Structural Features of Three Hetero-Galacturonans from Passiflora foetida Fruits and Their in Vitro Immunomodulatory Effects
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Chemicals
2.2. Separation and Purification of Salt-Eluted Polysaccharides
2.3. UV-Visible and FT-IR Spectra Analysis
2.4. Molecular Weight Analysis
2.5. Monosaccharide Composition Analysis
2.6. Molecular Morphological analysis
2.7. Methylation Analysis
2.8. NMR Analysis
2.9. Immunomodulatory Effect Analysis
2.10. Statistical Analysis
3. Results and Discussion
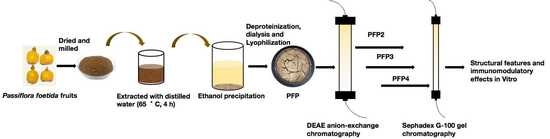
3.1. Extraction and Purification of the Polysaccharide Fractions
3.2. Analysis of UV–Vis and FT-IR Spectra
3.3. Morphological Properties
3.4. Molecular Weight and Monosaccharide Composition
3.5. Methylation Analysis
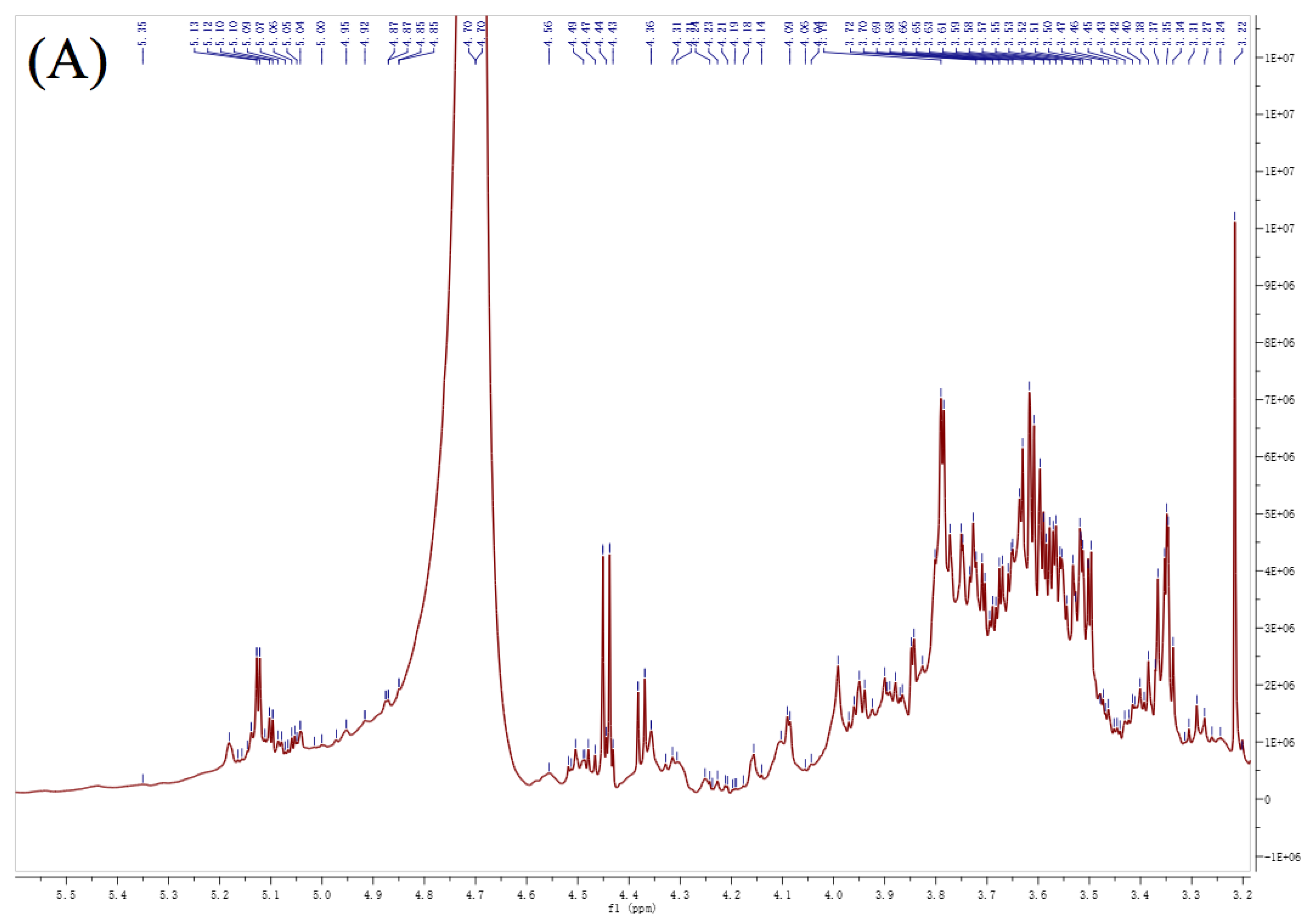
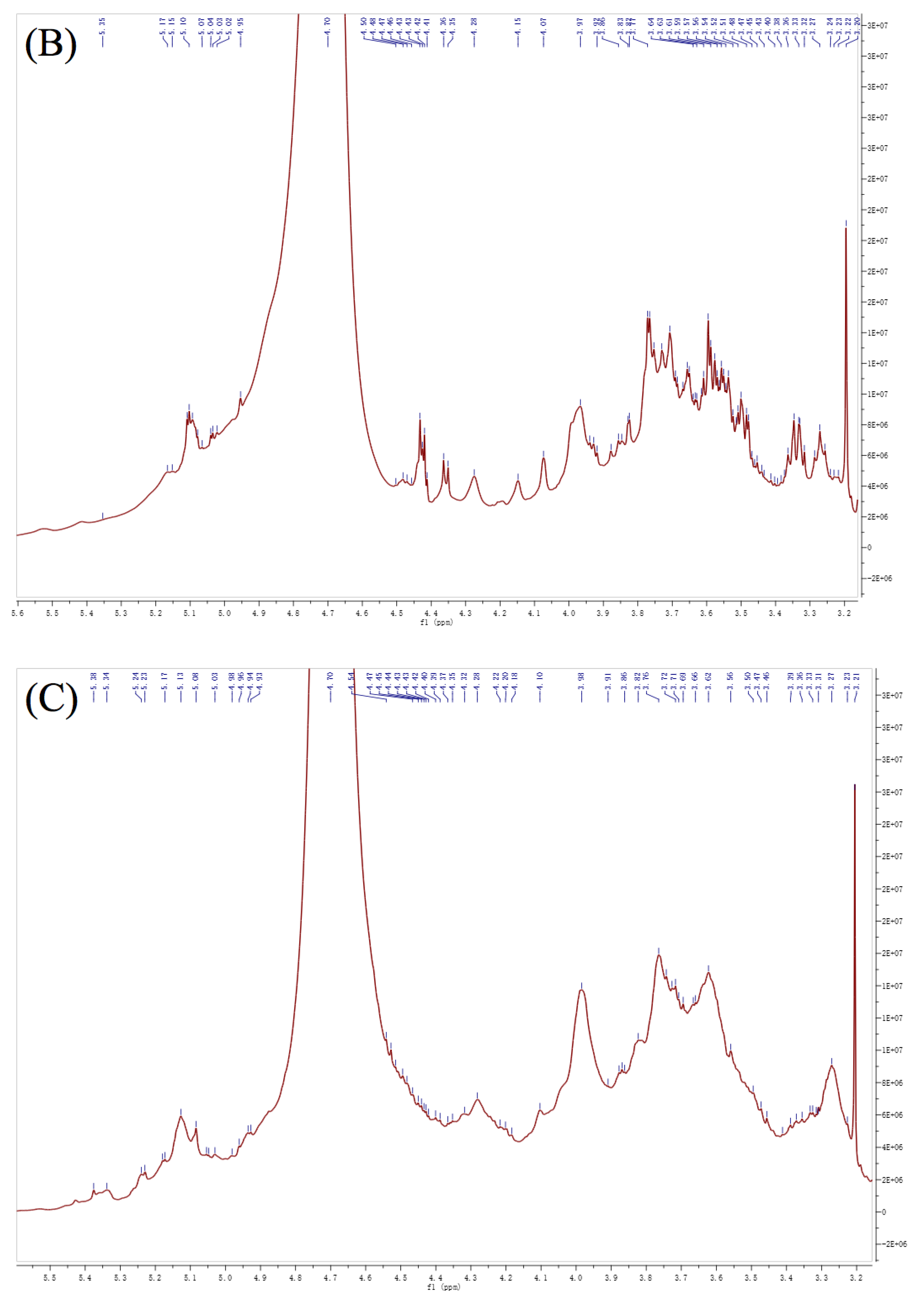
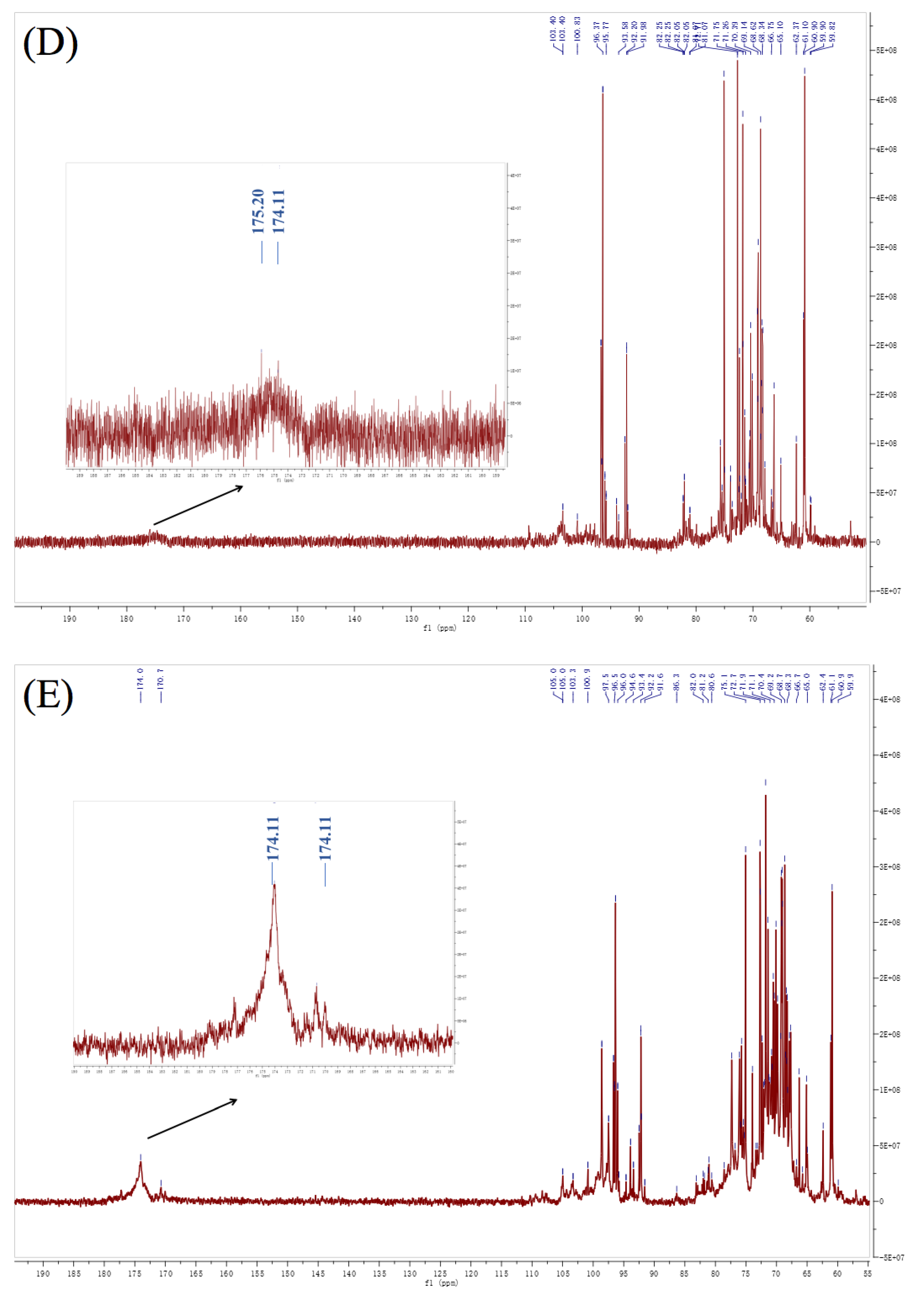
3.6. NMR Analysis
3.7. Immunomodulatory Effect
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mohanasundari, C.; Natarajan, D.; Srinivasan, K.; Umamaheswari, S.; Ramachandran, A. Antibacterial properties of Passiflora foetida L.—A common exotic medicinal plant. Afr. J. Biotechnol. 2007, 6, 2650–2653. [Google Scholar]
- Ahmad, N.; Chillara, R.; Kushwaha, P.; Khedgikar, V.; Karvande, A.; Choudhary, D.; Adhikary, S.; Maurya, R.; Trivedi, R. Evaluation of anti-osteoporotic activity of butanolic fraction from Passiflora foetida in ovariectomy-induced bone loss in mice. Biomed. Pharmacother. 2017, 88, 804–813. [Google Scholar] [CrossRef]
- Nguyen, T.Y.; To, D.C.; Tran, M.H.; Lee, J.S.; Lee, J.H.; Kim, J.A.; Woo, M.H.; Min, B.S. Anti-inflammatory flavonoids isolated from Passiflora foetida. Nat. Prod. Commun. 2015, 10, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Wei, X.Q.; Li, M.Y.; Duan, X.W.; Sun, Y.M.; Yang, R.L.; Huang, R.; Wang, H. Nutritional composition and antioxidant properties of the fruits of a Chinese wild Passiflora foetida. Molecules 2018, 23, 459. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Li, J.; Zhu, J.; Huang, C.; Bi, S.; Song, L.; Hu, X.; Yu, R. Structural characterization and immunomodulatory activity of a novel polysaccharide from Ficus carica. Food Funct. 2018, 9, 3930–3943. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wen, L.; Zhao, Y.; Jiang, Y.; Tian, M.; Liu, H.; Liu, J.; Yang, B. Structure identification of an arabinogalacturonan in Citrus reticulata Blanco ‘Chachiensis’ peel. Food Hydrocoll. 2018, 84, 481–488. [Google Scholar] [CrossRef]
- Yang, J.; Tu, J.; Liu, H.; Wen, L.; Jiang, Y.; Yang, B. Identification of an immunostimulatory polysaccharide in banana. Food Chem. 2019, 277, 46–53. [Google Scholar] [CrossRef]
- Yue, H.; Xu, Q.; Li, X.; Elango, J.; Wu, W.; Xu, J. Physicochemical characterization and immunomodulatory activity of a novel acid polysaccharide from Solanum muricatum. Polymers 2019, 11, 1972. [Google Scholar] [CrossRef]
- Cordeiro Caillot, A.R.; de Lacerda Bezerra, I.; Palhares, L.; Santana-Filho, A.P.; Chavante, S.F.; Sassaki, G.L. Structural characterization of blackberry wine polysaccharides and immunomodulatory effects on LPS-activated RAW 264.7 macrophages. Food Chem. 2018, 257, 143–149. [Google Scholar] [CrossRef]
- Cao, J.; Tang, D.; Wang, Y.; Li, X.; Hong, L.; Sun, C. Characteristics and immune-enhancing activity of pectic polysaccharides from sweet cherry (Prunus avium). Food Chem. 2018, 254, 47–54. [Google Scholar] [CrossRef]
- Nie, C.; Zhu, P.; Ma, S.; Wang, M.; Hu, Y. Purification, characterization and immunomodulatory activity of polysaccharides from stem lettuce. Carbohydr. Polym. 2018, 188, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhang, X.; Wang, S.; Guo, Q.; Li, Z.; Liu, H.; Wang, C. Structural characterisation and immunomodulatory activity of polysaccharides from white asparagus skin. Carbohydr. Polym. 2020, 227, 115314. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhu, M.; Hao, H.; Deng, J.; Li, M.; Sun, Y.; Yang, R.; Wang, H.; Huang, R. Structure characterization of a novel polysaccharide from Chinese wild fruits (Passiflora foetida) and its immune-enhancing activity. Int. J. Biol. Macromol. 2019, 136, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Mecozzi, M. Estimation of total carbohydrate amount in environmental samples by the phenol-sulphuric acid method assisted by multivariate calibration. Chemometr. Intell. Lab. 2005, 9, 84–90. [Google Scholar] [CrossRef]
- Taylor, R.L.; Conrad, H.E. Stoichiometric depolymerization of polyuronides and glycosaminoglycuronans to monosaccharides following reduction of their carbodiimide-activated carboxyl group. Biochemistry 1972, 11, 1383–1388. [Google Scholar] [CrossRef] [PubMed]
- Anumula, K.R.; Taylor, P.B. A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal. Biochem. 1992, 203, 101–108. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, Y.; Wu, Q.; John, A.; Jiang, Y.; Yang, J.; Liu, H.; Yang, B. Structure characterisation of polysaccharides in vegetable "okra" and evaluation of hypoglycemic activity. Food Chem. 2018, 242, 211–216. [Google Scholar] [CrossRef]
- Ji, X.; Liu, F.; Peng, Q.; Wang, M. Purification, structural characterization, and hypolipidemic effects of a neutral polysaccharide from Ziziphus Jujuba cv. Muzao. Food Chem. 2018, 245, 1124–1130. [Google Scholar] [CrossRef]
- Liang, X.X.; Gao, Y.Y.; Pan, Y.; Zou, Y.F.; He, M.; He, C.L.; Li, L.X.; Yin, Z.Q.; Lv, C. Purification, chemical characterization and antioxidant activities of polysaccharides isolated from Mycena dendrobii. Carbohydr. Polym. 2019, 203, 45–51. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, B.; Xiao, J.; Huang, Q.; Li, C.; Fu, X. Physicochemical, functional, and biological properties of water-soluble polysaccharides from Rosa roxburghii Tratt fruit. Food Chem. 2018, 249, 127–135. [Google Scholar] [CrossRef]
- Jiao, Y.; Hua, D.; Huang, D.; Zhang, Q.; Yan, C. Characterization of a new heteropolysaccharide from green guava and its application as an alpha-glucosidase inhibitor for the treatment of type II diabetes. Food Funct. 2018, 9, 3997–4007. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Min, T.; Li, X.; Li, L.; Lai, F.; Tang, Y.; Yang, X. Physicochemical properties and antioxidant activities of acidic polysaccharides from wampee seeds. Int. J. Biol. Macromol. 2013, 59, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.C.; Freitas, A.L.P.; Barros, F.C.N.; Lins, K.O.A.L.; Alves, A.P.N.N.; Alencar, N.M.N.; Figueiredo, I.S.T.; Pessoa, C.; Moraes, M.O.; Costa-Lotufo, L.V.; et al. Polysaccharide isolated from Passiflora edulis: Characterization and antitumor properties. Carbohydr. Polym. 2012, 87, 139–145. [Google Scholar] [CrossRef]
- Ji, X.; Zhang, F.; Zhang, R.; Liu, F.; Peng, Q.; Wang, M. An acidic polysaccharide from Ziziphus Jujuba cv. Muzao: Purification and structural characterization. Food Chem. 2019, 274, 494–499. [Google Scholar] [CrossRef] [PubMed]
- Molaei, H.; Jahanbin, K. Structural features of a new water-soluble polysaccharide from the gum exudates of Amygdalus scoparia Spach (Zedo gum). Carbohydr. Polym. 2018, 182, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, Y.; Liu, C.; Li, E.; Gao, Z.; Liu, C.; Gu, W.; Huang, Y.; Liu, J.; Wang, D.; et al. Structural characterization of an acidic Epimedium polysaccharide and its immune-enhancement activity. Carbohydr. Polym. 2016, 138, 134–142. [Google Scholar] [CrossRef]
- Yao, Y.; Yao, J.; Du, Z.; Wang, P.; Ding, K. Structural elucidation and immune-enhancing activity of an arabinogalactan from flowers of Carthamus tinctorius L. Carbohydr. Polym. 2018, 202, 134–142. [Google Scholar] [CrossRef]
- Zhang, H.; Li, W.J.; Nie, S.P.; Chen, Y.; Wang, Y.X.; Xie, M.Y. Structural characterisation of a novel bioactive polysaccharide from Ganoderma atrum. Carbohydr. Polym. 2012, 88, 1047–1054. [Google Scholar] [CrossRef]
- Wei, C.; Zhang, Y.; He, L.; Cheng, J.; Li, J.; Tao, W.; Mao, G.; Zhang, H.; Linhardt, R.J.; Ye, X.; et al. Structural characterization and anti-proliferative activities of partially degraded polysaccharides from peach gum. Carbohydr. Polym. 2019, 203, 193–202. [Google Scholar] [CrossRef]
- Guo, Q.; Du, J.; Jiang, Y.; Goff, H.D.; Cui, S.W. Pectic polysaccharides from hawthorn: Physicochemical and partial structural characterization. Food Hydrocoll. 2019, 90, 146–153. [Google Scholar] [CrossRef]
- Bi, S.; Jing, Y.; Zhou, Q.; Hu, X.; Zhu, J.; Guo, Z.; Song, L.; Yu, R. Structural elucidation and immunostimulatory activity of a new polysaccharide from Cordyceps militaris. Food Funct. 2018, 9, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Schepetkin, I.A.; Quinn, M.T. Immunomodulatory activity of acidic polysaccharides isolated from Tanacetum vulgare L. Int. Immunopharmcol. 2007, 7, 1639–1650. [Google Scholar] [CrossRef] [PubMed]
- Ho, G.T.T.; Zou, Y.; Aslaksen, T.H.; Wangensteen, H.; Barsett, H. Structural characterization of bioactive pectic polysaccharides from elderflowers (Sambuci flos). Carbohydr. Polym. 2016, 135, 128–137. [Google Scholar] [CrossRef] [PubMed]







| Parameters | PFP2 | PFP3 | PFP4 |
|---|---|---|---|
| Molecular and conformation parameters | |||
| Mn (g/mol) | (3.86 ± 0.01) × 104 | (1.15 ± 0.02) ×104 | (5.63 ± 0.01) ×104 |
| Mw (g/mol) | (6.11 ± 0.01) × 104 | (4.37 ± 0.01) ×104 | (3.48 ± 0.01) ×105 |
| Mw/Mn | 1.58 ± 0.01 | 3.82 (±0.02) | 6.18 (±0.01) |
| Monosaccharide compositions ratio (%) | |||
| Fucose | 1.61 | 0.99 | 1.82 |
| Arabinose | 17.94 | 14.68 | 15.78 |
| Galactose | 26.99 | 18.37 | 21.56 |
| Glucose | 5.75 | 2.81 | 7.37 |
| Xylose | 5.62 | 9.14 | 4.61 |
| Mannose | 5.99 | 3.15 | 4.75 |
| Ribose | 0.02 | NAa | NAa |
| Galacturonic acid | 30.76 | 47.34 | 36.96 |
| Glucuronic acid | 5.32 | 3.52 | 7.15 |
| Retention Time (min) | Methylated Sugars | Type of Linkage | Molar Ratios (%) | Mass Fragments | ||
|---|---|---|---|---|---|---|
| PFP2 | PFP3 | PFP4 | ||||
| 8.136 | 2, 3, 5-Me3-Araf | T-Araf | 1.46 | 2.21 | 3.32 | 43, 71, 88, 101, 118, 129, 161, 191 |
| 9.164 | 2, 3, 5-Me3-Xylp | T-Xylp | 8.03 | 16.45 | 13.83 | 43, 71, 87, 101, 102, 118, 129, 146, 162 |
| 13.675 | 2, 3, 4, 6-Me4-Galp | T-Galp | 16.42 | 20.37 | 15.32 | 43, 71, 87, 102, 118, 129, 145, 161, 205 |
| 15.544 | 3, 4, 6-Me3-Galp | 1, 2-Galp | ND | 7.11 | ND | 43, 71, 88, 100, 129, 145, 161, 190, 205 |
| 15.927 | 2, 3, 6-Me3-GalpAc | 1, 4-GalpAc | 12.96 | 30.21 | 15.69 | 43, 75, 99, 117, 159, 203, 233, 301, 318 |
| 16.143 | 2, 3, 6-Me3-Manp | 1, 4-Manp | 7.71 | 3.54 | 10.05 | 43, 71, 87, 102, 118, 162, 203, 233, 277 |
| 16.468 | 2, 4, 6-Me3-Galp | 1, 3-Galp | 15.46 | 5.36 | 12.77 | 43, 71, 87, 101, 118, 129, 161, 234, 277 |
| 16.635 | 2, 3, 4-Me3-Glcp | 1, 6-Glcp | 2.51 | ND | 6.58 | 43, 71, 87, 99, 102, 118, 129, 162, 189 |
| 17.607 | 2, 3, 4-Me3-Galp | 1, 6-Galp | 8.99 | ND | 6.91 | 43, 71, 87, 99, 102, 118, 129, 189, 233, 271 |
| 18.045 | 2, 6-Me2-GalpAc | 1, 3, 4-GalpAc | 9.69 | 12.98 | 7.49 | 43, 87, 118, 129, 161, 262, 305 |
| 18.104 | 4, 6-Me2-Galp | 1, 2, 3-Galp | 1.07 | ND | ND | 43, 86, 101, 129, 161, 202, 262 |
| 20.47 | 2, 4-Me2-Galp | 1, 3, 6-Galp | 15.70 | 1.77 | 8.04 | 43, 87, 101, 118, 129, 189, 234, 305 |
| Residues | Linkage | 1 | 2 | 3 | 4 | 5 | 6 | ||
|---|---|---|---|---|---|---|---|---|---|
| A | α-L-Araf | PFP2 | H | 5.39 | 4.26 | 4.09 | 4.14 | 3.81 | |
| C | 109.45 | 81.21 | 77.67 | 83.33 | 63.09 | ||||
| PFP3 | H | 5.36 | 4.26 | 4.11 | 4.14 | 3.81 | |||
| C | 109.16 | 81.40 | 77.71 | 83.33 | 62.84 | ||||
| PFP4 | H | 5.35 | 4.24 | 4.12 | 4.13 | 3.82 | |||
| C | 109.63 | 81.47 | 76.69 | 83.07 | 62.80 | ||||
| B | β-D-Xylp | PFP2 | H | 4.67 | 3.38 | 3.49 | 3.67 | 3.79 | |
| C | 105.24 | 74.91 | 76.26 | 70.93 | 66.26 | ||||
| PFP3 | H | 4.66 | 3.38 | 3.45 | 3.66 | 3.78 | |||
| C | 104.90 | 73.28 | 76.85 | 71.74 | 66.02 | ||||
| PFP4 | H | 4.64 | 3.38 | 3.50 | 3.66 | 3.76 | |||
| C | 105.27 | 74.70 | 76.20 | 72.60 | 66.12 | ||||
| C | β-D-Galp | PFP2 | H | 4.57 | 3.55 | 3.78 | 3.97 | 4.05 | 3.95 |
| C | 103.73 | 72.18 | 72.89 | 70.79 | 74.73 | 62.35 | |||
| PFP3 | H | 4.56 | 3.53 | 3.77 | 4.00 | 4.05 | 3.95 | ||
| C | 103.31 | 71.73 | 72.50 | 69.20 | 72.14 | 62.84 | |||
| PFP4 | H | 4.58 | 3.52 | 3.75 | 4.01 | 4.05 | 3.92 | ||
| C | 103.14 | 72.11 | 72.80 | 69.40 | 73.10 | 62.80 | |||
| D | →4)-α-L-GalpAc-(1→ | PFP2 | H | 5.07 | 3.78 | 4.42 | 4.05 | 4.76 | |
| C | 101.21 | 70.00 | 79.43 | 70.57 | 77.85 | 175.20 | |||
| PFP3 | H | 5.02 | 3.77 | 4.40 | 4.05 | 4.74 | |||
| C | 99.03 | 69.58 | 79.17 | 70.05 | 77.34 | 174.00 | |||
| PFP4 | H | 5.14 | 3.77 | 4.40 | 4.07 | 4.77 | |||
| C | 98.25 | 70.31 | 79.40 | 70.02 | 77.49 | 174.80 | |||
| E | →4)-α-L-Manp-(1→ | PFP2 | H | 5.33 | 4.17 | 3.67 | 3.84 | 3.75 | 4.04 |
| C | 107.21 | 69.48 | 75.69 | 80.87 | 71.33 | 73.75 | |||
| PFP3 | H | 5.32 | 4.17 | 3.67 | 3.84 | 3.75 | 4.00 | ||
| C | 106.34 | 67.90 | 75.43 | 80.13 | 71.50 | 71.60 | |||
| PFP4 | H | 5.30 | 4.17 | 3.67 | 3.83 | 3.75 | 3.99 | ||
| C | 107.14 | 68.90 | 75.78 | 80.89 | 71.61 | 70.39 | |||
| F | →3)-α-L-Galp-(1→ | PFP2 | H | 5.14 | 3.78 | 4.07 | 3.94 | 3.49 | 3.63 |
| C | 99.55 | 70.00 | 83.61 | 61.33 | 73.32 | 60.00 | |||
| PFP3 | H | 5.22 | 3.77 | 4.05 | 3.93 | 3.48 | 3.60 | ||
| C | 100.96 | 69.58 | 83.91 | 60.88 | 73.05 | 59.55 | |||
| PFP4 | H | 5.21 | 3.77 | 4.05 | 3.93 | 3.49 | 3.59 | ||
| C | 101.54 | 70.31 | 82.07 | 61.12 | 73.49 | 59.84 | |||
| G | →3, 4)-β-D-GalpAc-(1→ | PFP2 | H | 4.83 | 3.75 | 4.35 | 4.51 | 5.19 | |
| C | 103.99 | 71.33 | 68.61 | 79.92 | 70.88 | 174.11 | |||
| PFP3 | H | 4.92 | 3.75 | 4.34 | 4.49 | 5.18 | |||
| C | 101.22 | 71.50 | 68.22 | 79.35 | 70.48 | 170.70 | |||
| PFP4 | H | 4.97 | 3.75 | 4.34 | 4.47 | 5.17 | |||
| C | 102.64 | 71.61 | 68.60 | 81.77 | 70.81 | 173.20 | |||
| H | →3, 6)-β-D-Galp-(1→ | PFP2 | H | 4.90 | 3.67 | 3.73 | 4.17 | 3.85 | 3.89 |
| C | 100.91 | 70.93 | 79.29 | 69.48 | 76.28 | 71.98 | |||
| PFP3 | H | 4.81 | 3.66 | 3.74 | 4.17 | 3.80 | 3.88 | ||
| C | 103.58 | 71.74 | 79.95 | 69.48 | 76.42 | 71.96 | |||
| PFP4 | H | 4.82 | 3.66 | 3.74 | 4.17 | 3.81 | 3.88 | ||
| C | 104.12 | 72.60 | 81.47 | 68.90 | 76.08 | 72.30 | |||
| I | →6)-α-L-Glcp-(1→ | PFP2 | H | 4.96 | 3.89 | 3.47 | 3.32 | 3.67 | 3.97 |
| C | 104.01 | 71.98 | 75.13 | 68.55 | 75.69 | 70.79 | |||
| PFP4 | H | 4.94 | 3.88 | 3.45 | 3.32 | 3.67 | 4.01 | ||
| C | 104.05 | 72.30 | 74.90 | 69.21 | 75.78 | 69.40 | |||
| J | →6)-β-D-Galp-(1→ | PFP2 | H | 4.76 | 3.55 | 3.74 | 3.97 | 3.89 | 4.04 |
| C | 104.35 | 75.921 | 76.55 | 70.79 | 71.98 | 73.75 | |||
| PFP4 | H | 4.74 | 3.62 | 3.76 | 4.01 | 3.88 | 3.99 | ||
| C | 104.59 | 75.89 | 77.08 | 69.40 | 72.30 | 70.39 | |||
| K | →2, 3)-α-L-Galp-(1→ | PFP2 | H | 5.07 | 4.07 | 3.98 | 3.75 | 3.79 | 3.81 |
| C | 101.85 | 83.61 | 82.13 | 71.33 | 68.05 | 63.09 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Y.; Wen, P.; Hao, H.; Zhu, M.; Sun, Y.; Zou, Y.; Requena, T.; Huang, R.; Wang, H. Structural Features of Three Hetero-Galacturonans from Passiflora foetida Fruits and Their in Vitro Immunomodulatory Effects. Polymers 2020, 12, 615. https://doi.org/10.3390/polym12030615
Song Y, Wen P, Hao H, Zhu M, Sun Y, Zou Y, Requena T, Huang R, Wang H. Structural Features of Three Hetero-Galacturonans from Passiflora foetida Fruits and Their in Vitro Immunomodulatory Effects. Polymers. 2020; 12(3):615. https://doi.org/10.3390/polym12030615
Chicago/Turabian StyleSong, Ya, Peng Wen, Huili Hao, Minqian Zhu, Yuanming Sun, Yuxiao Zou, Teresa Requena, Riming Huang, and Hong Wang. 2020. "Structural Features of Three Hetero-Galacturonans from Passiflora foetida Fruits and Their in Vitro Immunomodulatory Effects" Polymers 12, no. 3: 615. https://doi.org/10.3390/polym12030615
APA StyleSong, Y., Wen, P., Hao, H., Zhu, M., Sun, Y., Zou, Y., Requena, T., Huang, R., & Wang, H. (2020). Structural Features of Three Hetero-Galacturonans from Passiflora foetida Fruits and Their in Vitro Immunomodulatory Effects. Polymers, 12(3), 615. https://doi.org/10.3390/polym12030615