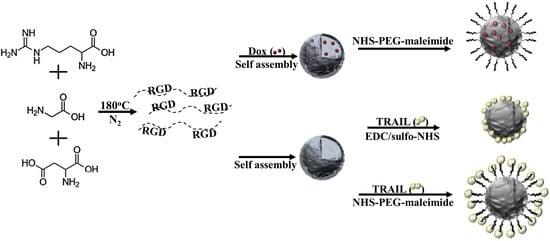
Engineering of Doxorubicin-Encapsulating and TRAIL-Conjugated Poly(RGD) Proteinoid Nanocapsules for Drug Delivery Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of P(RGD) Proteinoid by Step-Growth Polymerization Mechanism
2.3. Proteinoid Characterization and Analysis
2.4. Preparation of Hollow P(RGD) NCs
2.5. Preparation of Dox/P(RGD) NCs
2.6. PEGylation of Dox/P(RGD) NCs
2.7. Indirect Conjugation of TRAIL to P(RGD) NCs
2.8. Direct Conjugation of TRAIL to P(RGD) NCs
2.9. Diameter and Size Distribution Measurements
2.10. ζ-Potential Measurements
2.11. Quantification of Conjugated TRAIL
2.12. HPLC Analysis
2.13. Optical Characterization of Dox and Dox/P(RGD) NCs
2.14. Photostability of Free and Encapsulated Dox
2.15. Drug Release Model
2.16. In Vitro XTT Cell Viability Assay
3. Results and Discussion
3.1. Synthesis and Characterization of P(RGD) Proteinoid Nanocapsules (NCs)
3.2. Synthesis and Characterization of P(RGD) NCs
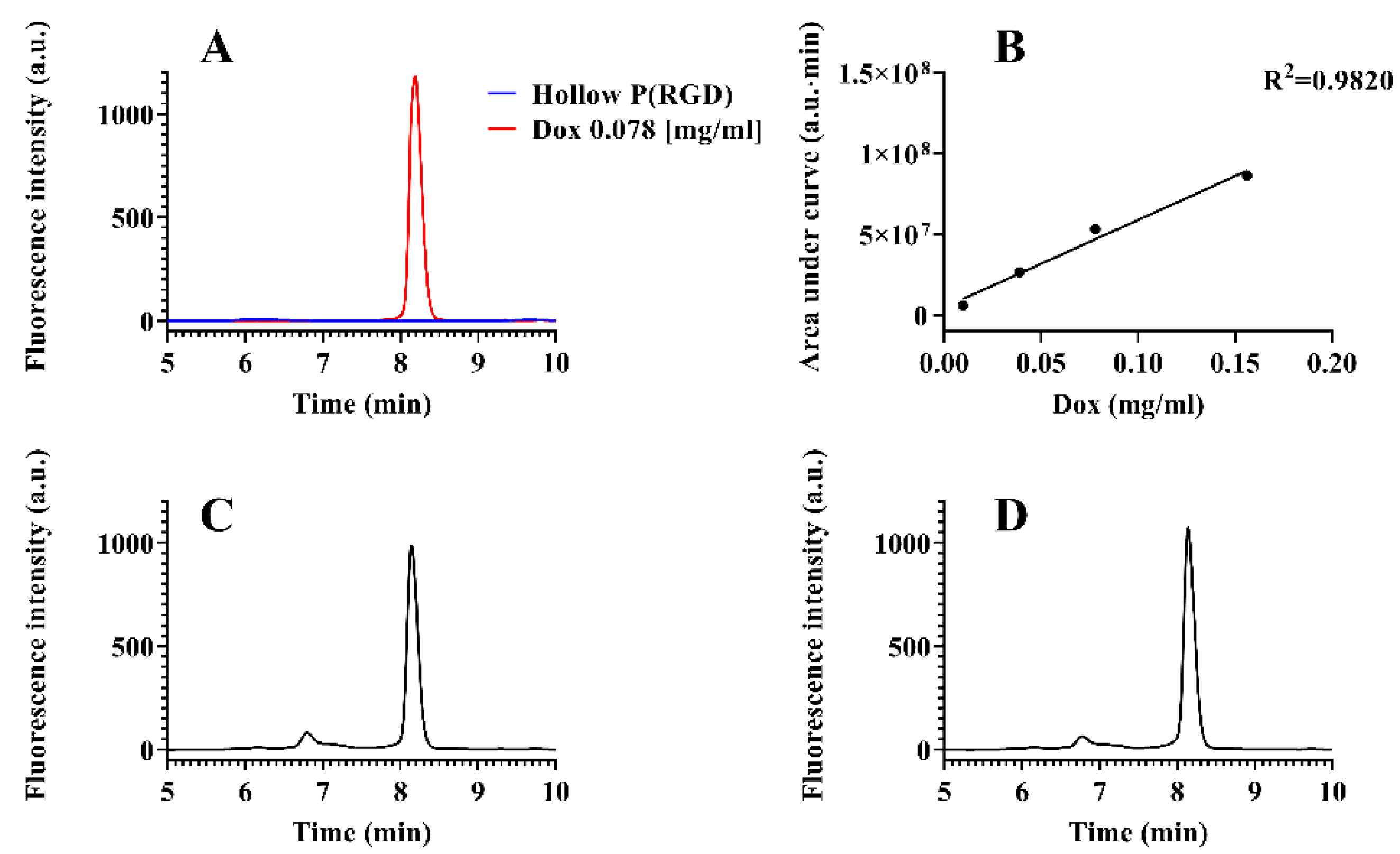
3.3. Loading Capacity of Dox and TRAIL in P(RGD) NCs
3.4. Optical Characterization of Free Dox and Dox/P(RGD) NCs
3.5. Photostability of Free Dox and Dox/P(RGD) NCs
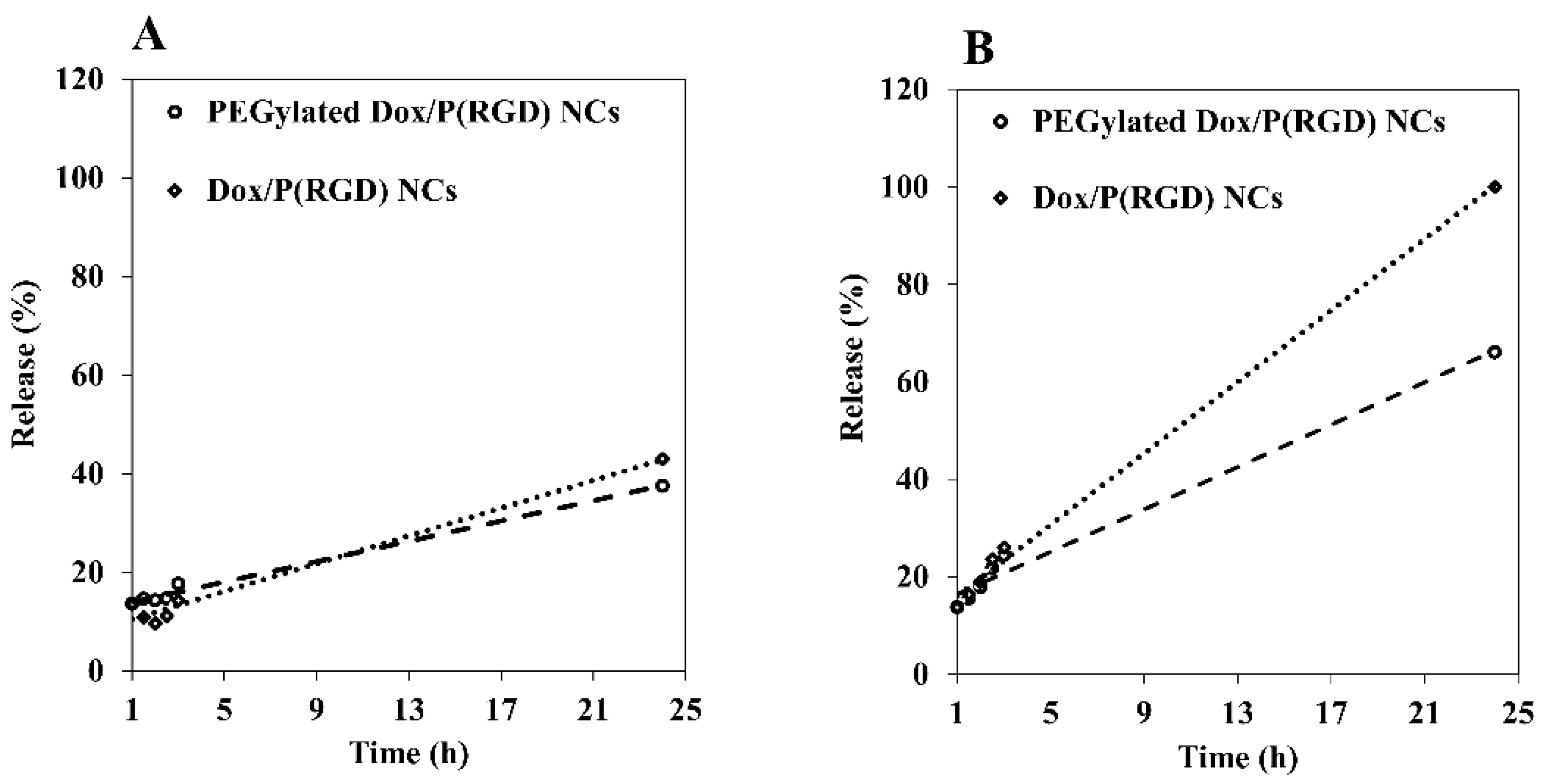
3.6. Controlled Release of Dox from P(RGD) Nanocapsules
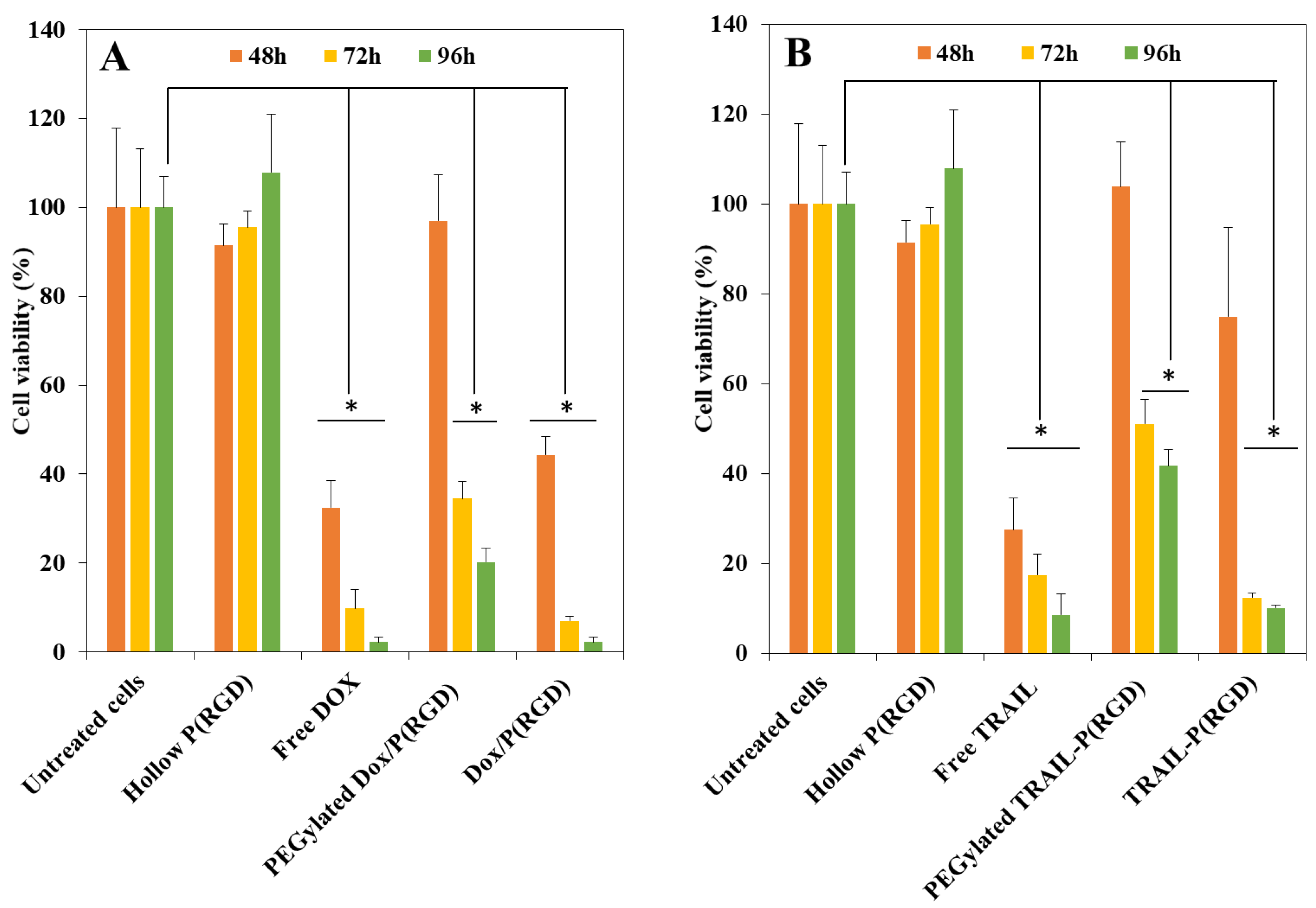
3.7. Cytotoxicity of Drugs and NCs
4. Summary and Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Menon, U. Ovarian cancer: Challenges of early detection. Nat. Clin. Pr. Oncol. 2007, 4, 498–499. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA A Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef]
- Faizi, N.; Kazmi, S. Universal health coverage-There is more to it than meets the eye. J. Fam. Med. Prim. Care 2017, 6, 169–170. [Google Scholar] [CrossRef] [PubMed]
- Weber, P.; Wagner, M.; Schneckenburger, H. Cholesterol dependent uptake and interaction of doxorubicin in MCF-7 breast cancer cells. Int. J. Mol. Sci. 2013, 14, 8358–8366. [Google Scholar] [CrossRef]
- Motlagh, N.S.H.; Parvin, P.; Ghasemi, F.; Atyabi, F. Fluorescence properties of several chemotherapy drugs: Doxorubicin, paclitaxel and bleomycin. Biomed. Opt. Express 2016, 7, 2400–2406. [Google Scholar] [CrossRef]
- Yeo, W.; Lau, T.K.; Li, L.; Lai, K.T.; Pang, E.; Cheung, M.; Chan, V.T.; Wong, A.; Soo, W.M.; Yeung, V.T.; et al. A randomized study of olanzapine-containing versus standard antiemetic regimens for the prevention of chemotherapy-induced nausea and vomiting in Chinese breast cancer patients. Breast 2020, 50, 30–38. [Google Scholar] [CrossRef]
- Pei, Z.; Hu, J.; Bai, Q.; Liu, B.; Cheng, N.; Liu, H.; Na, R.; Yu, Q. Thymoquinone protects against cardiac damage from doxorubicin-induced heart failure in Sprague-Dawley rats. RSC Adv. 2018, 8, 14633–14639. [Google Scholar] [CrossRef]
- Levy, I.; Grinberg, I.; Perlstein, B.; Corem-Salkmon, E.; Margel, S. Tumor necrosis factor related apoptosis inducing ligand-conjugated near IR fluorescent iron oxide/human serum albumin core-shell nanoparticles of narrow size distribution for cancer targeting and therapy. J. Nanomed. Nanotechnol. 2015, 6. [Google Scholar] [CrossRef]
- Levy, I.; Rudnick-glick, S.; Perlstein, B. Engineering of core and core/shell near infra-red fluorescent iron oxide and iron oxide/human serum albumin nanoparticles of narrow size distribution for cancer diagnosis and therapy. In Advances in Anti-Cancer Drugs; Avid Science: Hyderabad, India, 2017; pp. 2–55. [Google Scholar]
- Perlstein, B.; Finniss, S.A.; Miller, C.; Okhrimenko, H.; Kazimirsky, G.; Cazacu, S.; Lee, H.K.; Lemke, N.; Brodie, S.; Umansky, F.; et al. TRAIL conjugated to nanoparticles exhibits increased anti-tumor activities in glioma cells and glioma stem cells in vitro and in vivo. Neuro-Oncol. 2013, 15, 29–40. [Google Scholar] [CrossRef]
- Green-Sadan, T.; Kuttner, Y.; Lublin-Tennenbaum, T.; Kinor, N.; Boguslavsky, Y.; Margel, S.; Yadid, G. Glial cell line-derived neurotrophic factor-conjugated nanoparticles suppress acquisition of cocaine self-administration in rats. Exp. Neurol. 2005, 194, 97–105. [Google Scholar] [CrossRef]
- Skaat, H.; Ziv-Polat, O.; Shahar, A.; Margel, S. Enhancement of the growth and differentiation of nasal olfactory mucosa cells by the conjugation of growth factors to functional nanoparticles. Bioconjugate Chem. 2011, 22, 2600–2610. [Google Scholar] [CrossRef] [PubMed]
- Bikfalvi, A. Biological roles of fibroblast growth factor-2. Endocr. Rev. 1997, 18, 26–45. [Google Scholar] [CrossRef] [PubMed]
- Marcus, M.; Skaat, H.; Alon, N.; Margel, S.; Shefi, O. NGF-conjugated iron oxide nanoparticles promote differentiation and outgrowth of PC12 cells. Nanoscale 2015, 7, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Pierschbacher, M.D.; Ruoslahti, E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nat. Cell Biol. 1984, 309, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Dubey, P.K.; Mishra, V.; Jain, S.; Mahor, S.; Vyas, S. Liposomes modified with cyclic RGD peptide for tumor targeting. J. Drug Target. 2004, 12, 257–264. [Google Scholar] [CrossRef]
- Weis, S.M.; Cheresh, D.A. Tumor angiogenesis: Molecular pathways and therapeutic targets. Nat. Med. 2011, 17, 1359–1370. [Google Scholar] [CrossRef]
- Fu, X.; Yang, Y.; Li, X.; Lai, H.; Huang, Y.; He, L.; Zheng, W.; Chen, T. RGD peptide-conjugated selenium nanoparticles: Antiangiogenesis by suppressing VEGF-VEGFR2-ERK/AKT pathway. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 1627–1639. [Google Scholar] [CrossRef]
- Yin, H.-Q.; Mai, D.-S.; Gan, F.; Chen, X. One-step synthesis of linear and cyclic RGD conjugated gold nanoparticles for tumour targeting and imaging. RSC Adv. 2014, 4, 9078. [Google Scholar] [CrossRef]
- Fox, S.W. How did life begin? Science 1960, 132, 200–208. [Google Scholar] [CrossRef]
- Fox, S.W. The proteinoid theory of the origin of life and competing ideas. Am. Biol. Teach. 1974, 36, 161–181. [Google Scholar] [CrossRef]
- Fox, S.W. Thermal synthesis of amino acids and the origin of life. Geochim. Cosmochim. Acta 1995, 59, 1213–1214. [Google Scholar] [CrossRef]
- Fox, S.W.; Jungck, J.R.; Nakashima, T. From proteinoid microsphere to contemporary cell: Formation of internucleotide and peptide bonds by proteinoid particles. Orig. Life Evol. Biosphere 1974, 5, 227–237. [Google Scholar] [CrossRef]
- Fox, S.W.; Mccauley, R.J.; Fukushima, T.; Windsor, C.R.; Montgome, P.O. Selective action in boundaries of particles of thermal proteinoid. Fed. Proc. 1967, 26, 749. [Google Scholar]
- Fox, S.W.; Nakashima, T.; Przybylski, A.; Syren, R.M. The updated experimental proteinoid model. Int. J. Quantum Chem. 2009, 22, 195–204. [Google Scholar] [CrossRef]
- Kile, S.; Kolitz-Domb, M.; Corem-Salkmon, E.; Margel, S. Engineered doxorubicin delivery system using proteinoid-poly (L-lactic acid) polymeric nanoparticles of narrow size distribution and high molecular weight for cancer treatment. Int. J. Nanotechnol. Nanomed. 2017, 2, 1–11. [Google Scholar] [CrossRef]
- Kumar, A.B.M.; Rao, K.P. Preparation and characterization of pH-sensitive proteinoid microspheres for the oral delivery of methotrexate. Biomaterials 1998, 19, 725–732. [Google Scholar] [CrossRef]
- Kolitz-Domb, M.; Grinberg, I.; Corem-Salkmon, E.; Margel, S. Engineering of near infrared fluorescent proteinoid-poly (L-lactic acid) particles for in vivo colon cancer detection. J. Nanobiotechnology 2014, 12, 30. [Google Scholar] [CrossRef]
- Kwon, K.; Kim, J.-C. Monoolein cubic phase containing disulfide proteinoid and its reduction-responsive release property. J. Dispers. Sci. Technol. 2017, 39, 614–622. [Google Scholar] [CrossRef]
- Kwon, K.; Park, D.; Kim, J.-C. Disulfide proteinoid micelles responsive to reduction. J. Dispers. Sci. Technol. 2018, 40, 1413–1422. [Google Scholar] [CrossRef]
- Haham, H.; Margel, S.; Belostozky, A.; Kolitza-Domb, M.; Grinberg, I. Engineering of new UV-blocking hollow proteinoid nanoparticles of narrow size distribution containing all-trans retinoic acid for biomedical applications. J. Nanomed. Nanotechnol. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Sasson, E.; Pinhasi, R.V.O.; Margel, S.; Klipcan, L. Engineering and use of proteinoid polymers and nanocapsules containing agrochemicals. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hadad, E.; Rudnick-Glick, S.; Grinberg, I.; Kolitz-Domb, M.; Chill, J.H.; Margel, S. Synthesis and characterization of poly (RGD) proteinoid polymers and NIR fluorescent nanoparticles of optimal d,l-configuration for drug-delivery applications-In vitro study. ACS Omega 2020, 5, 23568–23577. [Google Scholar] [CrossRef] [PubMed]
- Harada, K. Thermal Polymerization of Amino Acid Mixtures Containing Aspartic Acid or a Thermal Precursor of Aspartic Acid. US Patent 3,052,655, 4 September 1962. [Google Scholar]
- Harada, K.; Matsuyama, M. Polycondensation of thermal precursors of amino acids and characterization of constituent amino acids. Biosystems 1979, 11, 47–53. [Google Scholar] [CrossRef]
- Hadad, E.; Rudnick-Glick, S.; Grinberg, I.; Yehuda, R.; Margel, S. Engineering of NIR fluorescent PEGylated poly (RGD) proteinoid polymers and nanoparticles for drug delivery applications in chicken embryo and mouse models. RSC Adv. 2020, 10, 34364–34372. [Google Scholar] [CrossRef]
- Lugasi, L.; Grinberg, I.; Margel, S. Targeted delivery of CBD-loaded poly (RGD) proteinoid nanoparticles for antitumor therapy. J. Nanomed. Nanotech. 2020, 11, 552. [Google Scholar] [CrossRef]
- Thi, T.T.H.; Pilkington, E.H.; Nguyen, D.H.; Lee, J.S.; Thi, T.T.H.; Truong, N.P. The Importance of Poly (ethylene glycol) alternatives for overcoming PEG immunogenicity in drug delivery and bioconjugation. Polymers 2020, 12, 298. [Google Scholar] [CrossRef]
- Aggarwal, P.; Hall, J.B.; McLeland, C.B.; Dobrovolskaia, M.A.; McNeil, S.E. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv. Drug Deliv. Rev. 2009, 61, 428–437. [Google Scholar] [CrossRef]
- Kazunori, K.; Kwon, G.S.; Masayuki, Y.; Teruo, O.; Yasuhisa, S. Block copolymer micelles as vehicles for drug delivery. J. Control. Release 1993, 24, 119–132. [Google Scholar] [CrossRef]
- Cohen, S.; Pellach, M.; Kam, Y.; Grinberg, I.; Corem-Salkmon, E.; Rubinstein, A.; Margel, S. Synthesis and characterization of near IR fluorescent albumin nanoparticles for optical detection of colon cancer. Mater. Sci. Eng. C 2013, 33, 923–931. [Google Scholar] [CrossRef]
- Roehm, N.W.; Rodgers, G.H.; Hatfield, S.M.; Glasebrook, A.L. An improved colorimetric assay for cell proliferation and viability utilizing the tetrazolium salt XTT. J. Immunol. Methods 1991, 142, 257–265. [Google Scholar] [CrossRef]
- Cohen, S.; Margel, S. Engineering of near IR fluorescent albumin nanoparticles for in vivo detection of colon cancer. J. Nanobiotechnology 2012, 10, 36. [Google Scholar] [CrossRef]
- Gaca, S.; Reichert, S.; Multhoff, G.; Wacker, M.; Hehlgans, S.; Botzler, C.; Gehrmann, M.; Rödel, C.; Kreuter, J.; Rodel, F. Targeting by cmHsp70.1-antibody coated and survivin miRNA plasmid loaded nanoparticles to radiosensitize glioblastoma cells. J. Control. Release 2013, 172, 201–206. [Google Scholar] [CrossRef]
- Abd-Allah, F. Development, characterization and ex vivo evaluation of various liposome-encapsulated aceclofenac formulations. Br. J. Pharm. Res. 2016, 9, 1–12. [Google Scholar] [CrossRef]
- Rahme, K.; Chen, L.; Hobbs, R.G.; Morris, M.A.; O’Driscoll, C.M.; Holmes, J.D. PEGylated gold nanoparticles: Polymer quantification as a function of PEG lengths and nanoparticle dimensions. RSC Adv. 2013, 3, 6085–6094. [Google Scholar] [CrossRef]
- Lugasi, L.; Grinberg, I.; Sabag, R.; Madar, R.; Einat, H.; Margel, S. Proteinoid nanocapsules as drug delivery system for improving antipsychotic activity of risperidone. Molecules 2020, 25, 4013. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ji, X.; Li, Z.; Huang, F. Fluorescent supramolecular polymeric materials. Adv. Mater. 2017, 29. [Google Scholar] [CrossRef]
- Haritoglou, C.; Freyer, W.; Priglinger, S.G.; Kampik, A. Light absorbing properties of indocyanine green (ICG) in solution and after adsorption to the retinal surface-an ex-vivo approach. Graefe’s Arch. Clin. Exp. Ophthalmol. 2006, 244, 1196–1202. [Google Scholar] [CrossRef]
- Zweck, J.; Penzkofer, A. Microstructure of indocyanine green J-aggregates in aqueous solution. Chem. Phys. 2001, 269, 399–409. [Google Scholar] [CrossRef]
- Kim, T.H.; Chen, Y.; Mount, C.W.; Gombotz, W.R.; Li, X.; Pun, S.H. Evaluation of temperature-sensitive, indocyanine green-encapsulating micelles for noninvasive near-infrared tumor imaging. Pharm. Res. 2010, 27, 1900–1913. [Google Scholar] [CrossRef]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]

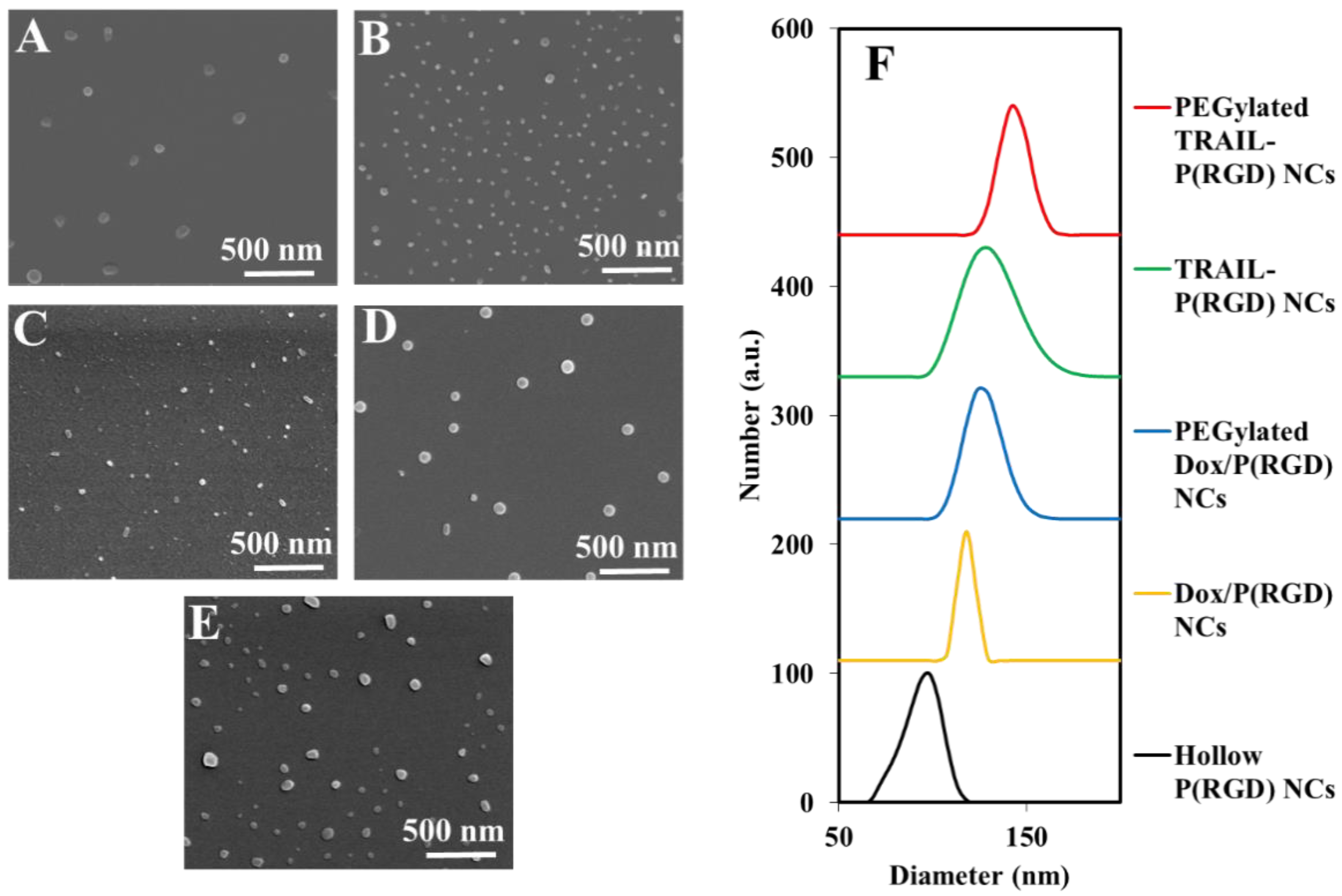


| Nanocapsule | Dry Diameter (nm) | Hydrodynamic Diameter (nm) | Zeta Potential (mV) |
|---|---|---|---|
| Hollow P(RGD) | 84 ± 16 | 90 ± 15 | −10 ± 7 |
| PEGylated Dox/P(RGD) | 46 ± 6 | 130 ± 17 | 2 ± 5 |
| Dox/P(RGD) | 44 ± 16 | 118 ± 7 | −15 ± 4 |
| PEGylated TRAIL-P(RGD) | 90 ± 12 | 146 ± 16 | −7 ± 3 |
| TRAIL-P(RGD) | 86 ± 10 | 134 ± 32 | −20 ± 8 |
| Nanocapsule | Initial Drug (w%) a | Entrapment/Conjugation Efficiency (%) |
|---|---|---|
| PEGylated Dox/P(RGD) | 1.25 | 62 |
| DOX/P(RGD) | 1.25 | 61 |
| PEGylated TRAIL-P(RGD) | 0.05 | 0.75 |
| TRAIL-P(RGD) | 0.05 | 0.025 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hadad, E.; Rudnick-Glick, S.; Itzhaki, E.; Avivi, M.Y.; Grinberg, I.; Elias, Y.; Margel, S. Engineering of Doxorubicin-Encapsulating and TRAIL-Conjugated Poly(RGD) Proteinoid Nanocapsules for Drug Delivery Applications. Polymers 2020, 12, 2996. https://doi.org/10.3390/polym12122996
Hadad E, Rudnick-Glick S, Itzhaki E, Avivi MY, Grinberg I, Elias Y, Margel S. Engineering of Doxorubicin-Encapsulating and TRAIL-Conjugated Poly(RGD) Proteinoid Nanocapsules for Drug Delivery Applications. Polymers. 2020; 12(12):2996. https://doi.org/10.3390/polym12122996
Chicago/Turabian StyleHadad, Elad, Safra Rudnick-Glick, Ella Itzhaki, Matan Y. Avivi, Igor Grinberg, Yuval Elias, and Shlomo Margel. 2020. "Engineering of Doxorubicin-Encapsulating and TRAIL-Conjugated Poly(RGD) Proteinoid Nanocapsules for Drug Delivery Applications" Polymers 12, no. 12: 2996. https://doi.org/10.3390/polym12122996
APA StyleHadad, E., Rudnick-Glick, S., Itzhaki, E., Avivi, M. Y., Grinberg, I., Elias, Y., & Margel, S. (2020). Engineering of Doxorubicin-Encapsulating and TRAIL-Conjugated Poly(RGD) Proteinoid Nanocapsules for Drug Delivery Applications. Polymers, 12(12), 2996. https://doi.org/10.3390/polym12122996