Molecular Mechanism of Xylogenesis in Moso Bamboo (Phyllostachys edulis) Shoots during Cold Storage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material, Treatment, and Storage
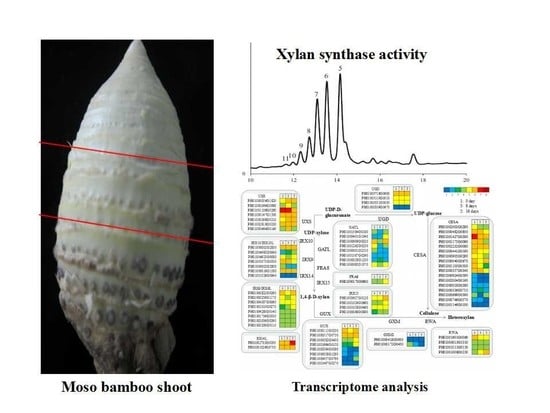
2.2. Sectioning of Stems
2.3. Measurements of Cellulose, Hemicellulose, and Lignin Contents
2.4. PAL, CAD, and POD Activity
2.5. Assay of Xylan XylT Activity
2.5.1. Preparation of Microsomal Membranes
2.5.2. Anthranilic Acid Labeling of Xyl5
2.5.3. Assay of Xylan XylT Activity
2.6. RNA Extraction, Library Construction, and RNA-Seq Analysis
2.7. Transcriptome Data Analysis
2.8. Validation of RNA-Seq Data by Quantitative Real-Time RT-PCR (qRT-PCR)
2.9. Statistical Analysis
3. Results
3.1. Changes in Cellulose, Hemicellulose, and Lignin Content of MBSes during Cold Storage
3.2. The Enzymatic Activities of PAL, CAD, and POD in MBSes during Cold Storage
3.3. A Comparison of the Xylan XylT Activity in MBSes during Cold Storage
3.4. Transcriptomic Analysis in MBSes during Cold Storage
3.5. Differentially Expressed Genes in MBSes during Cold Storage
3.6. Gene Ontology Annotation
3.7. Functional Genes Involved in Cellulose and Xylan Biosynthesis in MBSes during Cold Storage
3.8. Functional Genes Involved in Lignin Biosynthesis in MBSes
3.9. Functional Genes Involved in Candidate Transcription Factors in MBSes
3.10. Expressional Analysis of Functional Genes Involved in Cell Wall Biosynthesis
4. Discussion
4.1. The Effects of Cold Storage on Cell Wall Composition in Post-Harvest MBSes
4.2. Effects of Cold Storage on Molecular Mechanism of xylogenesis in MBSes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yeasmin, L.; Ali, M.N.; Gantait, S.; Chakraborty, S. Bamboo: An overview on its genetic diversity and characterization. 3 Biotech 2015, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; He, X.; Hu, Y.; Kuang, T.; Ceulemans, R. Lignification and lignin heterogeneity for various age classes of bamboo (phyllostachys pubescens) stems. Physiol. Plant. 2002, 114, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Xu, X.; Yan, B. Accumulation of lignin and involvement of enzymes in bamboo shoot during storage. Eur. Food Res. Technol. 2008, 226, 635–640. [Google Scholar] [CrossRef]
- Whetten, R.; Sederoff, R. Lignin biosynthesis. Plant Cell 1995, 7, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Imberty, A.; Goldberg, R.; Catesson, A.M. Isolation and characterization of populus isoperoxidases involved in the last step of lignin formation. Planta 1985, 164, 221–226. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Plant Biol. 2003, 54, 519–1146. [Google Scholar] [CrossRef]
- Donaldson, L.A. Lignification and lignin topochemistry—An ultrastructural view. Phytochemistry 2001, 57, 859–873. [Google Scholar] [CrossRef]
- Mellerowicz, E.J.; Sundberg, B. Wood cell walls: Biosynthesis, developmental dynamics and their implications for wood properties. Curr. Opin. Plant Biol. 2008, 11, 293–300. [Google Scholar] [CrossRef]
- Brown, D.M.; Zeef, L.A.; Ellis, J.; Goodacre, R.; Turner, S.R. Identification of novel genes in arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 2005, 17, 2281–2295. [Google Scholar] [CrossRef]
- Brown, D.M.; Zhang, Z.; Stephens, E.; Dupree, P.; Turner, S.R. Characterization of irx10 and irx10-like reveals an essential role in glucuronoxylan biosynthesis in arabidopsis. Plant J. Cell Mol. Biol. 2009, 57, 732–746. [Google Scholar] [CrossRef]
- Jensen, J.K.; Kim, H.; Cocuron, J.C.; Orler, R.; Ralph, J.; Wilkerson, C.G. The duf579 domain containing proteins irx15 and irx15-l affect xylan synthesis in arabidopsis. Plant J. Cell Mol. Biol. 2011, 66, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.M.; Rihouey, C.; Seveno, M.; Hornblad, E.; Singh, S.K.; Matsunaga, T.; Ishii, T.; Lerouge, P.; Marchant, A. The arabidopsis irx10 and irx10-like glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J. Cell Mol. Biol. 2009, 57, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Pena, M.J.; Zhong, R.; Zhou, G.K.; Richardson, E.A.; O’Neill, M.A.; Darvill, A.G.; York, W.S.; Ye, Z.H. Arabidopsis irregular xylem8 and irregular xylem9: Implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 2007, 19, 549–563. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Zhong, R.; Richardson, E.A.; Himmelsbach, D.S.; McPhail, B.T.; Ye, Z.H. The parvus gene is expressed in cells undergoing secondary wall thickening and is essential for glucuronoxylan biosynthesis. Plant Cell Physiol. 2007, 48, 1659–1672. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.; Wightman, R.; Zhang, Z.; Gomez, L.D.; Atanassov, I.; Bukowski, J.P.; Tryfona, T.; McQueen-Mason, S.J.; Dupree, P.; Turner, S. Arabidopsis genes irregular xylem (irx15) and irx15l encode duf579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant J. Cell Mol. Biol. 2011, 66, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.R.; Somerville, C.R. Collapsed xylem phenotype of arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 1997, 9, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Kim, W.C.; Kim, J.Y.; Ahn, S.J.; Han, K.H. Myb46-mediated transcriptional regulation of secondary wall biosynthesis. Mol. Plant 2012, 5, 961–963. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Demura, T.; Ye, Z.H. Snd1, a nac domain transcription factor, is a key regulator of secondary wall synthesis in fibers of arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Zhang, C.; Zhang, Y.; Hu, T.; Mu, S.; Li, X.; Gao, J. Transcriptome sequencing and analysis of the fast growing shoots of moso bamboo (phyllostachys edulis). PLoS ONE 2013, 8, e78944. [Google Scholar] [CrossRef]
- He, C.; Cui, K.; Zhang, J.; Duan, A.; Zeng, Y. Next-generation sequencing-based mrna and microrna expression profiling analysis revealed pathways involved in the rapid growth of developing culms in moso bamboo. BMC Plant Biol. 2013, 13, 1471–2229. [Google Scholar] [CrossRef]
- Gamuyao, R.; Nagai, K.; Ayano, M.; Mori, Y.; Minami, A.; Kojima, M.; Suzuki, T.; Sakakibara, H.; Higashiyama, T.; Ashikari, M.; et al. Hormone distribution and transcriptome profiles in bamboo shoots provide insights on bamboo stem emergence and growth. Plant Cell Physiol. 2017, 58, 702–716. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Zhou, M.; Yu, Z.; Zhang, J.; Ruan, R.; Wan, Y.; Liu, Y. Fractionation and characterization of hemicelluloses from young bamboo (phyllostachys pubescens mazel) leaves. Carbohydr. Polym. 2013, 95, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ying, Y.Q.; Wang, J.; Zhao, X.H.; Zeng, W.; Beahan, C.; He, J.B.; Chen, X.Y.; Bacic, A.; Song, L.L.; et al. Transcriptome analysis provides insights into xylogenesis formation in moso bamboo (phyllostachys edulis) shoot. Sci. Rep. 2018, 8, 3951. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Ouyang, K.; Gan, S.; Zeng, W.; Song, L.; Zhao, S.; Li, J.; Doblin, M.S.; Bacic, A.; Chen, X.Y.; et al. Biochemical and molecular changes associated with heteroxylan biosynthesis in neolamarckia cadamba (rubiaceae) during xylogenesis. Front. Plant Sci. 2014, 5, 602. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Joyce, D.C. Aba effects on ethylene production, pal activity, anthocyanin and phenolic contents of strawberry fruit. Plant Growth Regul. 2003, 39, 171–174. [Google Scholar] [CrossRef]
- Lurie, S.; Fallik, E.; Handros, A.; Shapira, R. The possible involvement of peroxidase in resistance to botrytis cinerea in heat treated tomato fruit. Physiol. Mol. Plant Pathol. 1997, 50, 141–149. [Google Scholar] [CrossRef]
- Zeng, W.; Chatterjee, M.; Faik, A. Udp-xylose-stimulated glucuronyltransferase activity in wheat microsomal membranes: Characterization and role in glucurono(arabino)xylan biosynthesis. Plant Physiol. 2008, 147, 78–91. [Google Scholar] [CrossRef]
- Alwael, H.; Connolly, D.; Paull, B. Liquid chromatographic profiling of monosaccharide concentrations in complex cell-culture media and fermentation broths. Anal. Methods 2010, 3, 1–228. [Google Scholar] [CrossRef]
- Zamboni, A.; Minoia, L.; Ferrarini, A.; Tornielli, G.B.; Zago, E.; Delledonne, M.; Pezzotti, M. Molecular analysis of post-harvest withering in grape by aflp transcriptional profiling. J. Exp. Bot. 2008, 59, 4145–4159. [Google Scholar] [CrossRef]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef]
- Storey, J.D. A direct approach to false discovery rates. J. R. Statist. Soc. B 2002, 64, 479–498. [Google Scholar] [CrossRef] [Green Version]
- Fan, C.; Ma, J.; Guo, Q.; Li, X.; Wang, H.; Lu, M. Selection of reference genes for quantitative real-time pcr in bamboo (phyllostachys edulis). PLoS ONE 2013, 8, e56573. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Ye, Z.H. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 2010, 15, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Ye, Z.H. Transcriptional regulation of lignin biosynthesis. Plant Signal. Behav. 2009, 4, 1028–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awano, T.; Takabe, K.; Fujita, M. Xylan deposition on secondary wall of fagus crenata fiber. Protoplasma 2002, 219, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Zeng, W.; Wu, A.; Picard, K.; Lampugnani, E.R.; Cheetamun, R.; Beahan, C.; Cassin, A.; Lonsdale, A.; Doblin, M.S.; et al. Asparagus spears as a model to study heteroxylan biosynthesis during secondary wall development. PLoS ONE 2015, 10, e0123878. [Google Scholar] [CrossRef]
- Boudet, A. Lignins and lignification: Selected issues. Plant Physiol. Biochem. 2000, 38, 81–96. [Google Scholar] [CrossRef]
- Huang, H.; Xu, L.; Tong, Z.; Lin, E.; Liu, Q.; Cheng, L.; Zhu, M. De novo characterization of the chinese fir (cunninghamia lanceolata) transcriptome and analysis of candidate genes involved in cellulose and lignin biosynthesis. BMC Genom. 2012, 13, 1471–2164. [Google Scholar] [CrossRef]
- Logacheva, M.D.; Kasianov, A.S.; Vinogradov, D.V.; Samigullin, T.H.; Gelfand, M.S.; Makeev, V.J.; Penin, A.A. De novo sequencing and characterization of floral transcriptome in two species of buckwheat (fagopyrum). BMC Genom. 2011, 12, 1471–2164. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Tong, T.T.; Shao, X.J.; Song, L.L.; Wu, A.M. Functions of PeIRX10 gene from Phyllostachys edulis in xylan synthesis. Scientia Silvae Sinicae 2016, 52, 79–87. [Google Scholar]
- Zhou, J.; Lee, C.; Zhong, R.; Ye, Z.H. Myb58 and myb63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. The nac transcription factors nst1 and nst2 of arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 2005, 17, 2993–3006. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Iwase, A.; Yamamoto, H.; Yoshida, M.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. Nac transcription factors, nst1 and nst3, are key regulators of the formation of secondary walls in woody tissues of arabidopsis. Plant Cell 2007, 19, 270–280. [Google Scholar] [CrossRef] [PubMed]









| Days | Cellulose (%) | Hemicellulose (%) | Lignin (%) |
|---|---|---|---|
| 0 | 24.10 ± 1.12 | 35.57 ± 0.09 | 11.85 ± 0.42 |
| 4 | 25.52 ± 1.00 | 33.76 ± 0.46 | 11.22 ± 0.26 |
| 8 | 26.24 ± 1.03 | 33.15 ± 1.12 | 13.70 ± 0.81 |
| 12 | 26.67 ± 1.04 | 31.67 ± 0.74 | 14.17 ± 0.34 |
| 16 | 28.90 ± 1.15 | 28.92 ± 0.61 | 15.62 ± 1.01 |
| Sample | ReadSum | BaseSum | GC guanine and cytosine (G+C) content (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|
| Stage 1 | 22,709,174 | 6,812,752,200 | 54.47 | 97.46 | 93.74 |
| Stage 3 | 32,068,870 | 9,620,661,000 | 53.60 | 97.38 | 93.63 |
| Stage 5 | 36,898,380 | 11,069,514,000 | 53.81 | 97.53 | 93.94 |
| Sample | Stage 1 | Stage 3 | Stage 5 | |||
|---|---|---|---|---|---|---|
| Statistical Content | Number | Percentage | Number | Percentage | Number | Percentage |
| Total reads | 41,529,916 | 100.00% | 44,048,144 | 100.00% | 46,984,844 | 100.00% |
| Mapped reads | 36,191,746 | 87.15% | 38,481,918 | 87.36% | 41,134,475 | 87.55% |
| Unique mapped reads | 33,702,857 | 81.15% | 36,257,017 | 82.31% | 38,784,805 | 82.55% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Xuan, L.; He, Y.; Wang, J.; Zhang, H.; Ying, Y.; Wu, A.; Bacic, A.; Zeng, W.; Song, L. Molecular Mechanism of Xylogenesis in Moso Bamboo (Phyllostachys edulis) Shoots during Cold Storage. Polymers 2019, 11, 38. https://doi.org/10.3390/polym11010038
Li C, Xuan L, He Y, Wang J, Zhang H, Ying Y, Wu A, Bacic A, Zeng W, Song L. Molecular Mechanism of Xylogenesis in Moso Bamboo (Phyllostachys edulis) Shoots during Cold Storage. Polymers. 2019; 11(1):38. https://doi.org/10.3390/polym11010038
Chicago/Turabian StyleLi, Changtao, Lingling Xuan, Yuming He, Jie Wang, Hui Zhang, Yeqing Ying, Aimin Wu, Antony Bacic, Wei Zeng, and Lili Song. 2019. "Molecular Mechanism of Xylogenesis in Moso Bamboo (Phyllostachys edulis) Shoots during Cold Storage" Polymers 11, no. 1: 38. https://doi.org/10.3390/polym11010038
APA StyleLi, C., Xuan, L., He, Y., Wang, J., Zhang, H., Ying, Y., Wu, A., Bacic, A., Zeng, W., & Song, L. (2019). Molecular Mechanism of Xylogenesis in Moso Bamboo (Phyllostachys edulis) Shoots during Cold Storage. Polymers, 11(1), 38. https://doi.org/10.3390/polym11010038