating the backbone 15N or 13C’ chemical shifts of multiple contiguous residues in highly resolved 3D spectra. J. Biomol. NMR 2015, 61, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Markley, J.L.; Bax, A.; Arata, Y.; Hilbers, C.W.; Kaptein, R.; Sykes, B.D.; Wright, P.E.; Wüthrich, K. Recommendations for the presentation of NMR structures of proteins and nucleic acids—(IUPAC Recommendations 1998). Pure. Appl. Chem. 1998, 70, 117–142. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Sugase, K.; Konuma, T.; Lansing, J.C.; Wright, P.E. Fast and accurate fitting of relaxation dispersion data using the flexible software package GLOVE. J. Biomol. NMR 2013, 56, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, M.; Katou, H.; Hagihara, Y.; Hasegawa, K.; Naiki, H.; Goto, Y. Mapping the core of the β2-microglobulin amyloid fibril by H/D exchange. Nat. Struct. Biol. 2002, 9, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Reyes-Turcu, F.; Licchesi, J.D.; Odenwaelder, P.; Wilkinson, K.D.; Barford, D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO. Rep. 2009, 10, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, D.; Walinda, E.; Iwakawa, N.; Nishizawa, M.; Kawata, Y.; Yamamoto, A.; Shirakawa, M.; Scheler, U.; Sugase, K. High-sensitivity Rheo-NMR spectroscopy for protein studies. Anal. Chem. 2017, 89, 7286–7290. [Google Scholar] [CrossRef] [PubMed]
- Isogai, S.; Morimoto, D.; Arita, K.; Unzai, S.; Tenno, T.; Hasegawa, J.; Sou, Y.S.; Komatsu, M.; Tanaka, K.; Shirakawa, M.; et al. Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J. Biol. Chem. 2011, 286, 31864–31874. [Google Scholar] [CrossRef] [PubMed]
- Walinda, E.; Morimoto, D.; Sugase, K.; Konuma, T.; Tochio, H.; Shirakawa, M. Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin. J. Biol. Chem. 2014, 289, 13890–13902. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Amyloid formation by globular proteins under native conditions. Nat. Chem. Biol. 2009, 5, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Elam, J.S.; Taylor, A.B.; Strange, R.; Antonyuk, S.; Doucette, P.A.; Rodriguez, J.A.; Hasnain, S.S.; Hayward, L.J.; Valentine, J.S.; Yeates, T.O.; et al. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat. Struct. Biol. 2003, 10, 461–467. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morimoto, D.; Nishizawa, R.; Walinda, E.; Takashima, S.; Sugase, K.; Shirakawa, M. Hydrogen-Deuterium Exchange Profiles of Polyubiquitin Fibrils. Polymers 2018, 10, 240. https://doi.org/10.3390/polym10030240
Morimoto D, Nishizawa R, Walinda E, Takashima S, Sugase K, Shirakawa M. Hydrogen-Deuterium Exchange Profiles of Polyubiquitin Fibrils. Polymers. 2018; 10(3):240. https://doi.org/10.3390/polym10030240
Chicago/Turabian StyleMorimoto, Daichi, Ryo Nishizawa, Erik Walinda, Shingo Takashima, Kenji Sugase, and Masahiro Shirakawa. 2018. "Hydrogen-Deuterium Exchange Profiles of Polyubiquitin Fibrils" Polymers 10, no. 3: 240. https://doi.org/10.3390/polym10030240
APA StyleMorimoto, D., Nishizawa, R., Walinda, E., Takashima, S., Sugase, K., & Shirakawa, M. (2018). Hydrogen-Deuterium Exchange Profiles of Polyubiquitin Fibrils. Polymers, 10(3), 240. https://doi.org/10.3390/polym10030240