Ice-Binding Protein from Shewanella frigidimarinas Inhibits Ice Crystal Growth in Highly Alkaline Solutions
Abstract
:1. Introduction
1.1. Ice-Binding Proteins
1.2. Scope of Work
2. Materials and Methods
2.1. Materials
2.2. Experimental Methods
2.2.1. CD Spectroscopy
2.2.2. SEC-UV
2.2.3. SDS-PAGE
2.2.4. IRI Activity
2.2.5. Statistical Analyses
3. Results
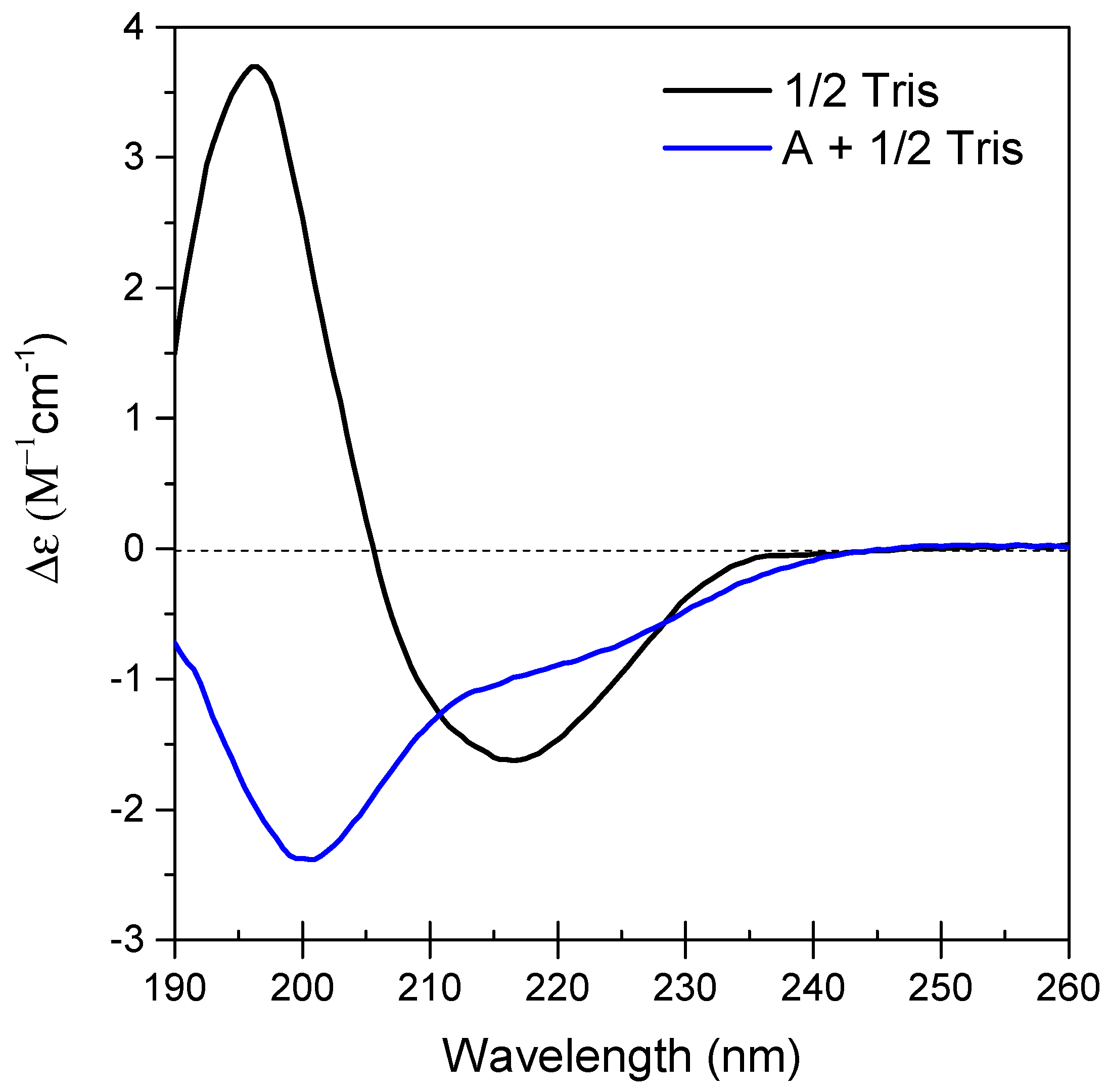
3.1. CD Spectroscopy
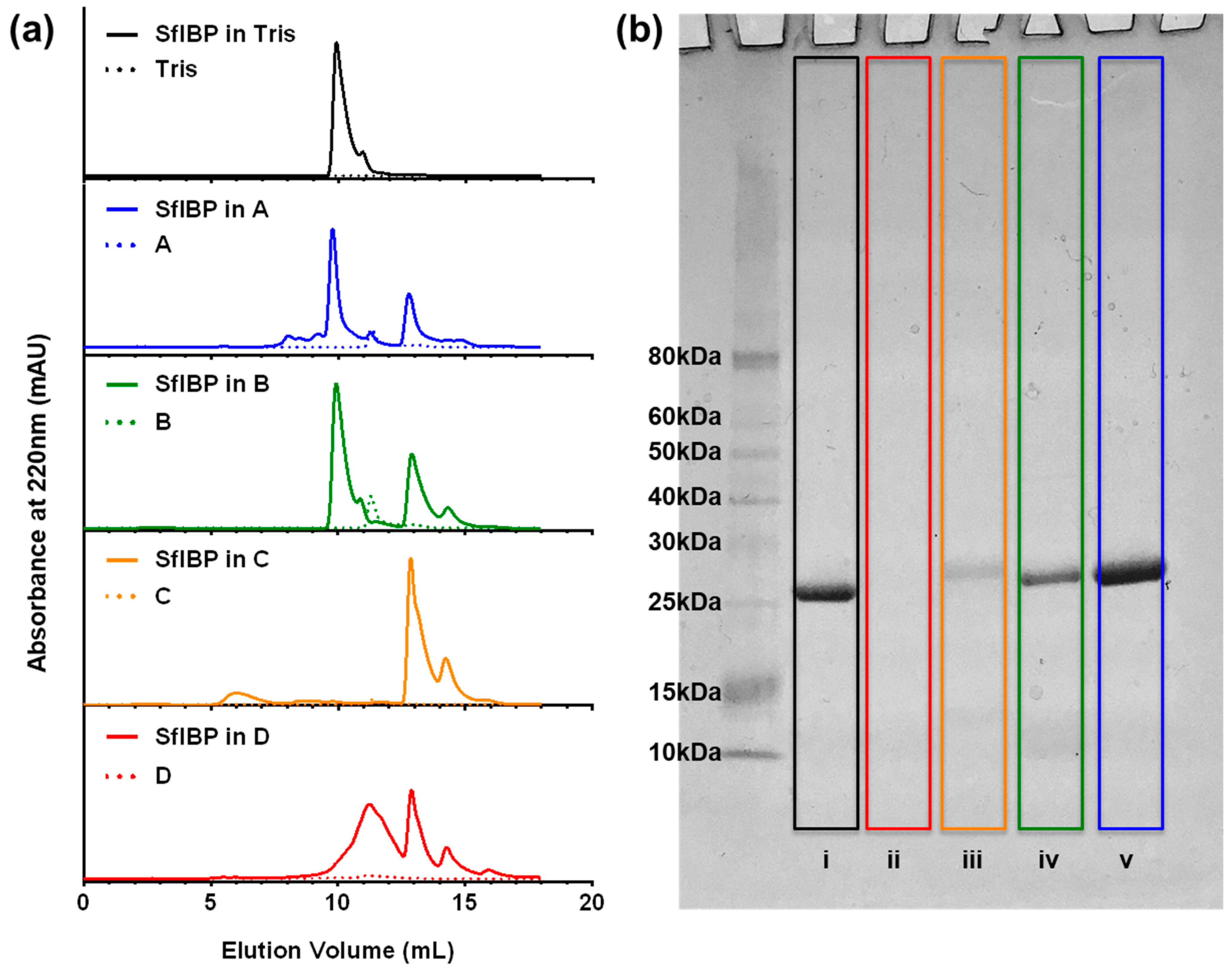
3.2. SEC-UV and SDS-PAGE
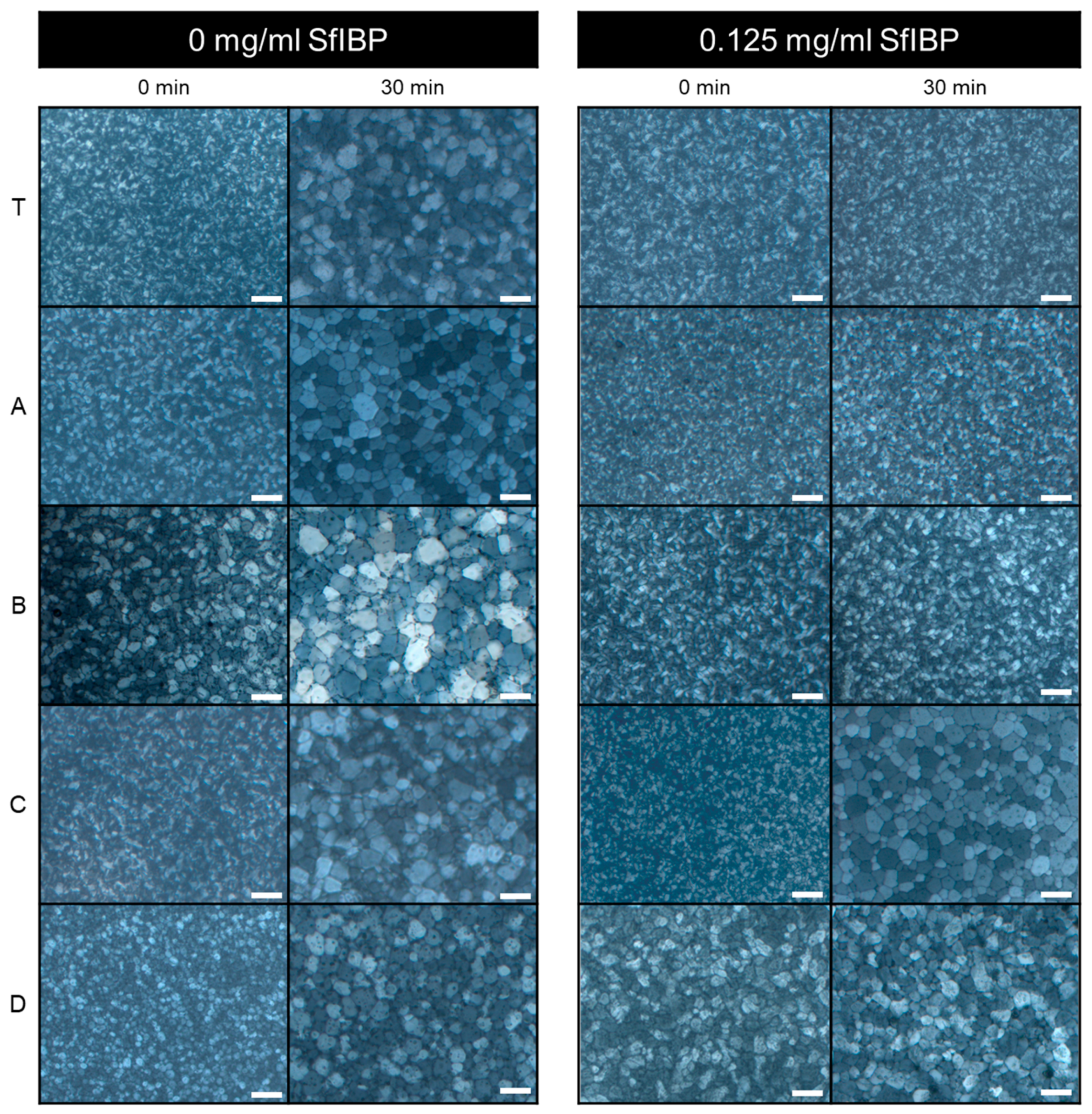
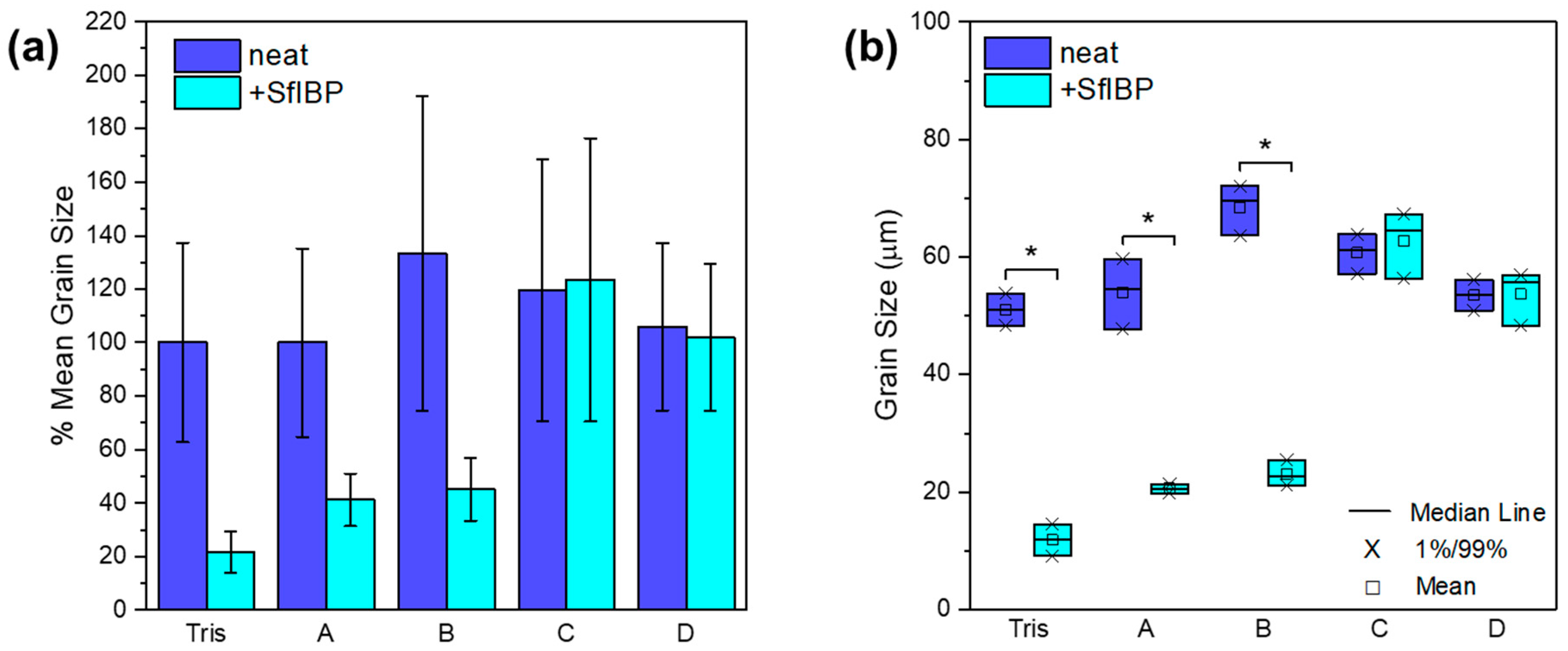
3.3. IRI Activity
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Davies, P.L.; Baardsnes, J.; Kuiper, M.J.; Walker, V.K. Structure and function of antifreeze proteins. Philosophi. Transac. Royal Soc. London B 2002, 357, 927–935. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.W.; Duman, J.G.; Xu, L. Enhancement of insect antifreeze protein activity by antibodies. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1991, 1076, 416–420. [Google Scholar] [CrossRef]
- Marshall, C.B.; Fletcher, G.L.; Davies, P.L. Hyperactive antifreeze protein in a fish. Nature 2004, 429, 153. [Google Scholar] [CrossRef] [PubMed]
- DeVries, A.L. The role of antifreeze glycopeptides and peptides in the freezing avoidance of Antarctic fishes. Compara. Biochem. Physiol. Part B 1988, 90, 611–621. [Google Scholar] [CrossRef]
- Davies, P.L. Ice-binding proteins: a remarkable diversity of structures for stopping and starting ice growth. Trends Biochem. Sci. 2014, 39, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Sally, O.Y.; Brown, A.; Middleton, A.J.; Tomczak, M.M.; Walker, V.K.; Davies, P.L. Ice restructuring inhibition activities in antifreeze proteins with distinct differences in thermal hysteresis. Cryobiology 2010, 61, 327–334. [Google Scholar]
- Martino, M.N.; Zaritzky, N. Ice recrystallization in a model system and in frozen muscle tissue. Cryobiology 1989, 26, 138–148. [Google Scholar] [CrossRef]
- Kurtz, S.K.; Carpay, F.M.A. Microstructure and normal grain growth in metals and ceramics. Part I. Theory. J. Appl. Phys. 1980, 51, 5725–5744. [Google Scholar] [CrossRef]
- Liou, Y.C.; Tocilj, A.; Davies, P.L.; Jia, Z. Mimicry of ice structure by surface hydroxyls and water of a β-helix antifreeze protein. Nature 2000, 406, 322–324. [Google Scholar] [CrossRef]
- Graether, S.P.; Kuiper, M.J.; Gagne, S.M.; Walker, V.K.; Jia, Z.; Sykes, B.D.; Davies, P.L. β-Helix structure and ice-binding properties of a hyperactive antifreeze protein from an insect. Nature 2000, 406, 325–328. [Google Scholar] [CrossRef]
- Jia, Z.; De Luca, C.I.; Chao, H.; Davies, P.L. Structural basis for the binding of a globular antifreeze protein to ice. Nature 1996, 384, 285. [Google Scholar] [CrossRef] [PubMed]
- Knight, C.A.; Cheng, C.C.; De Vries, A.L. Adsorption of alpha-helical antifreeze peptides on specific ice crystal surface planes. Biophys. J. 1991, 59, 409–418. [Google Scholar] [CrossRef] [Green Version]
- Liang, S.; Yuan, B.; Kwon, J.W.; Ahn, M.; Cui, X.S.; Bang, J.K.; Kim, N.H. Effect of antifreeze glycoprotein 8 supplementation during vitrification on the developmental competence of bovine oocytes. Theriogenology 2016, 86, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Koo, B.W.; Kim, D.; Seo, Y.S.; Nam, Y.K. Effect of Marine-Derived Ice-Binding Proteins on the Cryopreservation of Marine Microalgae. Marine Drugs 2017, 15, 372. [Google Scholar] [CrossRef] [PubMed]
- Tomalty, H.E.; Hamilton, E.F.; Hamilton, A.; Kukal, O.; Allen, T.; Walker, V.K. Kidney Preservation at Subzero Temperatures Using a Novel Storage Solution and Insect Ice-Binding Proteins. CryoLetters 2017, 38, 100–107. [Google Scholar] [PubMed]
- Kim, M.K.; Kong, H.S.; Youm, H.W.; Jee, B.C. Effects of supplementation with antifreeze proteins on the follicular integrity of vitrified-warmed mouse ovaries: Comparison of two types of antifreeze proteins alone and in combination. Clin. Exp. Reprod. Med. 2017, 44, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.G.; Koh, H.Y.; Lee, J.H.; Kang, S.H.; Kim, H.J. Cryopreservative effects of the recombinant ice-binding protein from the arctic yeast Leucosporidium sp. on red blood cells. Appl. Biochem. Biotechnol. 2012, 167, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.; Davies, P.L.; Carpenter, J.F. Effects of antifreeze proteins on red blood cell survival during cryopreservation. J. Exp. Biol. 1996, 199, 2071–2076. [Google Scholar] [PubMed]
- Wang, J.H. A comprehensive evaluation of the effects and mechanisms of antifreeze proteins during low-temperature preservation. Cryobiology 2000, 41, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ptitsyn, O.B. Protein folding: hypotheses and experiments. J. Protein Chem. 1987, 6, 273–293. [Google Scholar] [CrossRef]
- Congdon, T.; Dean, B.T.; Kasperczak-Wright, J.; Biggs, C.I.; Notman, R.; Gibson, M.I. Probing the biomimetic ice nucleation inhibition activity of poly (vinyl alcohol) and comparison to synthetic and biological polymers. Biomacromolecules 2015, 16, 2820–2826. [Google Scholar] [CrossRef] [PubMed]
- Vance, T.D.; Graham, L.A.; Davies, P.L. An ice-binding and tandem beta-sandwich domain-containing protein in Shewanella frigidimarina is a potential new type of ice adhesin. FEBS J. 2018, 285, 1511–1527. [Google Scholar] [CrossRef]
- Kristiansen, E.; Pedersen, S.A.; Zachariassen, K.E. Salt-induced enhancement of antifreeze protein activity: A salting-out effect. Cryobiology 2008, 57, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Leiter, A.; Rau, S.; Winger, S.; Muhle-Goll, C.; Luy, B.; Gaukel, V. Influence of heating temperature, pressure and pH on recrystallization inhibition activity of antifreeze protein type III. J. Food Eng. 2016, 187, 53–61. [Google Scholar] [CrossRef]
- Czechura, P.; Tam, R.Y.; Dimitrijevic, E.; Murphy, A.V.; Ben, R.N. The importance of hydration for inhibiting ice recrystallization with C-linked antifreeze glycoproteins. JACS 2008, 130, 2928–2929. [Google Scholar] [CrossRef] [PubMed]
- Knight, C.A.; Hallett, J.; De Vries, A.L. Solute effects on ice recrystallization: an assessment technique. Cryobiology 1988, 25, 55–60. [Google Scholar] [CrossRef]
- Budke, C.; Dreyer, A.; Jaeger, J.; Gimpel, K.; Berkemeier, T.; Bonin, A.S.; Koop, T. Quantitative efficacy classification of ice recrystallization inhibition agents. Crystal Grow. Des. 2014, 14, 4285–4294. [Google Scholar] [CrossRef]
- Middleton, A.J.; Vanderbeld, B.; Bredow, M.; Tomalty, H.; Davies, P.L.; Walker, V.K. Isolation and characterization of ice-binding proteins from higher plants. In Plant Cold Acclimation; Humana Press: New York, NY, USA, 2014; pp. 255–277. [Google Scholar]
- Ghods, P.; Isgor, O.B.; McRae, G.; Miller, T. The effect of concrete pore solution composition on the quality of passive oxide films on black steel reinforcement. Cement Concrete Comp. 2009, 31, 2–11. [Google Scholar] [CrossRef]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta Proteins Proteom. 2005, 1751, 119–139. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef]
- Song, B.; Cho, J.H.; Raleigh, D.P. Ionic-strength-dependent effects in protein folding: Analysis of rate equilibrium free-energy relationships and their interpretation. Biochemistry 2007, 46, 14206–14214. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.A. Dominant forces in protein folding. Biochemistry 1990, 29, 7133–7155. [Google Scholar] [CrossRef] [PubMed]
- Barth, H.G.; Boyes, B.E.; Jackson, C. Size exclusion chromatography and related separation techniques. Anal. Chem. 1998, 70, 251–278. [Google Scholar] [CrossRef]
- Radzicka, A.; Wolfenden, R. Rates of uncatalyzed peptide bond hydrolysis in neutral solution and the transition state affinities of proteases. JACS 1996, 118, 6105–6109. [Google Scholar] [CrossRef]
- Lawrence, L.; Moore, W.J. Kinetics of the hydrolysis of simple glycine peptides. JACS 1951, 73, 3973–3977. [Google Scholar] [CrossRef]
- Feeney, E.P.; Guinee, T.P.; Fox, P.F. Effect of pH and calcium concentration on proteolysis in Mozzarella cheese. J. Dairy Sci. 2002, 85, 1646–1654. [Google Scholar] [CrossRef]
- Bar, M.; Bar-Ziv, R.; Scherf, T.; Fass, D. Efficient production of a folded and functional, highly disulfide-bonded β-helix antifreeze protein in bacteria. Protein Expres. Purif. 2006, 48, 243–252. [Google Scholar] [CrossRef]
- Garnham, C.P.; Gilbert, J.A.; Hartman, C.P.; Campbell, R.L.; Laybourn-Parry, J.; Davies, P.L. A Ca2+-dependent bacterial antifreeze protein domain has a novel β-helical ice-binding fold. Biochem. J. 2008, 411, 171–180. [Google Scholar] [CrossRef]
- Congdon, T.; Notman, R.; Gibson, M.I. Antifreeze (glyco) protein mimetic behavior of poly (vinyl alcohol): detailed structure ice recrystallization inhibition activity study. Biomacromolecules 2013, 14, 1578–1586. [Google Scholar] [CrossRef]
- Mitchell, D.E.; Lilliman, M.; Spain, S.G.; Gibson, M.I. Quantitative study on the antifreeze protein mimetic ice growth inhibition properties of poly (ampholytes) derived from vinyl-based polymers. Biomat. Sci. 2014, 2, 1787–1795. [Google Scholar] [CrossRef]
- Voets, I.K. From ice-binding proteins to bio-inspired antifreeze materials. Soft Matter 2017, 13, 4808–4823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Solution | I (mol/L) | pH | OH (mM) | Tris (mM) | Ca (mM) | Na (mM) | K (mM) | S (mM) | Al (mM) | Mg (mM) | Si (mM) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 0.03 | 12.4 ± 0.1 | 26.71 | 20 | 0.003 | 6.08 | 9.85 | 5.02 | - | 0.001 | 0.005 |
| A + 1/2 Tris* | 0.03 | 12.4 ± 0.1 | 23.62 | 10 | 0.003 | 6.08 | 9.85 | 5.02 | - | 0.001 | 0.005 |
| B | 0.05 | 12.7 ± 0.1 | 48.60 | 20 | 0.003 | 9.70 | 15.5 | 8.08 | - | 0.001 | 0.005 |
| C | 0.16 | 13.2 ± 0.1 | 168.5 | 20 | 0.007 | 23.9 | 37.3 | 20.6 | 0.001 | 0.002 | 0.005 |
| D | 0.69 | 13.9 ± 0.2 | 857.7 | 20 | 0.761 | 90.5 | 132.0 | 76.4 | 0.001 | - | 0.008 |
| Tris | 0.01 | 8.90 ± 0.1 | 0.008 | 20 | - | - | - | - | - | - | - |
| 1/2 Tris * | 0.005 | 8.40 ± 0.2 | 0.003 | 10 | - | - | - | - | - | - | - |
| Secondary Structure | 1/2 Tris | A + 1/2 Tris | Difference |
|---|---|---|---|
| Regular α-helix | 3.8% | 2.1% | −1.7% |
| Distorted α-helix | 5.1% | 2.3% | −2.8% |
| Left β-helix | 7.5% | 1.0% | −6.5% |
| Relaxed β-helix | 18.3% | 10.4% | −7.9% |
| Right β-helix | 13.2% | 21.1% | +7.9% |
| Parallel β-strand | 0.1% | 0.8% | +0.7% |
| Turn | 10.3% | 15.3% | +5.0% |
| Other | 41.7% | 46.9% | +5.2% |
| Solution | SfIBP Loading (mg/mL) | Mean Crystal Size (µm) | % Change in Mean Crystal Size |
|---|---|---|---|
| Tris | 0 | 51 ± 19 | - |
| Tris | 0.125 | 11 ± 4 | −78% |
| A | 0 | 51 ± 18 | - |
| A | 0.125 | 21 ± 5 | −59% |
| B | 0 | 68 ± 30 | - |
| B | 0.125 | 23 ± 6 | −66% |
| C | 0 | 61 ± 25 | - |
| C | 0.125 | 63 ± 27 | +3% |
| D | 0 | 54 ± 16 | - |
| D | 0.125 | 52 ± 14 | +4% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delesky, E.A.; Frazier, S.D.; Wallat, J.D.; Bannister, K.L.; Heveran, C.M.; Srubar, W.V., III. Ice-Binding Protein from Shewanella frigidimarinas Inhibits Ice Crystal Growth in Highly Alkaline Solutions. Polymers 2019, 11, 299. https://doi.org/10.3390/polym11020299
Delesky EA, Frazier SD, Wallat JD, Bannister KL, Heveran CM, Srubar WV III. Ice-Binding Protein from Shewanella frigidimarinas Inhibits Ice Crystal Growth in Highly Alkaline Solutions. Polymers. 2019; 11(2):299. https://doi.org/10.3390/polym11020299
Chicago/Turabian StyleDelesky, Elizabeth A., Shane D. Frazier, Jaqueline D. Wallat, Kendra L. Bannister, Chelsea M. Heveran, and Wil V. Srubar, III. 2019. "Ice-Binding Protein from Shewanella frigidimarinas Inhibits Ice Crystal Growth in Highly Alkaline Solutions" Polymers 11, no. 2: 299. https://doi.org/10.3390/polym11020299
APA StyleDelesky, E. A., Frazier, S. D., Wallat, J. D., Bannister, K. L., Heveran, C. M., & Srubar, W. V., III. (2019). Ice-Binding Protein from Shewanella frigidimarinas Inhibits Ice Crystal Growth in Highly Alkaline Solutions. Polymers, 11(2), 299. https://doi.org/10.3390/polym11020299






