Effect of Pollyallylamine on Alcoholdehydrogenase Structure and Activity
Abstract
1. Introduction
2. Materials
2.1. Registration of the Kinetics of Enzymatic Reactions
2.2. Determination of ADH Activity
2.3. Determination of Fluorescence Intensity of ADH
2.4. Preparation of PMCs
2.5. Adsorption Protein Encapsulation
2.6. Statistical Analysis of Data
3. Results
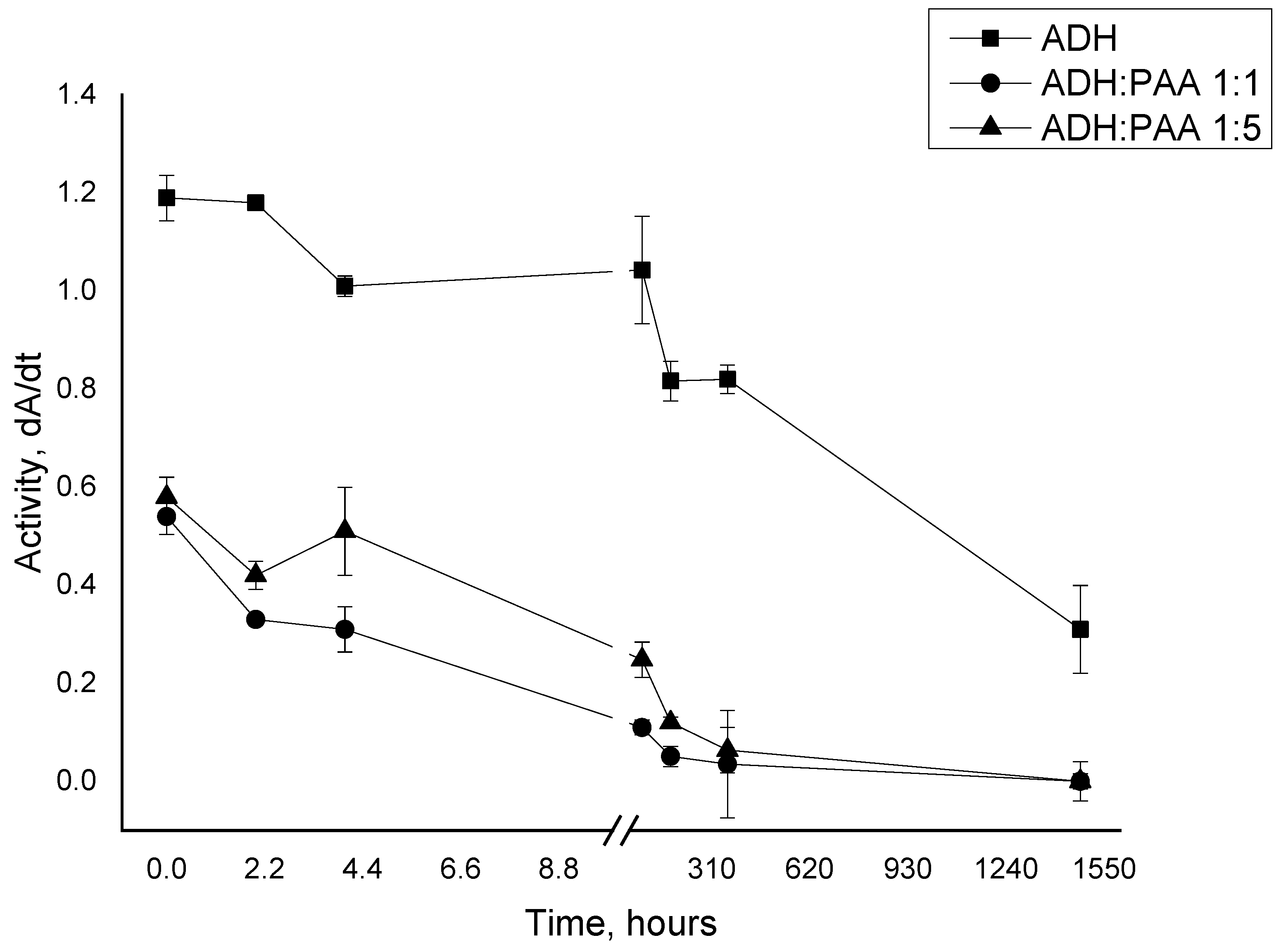
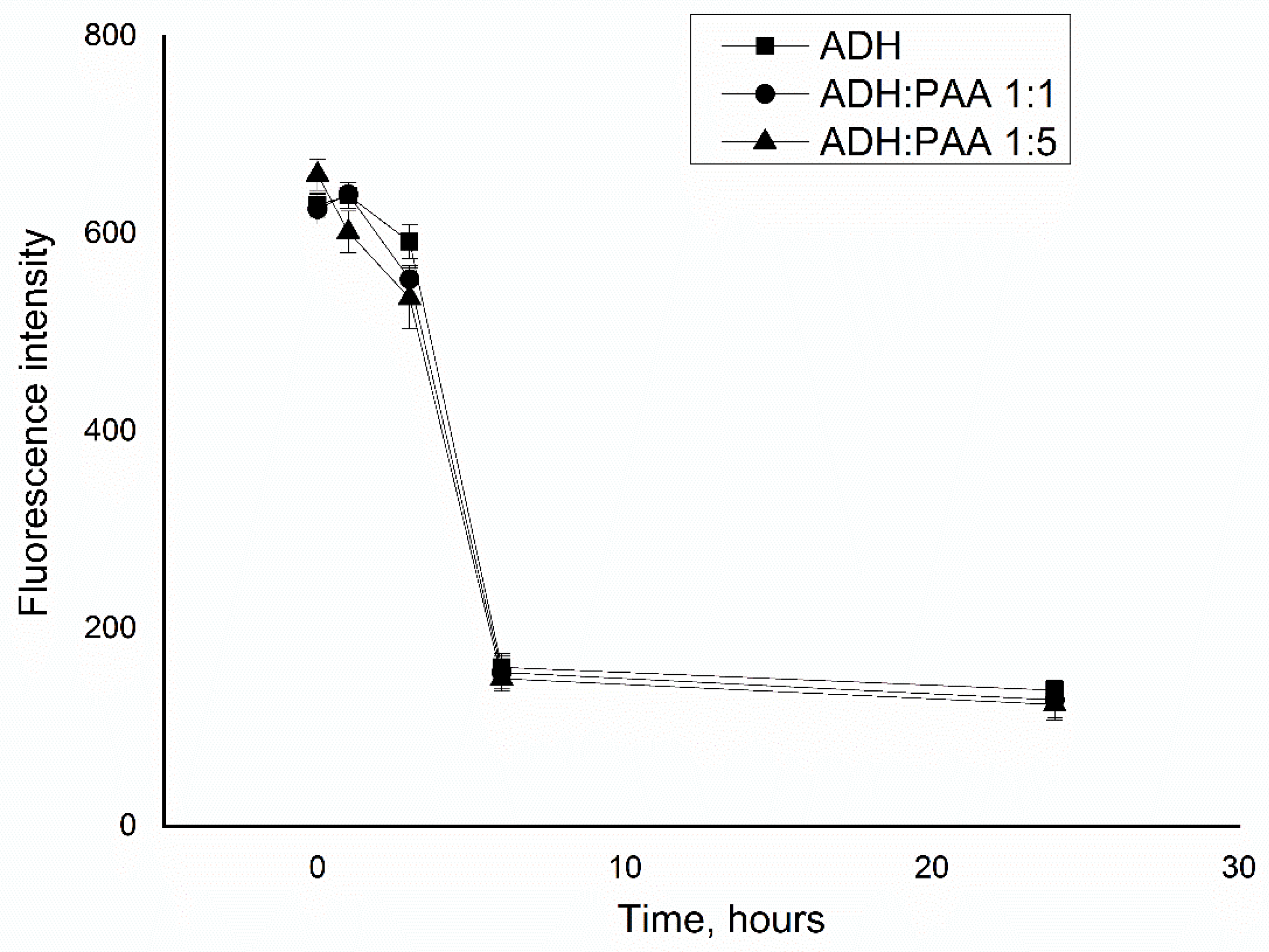
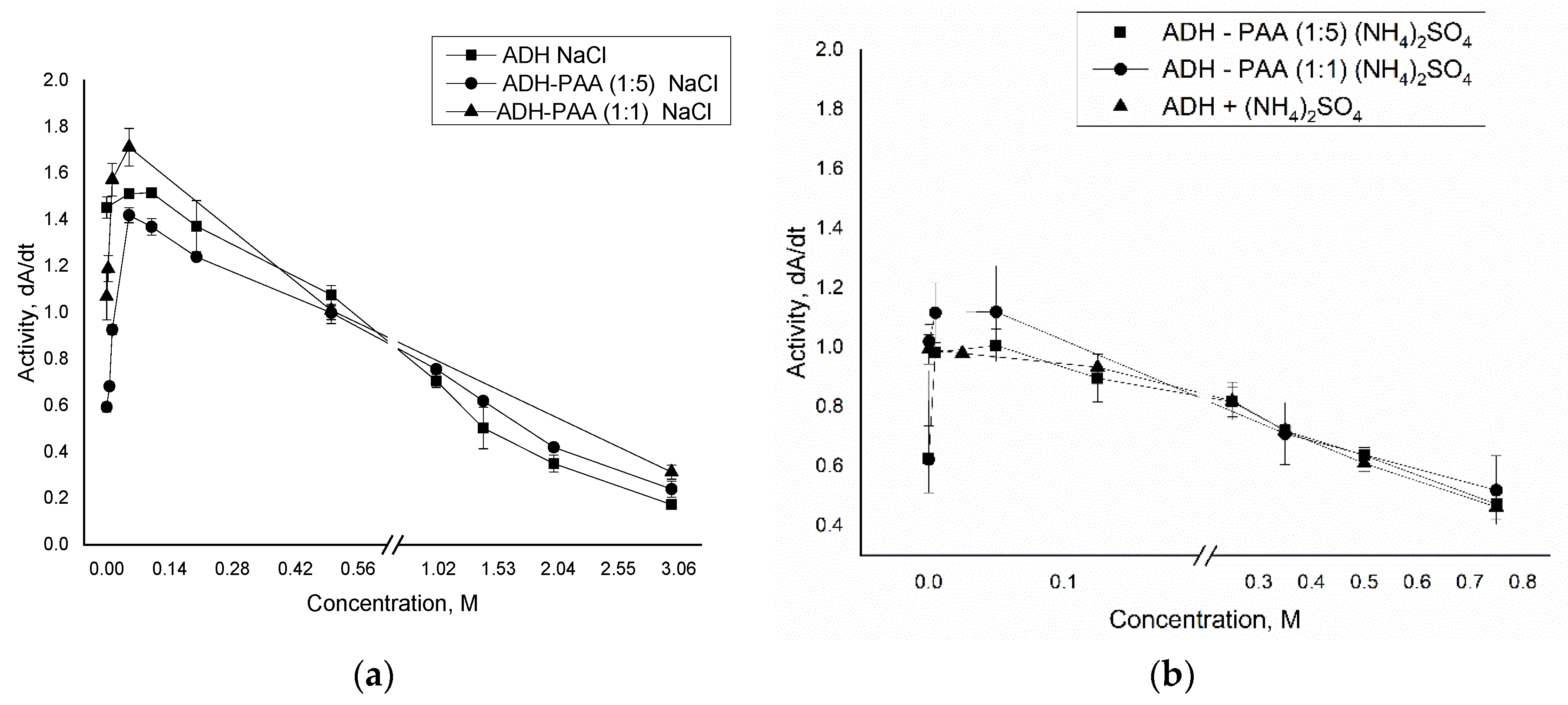
3.1. Study of the Interaction of Polyallylamine with Alcohol Dehydrogenase
3.2. Encapsulation of ADH in Polyelectrolyte Microcapsules
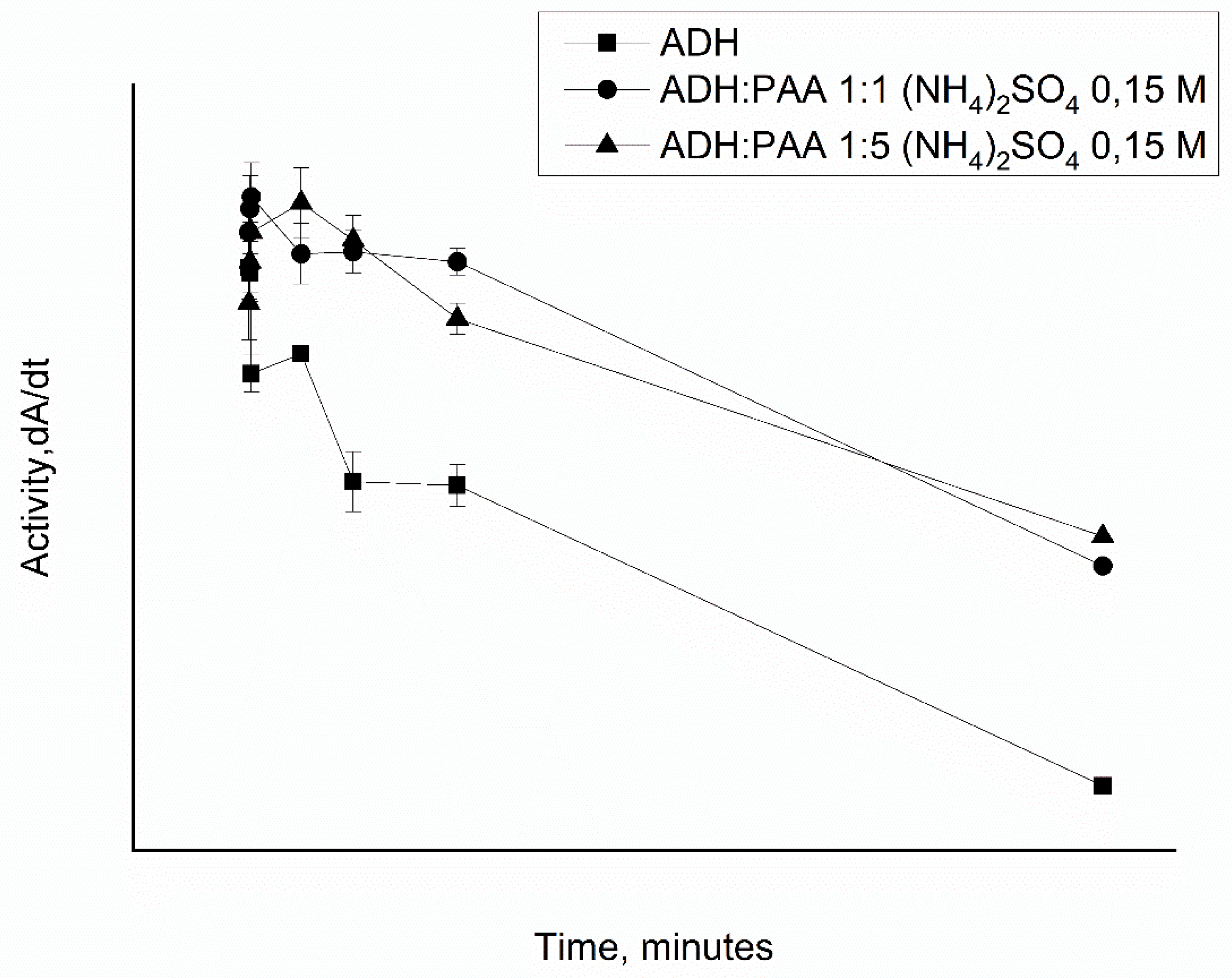
3.3. Comparison of Alcohol Dehydrogenase Affinity in Complex with Polyallylamine and in a Polyelectrolyte Microcapsule
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Price, C.P. Enzymes as reagents in clinical chemistry. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1983, 300, 411–422. [Google Scholar] [PubMed]
- McClatchey, K.D. Clinical laboratory medicine; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2002. [Google Scholar]
- Bobreshova, M.; Sukhorukov, G.B.; Saburova, E.A.; Elfimova, L.I.; Shabarchina, L.I.; Sukhorukov, B.I. Lactate dehydrogenase in an interpolyelectrolyte complex. Function and stability. Biofizika 1999, 44, 813–820. [Google Scholar] [PubMed]
- Donath, E.; Sukhorukov, G.B.; Caruso, F.; Davis, S.A.; Möhwald, H. Novel Hollow Polymer Shells by Colloid-Templated Assembly of Polyelectrolytes. Angew. Chemie Int. Ed. 1998, 37, 2201–2205. [Google Scholar] [CrossRef]
- Mayya, K.S.; Schoeler, B.; Caruso, F. Preparation and Organization of Nanoscale Polyelectrolyte-Coated Gold Nanoparticles. Adv. Funct. Mater. 2003, 13, 183–188. [Google Scholar] [CrossRef]
- Shenoy, D.B.; Antipov, A.A.; Sukhorukov, G.B.; Möhwald, H. Layer-by-Layer Engineering of Biocompatible, Decomposable Core−Shell Structures. Biomacromolecules 2003, 4, 265–272. [Google Scholar] [CrossRef]
- Sukhorukov, G.; Fery, A.; Möhwald, H. Intelligent micro- and nanocapsules. Prog. Polym. Sci. 2005, 30, 885–897. [Google Scholar] [CrossRef]
- Sukhorukov, B.I.; Tikhonenko, S.A.; Saburova, E.A.; Dubrovskiĭ, A.V.; Dybovskaia, I.N.; Shabarchina, L.I. Incapsulation of enzymes into polyelectrolyte nano- and microcapsules and the problem of the development of enzymatic microdiagnostics. Biofizika 2007, 52, 1041–1048. [Google Scholar]
- Reshetilov, A.N.; Plekhanova, Y.V.; Tikhonenko, S.A.; Dubrovskii, A.V. Polyelectrolyte microcapsules with urease and paramagnetic particles as a basis for a potentiometric biosensor for determining urea. J. Anal. Chem. 2015, 70, 1368–1372. [Google Scholar] [CrossRef]
- Horn, J.; Kapelner, R.; Obermeyer, A. Macro- and Microphase Separated Protein-Polyelectrolyte Complexes: Design Parameters and Current Progress. Polymers 2019, 11, 578. [Google Scholar] [CrossRef]
- Acar, H.; Ting, J.M.; Srivastava, S.; LaBelle, J.L.; Tirrell, M.V. Molecular Engineering Solutions for Therapeutic Peptide Delivery. Chem. Soc. Rev. 2017, 46, 6553–6569. [Google Scholar] [CrossRef]
- De Kruif, C.G.; Weinbreck, F.; de Vries, R. Complex Coacervation of Proteins and Anionic Polysaccharides. Curr. Opin. Colloid Interface Sci. 2004, 9, 340–349. [Google Scholar] [CrossRef]
- Tikhonenko, S.A.; Saburova, E.A.; Durdenko, E.N.; Sukhorukov, B.I. Enzyme-polyelectrolyte complex: Salt effects on the reaction of urease with polyallylamine. Russ. J. Phys. Chem. A 2009, 83, 1781–1788. [Google Scholar] [CrossRef]
- Saburova, E.A.; Bobreshova, M.E.; Elphimova, L.I.; Sukhorukov, B.I. Inhibitory Effect of Polyelectrolytes on Oligomeric Enzymes. Biochemistry 2000, 65, 976–985. [Google Scholar] [PubMed]
- Xu, Y.; Seeman, D.; Yan, Y.; Sun, L.; Post, J.; Dubin, P.L. Effect of Heparin on Protein Aggregation: Inhibition versus Promotion. Biomacromolecules 2012, 13, 1642–1651. [Google Scholar] [CrossRef]
- Du, X.; Dubin, P.L.; Hoagland, D.A.; Sun, L. Protein-Selective Coacervation with Hyaluronic Acid. Biomacromolecules 2014, 15, 726–734. [Google Scholar] [CrossRef]
- Seyrek, E.; Dubin, P.L.; Tribet, C.; Gamble, E.A. Ionic Strength Dependence of Protein-Polyelectrolyte Interactions. Biomacromolecules 2003, 4, 273–282. [Google Scholar] [CrossRef]
- Xu, Y.; Mazzawi, M.; Chen, K.; Sun, L.; Dubin, P.L. Protein Purification by Polyelectrolyte Coacervation: Influence of Protein Charge Anisotropy on Selectivity. Biomacromolecules 2011, 12, 1512–1522. [Google Scholar] [CrossRef]
- Kayitmazer, A.B.; Strand, S.P.; Tribet, C.; Jaeger, W.; Dubin, P.L. Effect of polyelectrolyte structure on protein-polyelectrolyte coacervates: Coacervates of bovine serum albumin with poly(diallyldimethylammonium chloride) versus chitosan. Biomacromolecules 2007, 8, 3568–3577. [Google Scholar] [CrossRef]
- Yu, S.; Xu, X.; Yigit, C.; van der Giet, M.; Zidek, W.; Jankowski, J.; Dzubiella, J.; Ballauff, M. Interaction of human serum albumin with short polyelectrolytes: A study by calorimetry and computer simulations. Soft Matter 2015, 11, 4630–4639. [Google Scholar] [CrossRef]
- Da Silva, F.L.B.; Jönsson, B. Polyelectrolyte–protein complexation driven by charge regulation. Soft Matter 2009, 5, 2862. [Google Scholar] [CrossRef]
- Dubrovsky, A.V.; Musin, E.V.; Kim, A.L.; Tikhonenko, S.A. Effect Of Polyelectrolytes on Catalytic Activity of Alcohol Dehydrogenase. Prikl. Biokhim. Mikrobiol. 2016, 52, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Heitz, J.R.; Brand, L. Fluorescence Changes Alcohol Associated with Denaturation of Dehydrogenase. Archives Biochem. Biophys. 1971, 144, 286–291. [Google Scholar] [CrossRef]
- Fluorescence Spectroscopy: A Tool for Protein Folding/Unfolding Study. Available online: https://www.semanticscholar.org/paper/Fluorescence-Spectroscopy%3A-A-tool-for-Protein-Study/9cbe89466a4e30fb925fa7fc51258d06f9da4740#paper-header (accessed on 23 March 2020).
- Arakawa, T.; Timasheff, S.N. Mechanism of Protein Salting In and Salting Out by Divalent Cation Salts: Balance between Hydration and Salt Binding. Biochemistry 1984, 23, 5912–5923. [Google Scholar] [CrossRef] [PubMed]
- Magonet, E.; Hayen, P.; Delforge, D.; Delaive, E.; Remacle, J. Importance of the structural zinc atom for the stability of yeast alcohol dehydrogenase. Biochem. J. 1992, 287, 361–365. [Google Scholar] [CrossRef]
- Volodkin, D.V.; Larionova, N.I.; Sukhorukov, G.B. Protein Encapsulation via Porous CaCO 3 Microparticles Templating. Biomacromolecules 2004, 5, 1962–1972. [Google Scholar] [CrossRef]
- Kochetkova, O.Y.; Kazakova, L.I.; Moshkov, D.A.; Vinokurov, M.G.; Shabarchina, L.I. Incorporation of proteins into polyelectrolyte microcapsules by coprecipitation and adsorption. Russ. J. Bioorganic Chem. 2013, 39, 504–509. [Google Scholar] [CrossRef]
- Kazakova, L.I.; Dubrovskiĭ, A.V.; Moshkov, D.A.; Shabarchina, L.I.; Sukhorukov, B.I. An electron microscopy study of the structure of polyelectrolyte microcapsules containing protein and containing no protein. Biofizika 2007, 52, 850–854. [Google Scholar]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, A.L.; Musin, E.V.; Dubrovskii, A.V.; Tikhonenko, S.A. Effect of Pollyallylamine on Alcoholdehydrogenase Structure and Activity. Polymers 2020, 12, 832. https://doi.org/10.3390/polym12040832
Kim AL, Musin EV, Dubrovskii AV, Tikhonenko SA. Effect of Pollyallylamine on Alcoholdehydrogenase Structure and Activity. Polymers. 2020; 12(4):832. https://doi.org/10.3390/polym12040832
Chicago/Turabian StyleKim, Aleksandr L., Egor V. Musin, Alexey V. Dubrovskii, and Sergey A. Tikhonenko. 2020. "Effect of Pollyallylamine on Alcoholdehydrogenase Structure and Activity" Polymers 12, no. 4: 832. https://doi.org/10.3390/polym12040832
APA StyleKim, A. L., Musin, E. V., Dubrovskii, A. V., & Tikhonenko, S. A. (2020). Effect of Pollyallylamine on Alcoholdehydrogenase Structure and Activity. Polymers, 12(4), 832. https://doi.org/10.3390/polym12040832




