PEGylated Thermo-Sensitive Bionic Magnetic Core-Shell Structure Molecularly Imprinted Polymers Based on Halloysite Nanotubes for Specific Adsorption and Separation of Bovine Serum Albumin
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials
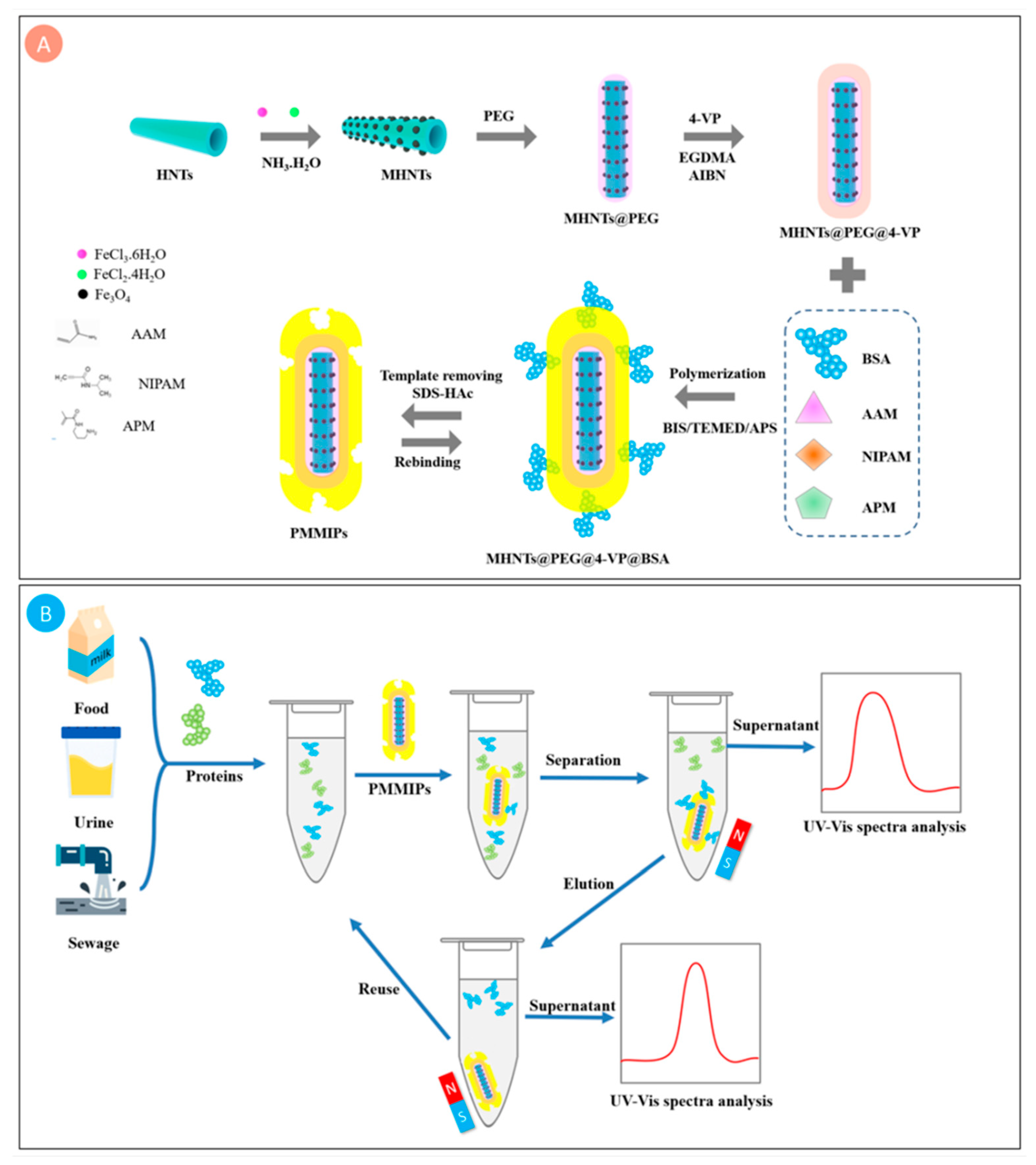
2.2. Preparation of PMMIPs and PMNIPs
2.3. Adsorption Experiments
2.3.1. Adsorption Isotherms
2.3.2. Adsorption Kinetics
2.4. Adsorption Selectivity
2.5. Removal of BSA from Real Samples
2.6. Regeneration and Reproducibility
3. Results and Discussion
3.1. Preparation of PMMIPs
3.2. Characterization of PMMIPs
3.2.1. Composition and Morphological Characterization
3.2.2. FT-IR Spectra
3.2.3. X-ray Diffraction
3.2.4. Magnetic Properties
3.2.5. TGA and DTG Analysis
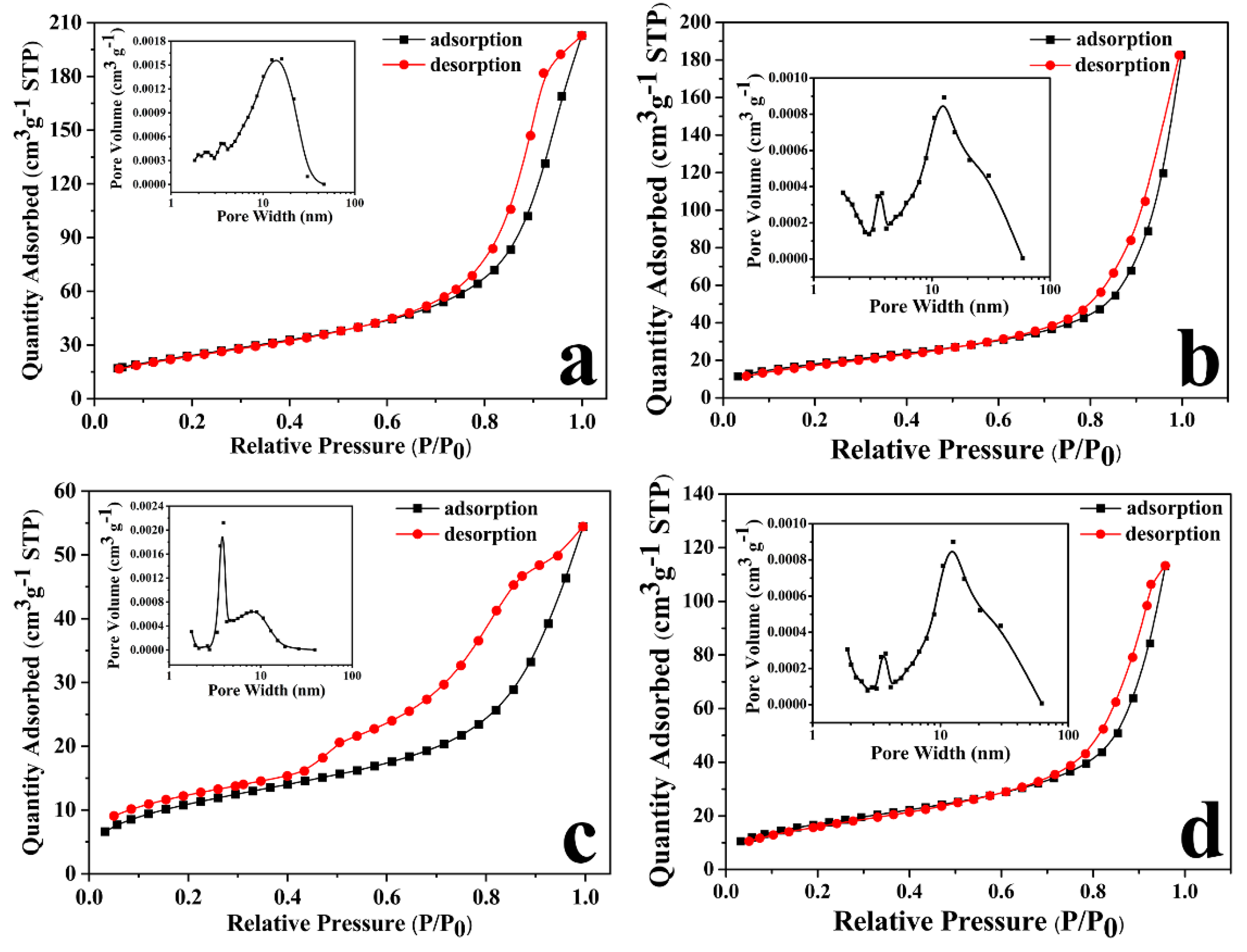
3.2.6. BET Analysis
3.2.7. Experiments to Optimize the Reaction Parameters of MIPs Based on the MHNTs@PEG@4-VP Nanotubes
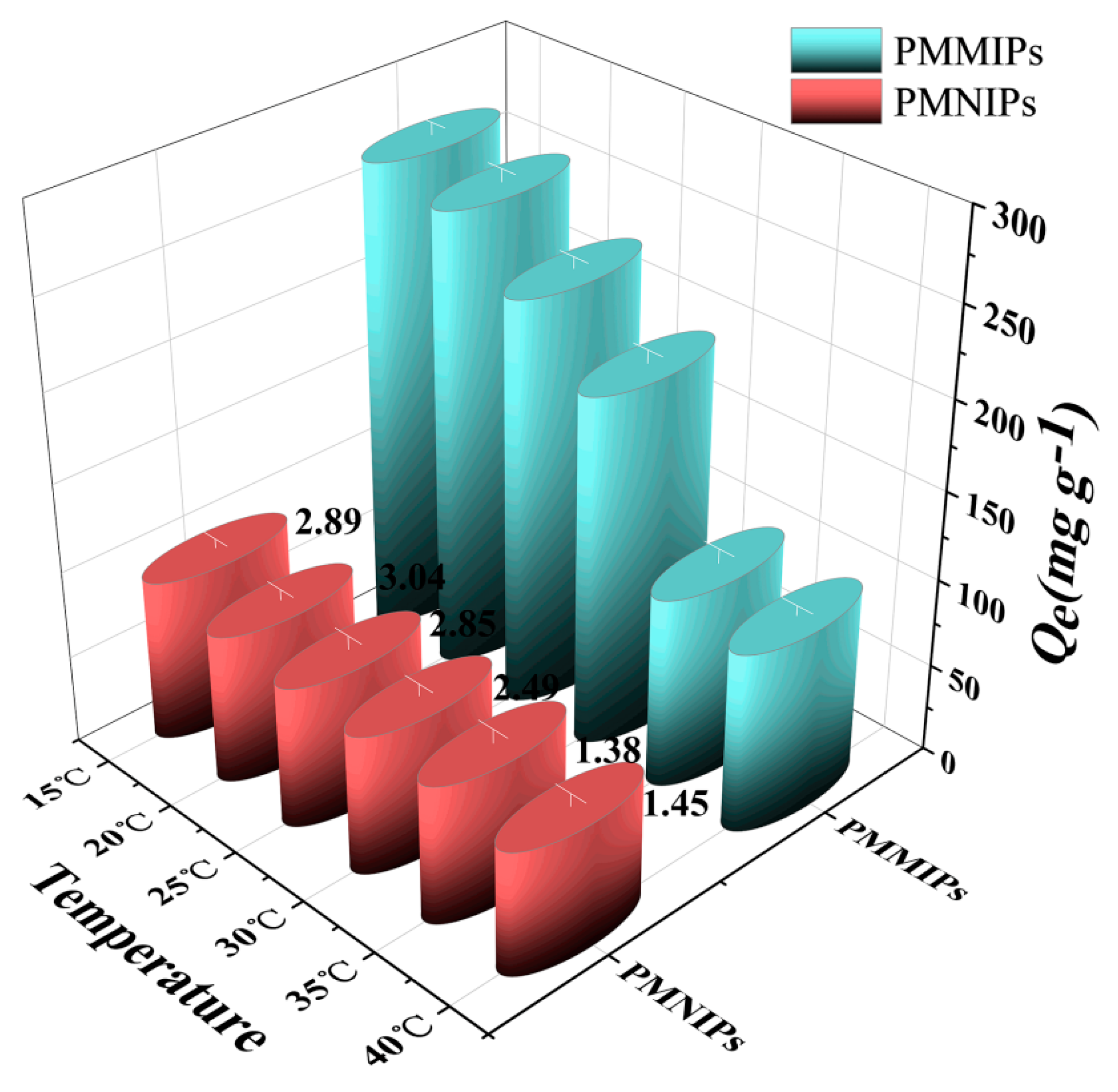
3.2.8. Thermo-Sensitivity Analysis
3.3. Adsorption Property
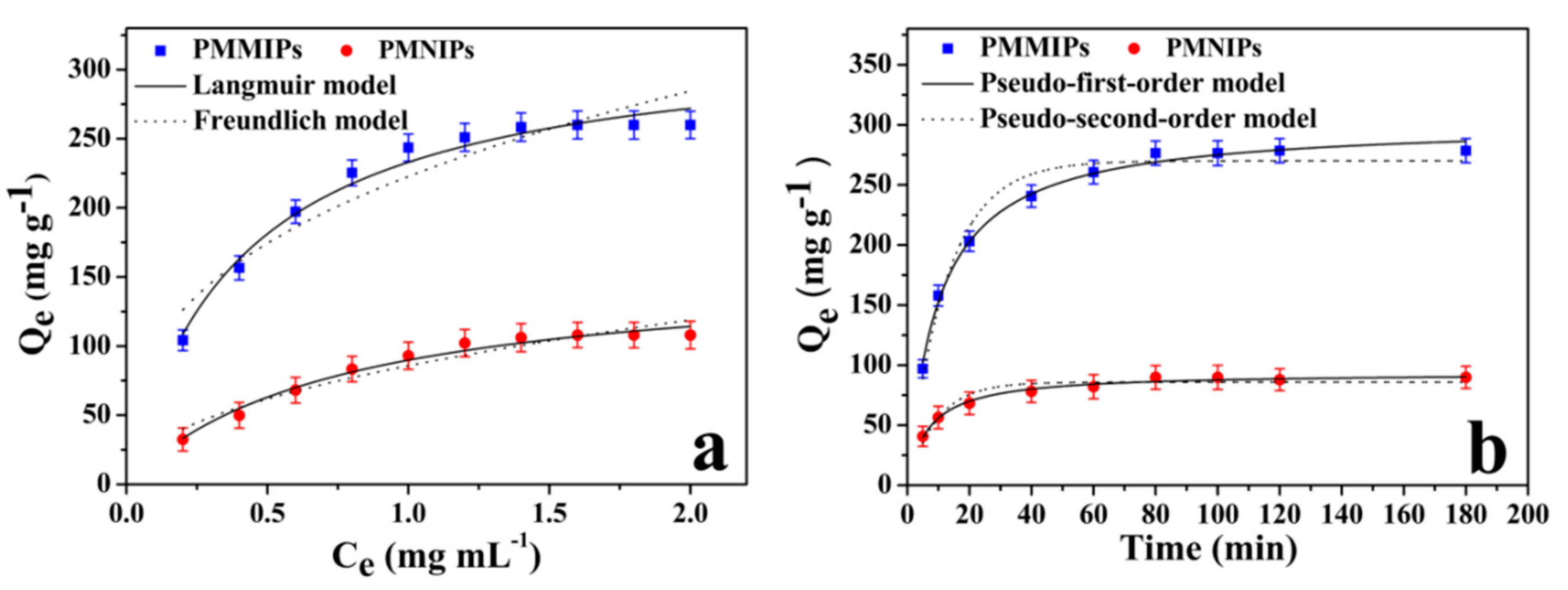
3.3.1. Adsorption Isotherms
3.3.2. Adsorption Kinetics
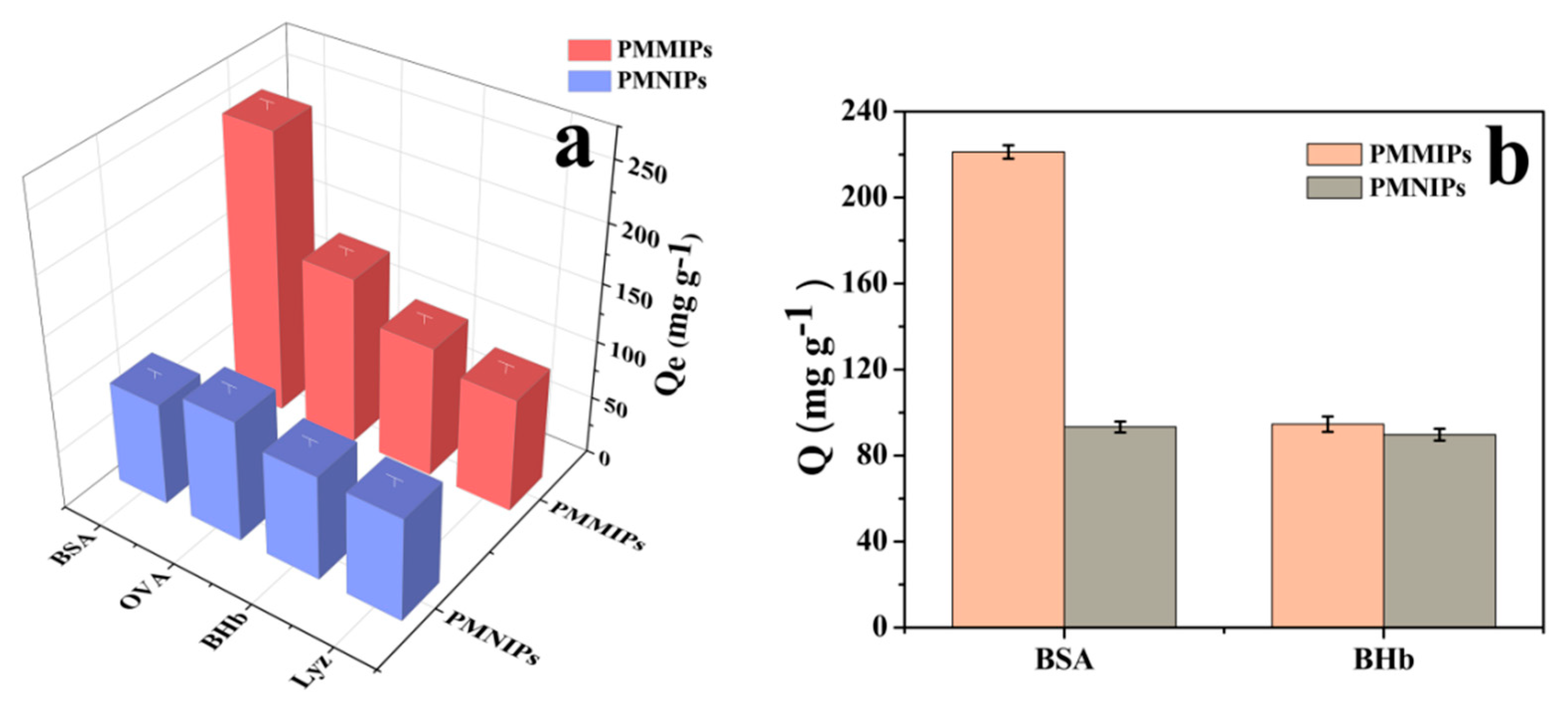
3.4. Adsorption Selectivity
3.5. Competitive Adsorption Tests
3.6. Removal of BSA from Real Samples
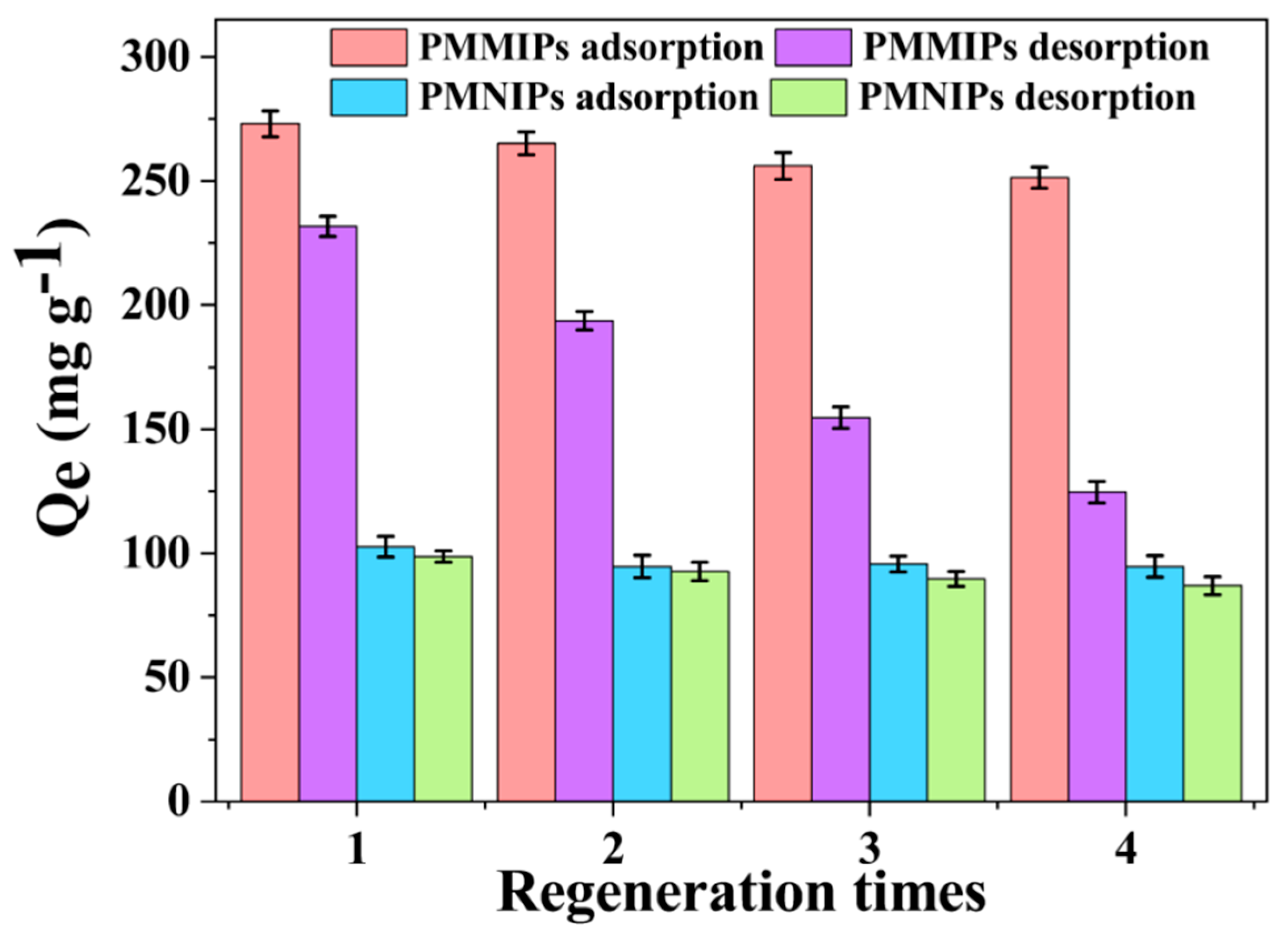
3.7. Regeneration and Reusability of PMMIPs
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Xu, K.; Wang, Y.; Wei, X.; Chen, J.; Xu, P.; Zhou, Y. Preparation of magnetic molecularly imprinted polymers based on a deep eutectic solvent as the functional monomer for specific recognition of lysozyme. Microchim. Acta 2018, 185, 146. [Google Scholar] [CrossRef]
- Niu, M.; Pham-Huy, C.; He, H. Core-shell nanoparticles coated with molecularly imprinted polymers: A review. Microchim. Acta 2016, 183, 2677–2695. [Google Scholar] [CrossRef]
- Chen, L.; Wang, X.; Lu, W.; Wu, X.; Li, J. Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 2016, 45, 2137–2211. [Google Scholar] [CrossRef] [PubMed]
- Saylan, Y.; Yilmaz, F.; Özgür, E.; Derazshamshir, A.; Yavuz, H.; Denizli, A. Molecular imprinting of macromolecules for sensor applications. Sensors 2017, 17, 898. [Google Scholar] [CrossRef] [PubMed]
- Viveiros, R.; Rebocho, S.; Casimiro, T. Green strategies for molecularly imprinted polymer development. Polymer 2018, 10, 306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, J.; Chen, W.; Ma, Y.; Pan, G. Molecularly imprinted polymers as receptor mimics for selective cell recognition. Chem. Soc. Rev. 2018, 47, 5574–5587. [Google Scholar] [CrossRef] [PubMed]
- Viveiros, R.; Lopes, M.I.; Heggie, W.; Casimiro, T. Green approach on the development of lock-and-key polymers for API purification. Chem. Eng. J. 2017, 308, 229–239. [Google Scholar] [CrossRef]
- Xu, J.; Merlier, F.; Avalle, B.; Vieillard, V.; Debré, P.; Haupt, K.; Tse Sum Bui, B. Molecularly Imprinted Polymer Nanoparticles as Potential Synthetic Antibodies for Immunoprotection against HIV. ACS Appl. Mater. Interfaces 2019, 11, 9824–9831. [Google Scholar] [CrossRef]
- Piletska, E.V.; Abd, B.H.; Krakowiak, A.S.; Parmar, A.; Pink, D.L.; Wall, K.S.; Piletsky, S.A. Magnetic high throughput screening system for the development of nano-sized molecularly imprinted polymers for controlled delivery of curcumin. Analyst 2015, 140, 3113–3120. [Google Scholar] [CrossRef]
- Peng, S.; Wang, Y.; Li, N.; Li, C. Enhanced cellular uptake and tumor penetration of nanoparticles by imprinting the “hidden” part of membrane receptors for targeted drug delivery. Chem. Commun. 2017, 53, 11114–11117. [Google Scholar] [CrossRef]
- Wang, C.; Ye, X.; Wang, Z.; Wu, T.; Wang, Y.; Li, C. Molecularly imprinted photo-electrochemical sensor for human epididymis protein 4 based on polymerized ionic liquid hydrogel and gold nanoparticle/ZnCdHgSe quantum dots composite film. Anal. Chem. 2017, 89, 12391–12398. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Pi, F.; Zhang, H.; Sun, J.; Zhang, Y.; Sun, X. A novel molecularly imprinted electrochemical sensor modified with carbon dots, chitosan, gold nanoparticles for the determination of patulin. Biosens. Bioelectron. 2017, 98, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, C.; Yan, Y.; Pan, J.; Xu, P.; Zhao, X. A Ce3+-imprinted functionalized potassium tetratitanate whisker sorbent prepared by surface molecularly imprinting technique for selective separation and determination of Ce3+. Microchim. Acta 2010, 169, 289–296. [Google Scholar] [CrossRef]
- Li, H.; Xie, T.; Ye, L.; Wang, Y.; Xie, C. Core-shell magnetic molecularly imprinted polymer nanoparticles for the extraction of triazophos residues from vegetables. Microchim. Acta 2017, 184, 1011–1019. [Google Scholar] [CrossRef]
- Wu, T.; Wei, X.; Ma, X.; Li, J. Amperometric sensing of L-phenylalanine using a gold electrode modified with a metal organic framework, a molecularly imprinted polymer, and β-cyclodextrin-functionalized gold nanoparticles. Microchim. Acta 2017, 184, 2901–2907. [Google Scholar] [CrossRef]
- Guo, J.; Wang, Y.; Liu, Y.; Zhang, C.; Zhou, Y. Magnetic-graphene based molecularly imprinted polymer nanocomposite for the recognition of bovine hemoglobin. Talanta 2015, 144, 411–419. [Google Scholar] [CrossRef]
- Wei, J.R.; Ni, Y.L.; Zhang, W.; Zhang, Z.Q.; Zhang, J. Detection of glycoprotein through fluorescent boronic acid-based molecularly imprinted polymer. Anal. Chim. Acta 2017, 960, 110–116. [Google Scholar] [CrossRef]
- Guo, L.; Hu, X.; Guan, P.; Du, C.; Wang, D.; Song, D.; Gao, X.; Song, R. Facile preparation of superparamagnetic surface-imprinted microspheres using amino acid as template for specific capture of thymopentin. Appl. Surf. Sci. 2015, 357, 1490–1498. [Google Scholar] [CrossRef]
- Qin, Y.P.; Li, D.Y.; He, X.W.; Li, W.Y.; Zhang, Y.K. Preparation of high-efficiency cytochrome c-imprinted polymer on the surface of magnetic carbon nanotubes by epitope approach via metal chelation and six-membered ring. ACS Appl. Mater. Interfaces 2016, 8, 10155–10163. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, N.; Du, C.; Guan, P.; Gao, X.; Wang, C.; Du, Y.; Ding, S.; Hu, X. Preparation of magnetic epitope imprinted polymer microspheres using cyclodextrin-based ionic liquids as functional monomer for highly selective and effective enrichment of cytochrome c. Chem. Eng. J. 2017, 317, 988–998. [Google Scholar] [CrossRef]
- Zamora-Gálvez, A.; Ait-Lahcen, A.; Mercante, L.A.; Morales-Narváez, E.; Amine, A.; Merkoçi, A. Molecularly Imprinted Polymer-decorated Magnetite Nanoparticles for Selective Sulfonamide Detection. Anal. Chem. 2016, 88, 3578–3584. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Wang, Y.; Wei, X.; Liu, Y.; Zhou, Y. Adsorption and specific recognition of DNA by using imprinted polymer layers grafted onto ionic liquid functionalized magnetic microspheres. Microchim. Acta 2017, 184, 1–9. [Google Scholar] [CrossRef]
- Gai, Q.Q.; Qu, F.; Zhang, T.; Zhang, Y.K. The preparation of bovine serum albumin surface-imprinted superparamagnetic polymer with the assistance of basic functional monomer and its application for protein separation. J. Chromatogr. A 2011, 1218, 3489–3495. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, X.; He, X.; Chen, L.; Zhang, Y. A self-assembled polydopamine film on the surface of magnetic nanoparticles for specific capture of protein. Nanoscale 2012, 4, 3141–3147. [Google Scholar] [CrossRef]
- Ma, F.X.; Hu, H.; Wu, H.B.; Xu, C.Y.; Lou, X.W.D. Formation of Uniform Fe3O4 Hollow Spheres Organized by Ultrathin Nanosheets and Their Excellent Lithium Storage Properties. Adv. Mater. 2015, 27, 4097–4101. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, H.; Li, G.; Liu, Y.; Huang, J.; Yang, D.P. Hydrothermal synthesis of 3D hollow porous Fe3O4 microspheres towards catalytic removal of organic pollutants. Nanoscale Res. Lett. 2014, 9, 648. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wang, Y.; Ma, Y.; Zhang, B.; Zhang, Q. Surface molecularly imprinted thermo-sensitive polymers based on light-weight hollow magnetic microspheres for specific recognition of BSA. Appl. Surf. Sci. 2019, 486, 265–273. [Google Scholar] [CrossRef]
- Fan, J.P.; Yu, J.X.; Yang, X.M.; Zhang, X.H.; Yuan, T.T.; Peng, H.L. Preparation, characterization, and application of multiple stimuli-responsive rattle-type magnetic hollow molecular imprinted poly (ionic liquids) nanospheres (Fe3O4@void@PILMIP) for specific recognition of protein. Chem. Eng. J. 2018, 337, 722–732. [Google Scholar] [CrossRef]
- Fan, D.; Jia, L.; Xiang, H.; Peng, M.; Li, H.; Shi, S. Synthesis and characterization of hollow porous molecular imprinted polymers for the selective extraction and determination of caffeic acid in fruit samples. Food Chem. 2017, 224, 32–36. [Google Scholar] [CrossRef]
- Zhao, T.; Guan, X.; Tang, W.; Ma, Y.; Zhang, H. Preparation of temperature sensitive molecularly imprinted polymer for solid-phase microextraction coatings on stainless steel fiber to measure ofloxacin. Anal. Chim. Acta 2015, 853, 668–675. [Google Scholar] [CrossRef]
- Zhang, X.; Li, H.; Kang, J.; Zhu, X.; Peng, W.; Zhou, H.; Wang, Y. The synthesis of temperature-sensitive molecularly imprinted film on support beads and its application for bovine serum albumin separation. Colloids Surf. A 2016, 504, 367–375. [Google Scholar] [CrossRef]
- Li, X.; Zhang, B.; Li, W.; Lei, X.; Fan, X.; Tian, L.; Zhang, Q. Preparation and characterization of bovine serum albumin surface-imprinted thermosensitive magnetic polymer microsphere and its application for protein recognition. Biosens. Bioelectron. 2014, 51, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Hu, X.; Guan, P.; Gao, B.; Li, J.; Wang, C.; Tang, Y. Preparation of bovine serum albumin imprinting sensitive hydrogels using ionic liquid as co-monomer and stabilizer. Talanta 2014, 121, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhou, J.; Tian, L.; Wang, Y.; Zhang, B.; Zhang, H.; Zhang, Q. Preparation of anti-nonspecific adsorption polydopamine-based surface protein-imprinted magnetic microspheres with the assistance of 2-methacryloyloxyethyl phosphorylcholine and its application for protein recognition. Sens. Actuators B 2017, 241, 413–421. [Google Scholar] [CrossRef]
- Pan, G.; Guo, Q.; Cao, C.; Yang, H.; Li, B. Thermo-responsive molecularly imprinted nanogels for specific recognition and controlled release of proteins. Soft Mater. 2013, 9, 3840–3850. [Google Scholar] [CrossRef]
- Demirel, G.; Özçetin, G.; Turan, E.; Çaykara, E. pH/temperature-sensitive imprinted ionic poly (N-tert-butylacrylamide-co-acrylamide/maleic acid) hydrogels for bovine serum albumin. Macromol. Biosci. 2005, 5, 1032–1037. [Google Scholar] [CrossRef]
- Fu, J.; Chen, Z.; Wang, M.; Liu, S.; Zhang, J.; Zhang, J.; Han, R.; Xu, Q. Adsorption of methylene blue by a high-efficiency adsorbent (polydopamine microspheres): Kinetics, isotherm, thermodynamics and mechanism analysis. Chem. Eng. J. 2015, 259, 53–61. [Google Scholar] [CrossRef]
- Du, P.; Wang, T.; Liu, P. Double-walled hollow polymeric microspheres with independent pH and temperature dual-responsive and magnetic-targeting function from onion-shaped core-shell structures. Colloids Surf. B 2013, 102, 1–8. [Google Scholar] [CrossRef]
- Fu, G.; Zhao, J.; Yu, H.; Liu, L.; He, B. Bovine serum albumin-imprinted polymer gels prepared by graft copolymerization of acrylamide on chitosan. React. Funct. Polym. 2017, 67, 442–450. [Google Scholar] [CrossRef]
- Zhu, X.; Li, H.; Zhou, H.; Zhong, S. Fabrication and evaluation of protein imprinted polymer based on magnetic halloysite nanotubes. RSC Adv. 2015, 5, 66147–66154. [Google Scholar] [CrossRef]
- He, H.; Fu, G.; Wang, Y.; Chai, Z.; Jiang, Y.; Chen, Z. Imprinting of protein over silica nanoparticles via surface graft copolymerization using low monomer concentration. Biosens. Bioelectron. 2010, 26, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Kong, X.; Wang, X.; He, X.; Chen, L.; Zhang, Y. Preparation and characterization of uniformly sized molecularly imprinted polymers functionalized with core-shell magnetic nanoparticles for the recognition and enrichment of protein. J. Mater. Chem. 2011, 21, 17863–17871. [Google Scholar] [CrossRef]
- Lin, Z.; Xia, Z.; Zheng, J.; Zheng, D.; Chen, G. Synthesis of uniformly sized molecularly imprinted polymer-coated silica nanoparticles for selective recognition and enrichment of lysozyme. J. Mater. Chem. 2012, 22, 17914–17922. [Google Scholar] [CrossRef]
- Li, X.; Zhou, J.; Tian, L.; Li, W.; Ali, Z.; Ali, N.; Zhang, B.; Zhang, H.; Zhang, Q. Effect of crosslinking degree and thickness of thermosensitive imprinted layers on recognition and elution efficiency of protein imprinted magnetic microspheres. Sens. Actuators B 2015, 225, 436–445. [Google Scholar] [CrossRef]
- Dong, X.; Ma, Y.; Hou, C.; Zhang, B.; Zhang, H.; Zhang, Q. Preparation of pH and temperature dual-sensitive molecularly imprinted polymers based on chitosan and N-isopropylacrylamide for recognition of bovine serum albumin. Polym. Int. 2019, 68, 955–963. [Google Scholar] [CrossRef]
- Simonin, J.P. On the comparison of pseudo-first order and pseudo-second order rate laws in the modeling of adsorption kinetics. Chem. Eng. J. 2016, 300, 254–263. [Google Scholar] [CrossRef] [Green Version]
- Jang, H.M.; Yoo, S.; Choi, Y.K.; Park, S.; Kan, E. Adsorption isotherm, kinetic modeling and mechanism of tetracycline on Pinus taeda-derived activated biochar. Bioresour. Technol. 2018, 259, 24–31. [Google Scholar] [CrossRef]
- Fallah, N.; Taghizadeh, M.; Hassanpour, S. Selective adsorption of Mo(VI) ions from aqueous solution using a surface-grafted Mo(VI) ion imprinted polymer. Polymer 2018, 144, 80–91. [Google Scholar] [CrossRef]
- Yuan, Y.; Yang, C.; Lv, T.; Qiao, F.; Zhou, Y.; Yan, H. Green synthesis of hydrophilic protein-imprinted resin with specific recognition of bovine serum albumin in aqueous matrix. Anal. Chim. Act. 2018, 1033, 213–220. [Google Scholar] [CrossRef]
- Yang, Y.Q.; He, X.W.; Wang, Y.Z.; Li, W.Y.; Zhang, Y.K. Epitope imprinted polymer coating CdTe quantum dots for specific recognition and direct fluorescent quantification of the target protein bovine serum albumin. Biosens. Bioelectron. 2014, 54, 266–272. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Liu, H.; Deng, Z.; Chen, W.; Li, T.; Zhang, Y.; Zhang, Z.; He, Y.; Tan, Z.; Zhong, S. PEGylated Thermo-Sensitive Bionic Magnetic Core-Shell Structure Molecularly Imprinted Polymers Based on Halloysite Nanotubes for Specific Adsorption and Separation of Bovine Serum Albumin. Polymers 2020, 12, 536. https://doi.org/10.3390/polym12030536
Li X, Liu H, Deng Z, Chen W, Li T, Zhang Y, Zhang Z, He Y, Tan Z, Zhong S. PEGylated Thermo-Sensitive Bionic Magnetic Core-Shell Structure Molecularly Imprinted Polymers Based on Halloysite Nanotubes for Specific Adsorption and Separation of Bovine Serum Albumin. Polymers. 2020; 12(3):536. https://doi.org/10.3390/polym12030536
Chicago/Turabian StyleLi, Xiufang, Hui Liu, Zhiwei Deng, Wenqing Chen, Tianhao Li, Yunshan Zhang, Zhuomin Zhang, Yao He, Zhijian Tan, and Shian Zhong. 2020. "PEGylated Thermo-Sensitive Bionic Magnetic Core-Shell Structure Molecularly Imprinted Polymers Based on Halloysite Nanotubes for Specific Adsorption and Separation of Bovine Serum Albumin" Polymers 12, no. 3: 536. https://doi.org/10.3390/polym12030536





