N-Glycan Modifications with Negative Charge in a Natural Polymer Mucin from Bovine Submaxillary Glands, and Their Structural Role
Abstract
:1. Introduction
2. Materials and Methods
2.1. N-Glycan Preparation
2.2. N-Glycan Analysis Using UPLC
2.3. N-Glycan Analysis Using LC-ESI-HCD-MS/MS
2.4. Relative Quantity of N-Glycans
2.5. Absolute Concentration of N-Glycans
3. Results
3.1. UPLC and LC-ESI-HCD-MS/MS Analysis of N-Glycans
3.2. Structural Analysis of N-Glycans
3.3. Structure and Quantification of N-Glycans
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, J.R.; Gao, W.N.; Grimm, R.; Jiang, S.; Liang, Y.; Ye, H.; Li, Z.G.; Yau, L.F.; Huang, H.; Liu, J.; et al. A method to identify trace sulfated IgG N-glycans as biomarkers for rheumatoid arthritis. Nat. Commun. 2017, 8, 631. [Google Scholar] [CrossRef] [PubMed]
- Sola, R.J.; Griebenow, K. Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 2009, 98, 1223–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.; Kim, J.; Lee, Y.K.; Kim, W.; You, S.K.; Do, J.; Jang, Y.; Oh, D.B.; Kim, J.I.; Kim, H.H. Four unreported types of glycans containing mannose-6-phosphate are heterogeneously attached at three sites (including newly found Asn 233) to recombinant human acid alpha-glucosidase that is the only approved treatment for Pompe disease. Biochem. Biophys. Res. Commun. 2018, 495, 2418–2424. [Google Scholar] [CrossRef] [PubMed]
- Kovensky, J. Sulfated oligosaccharides: New targets for drug development? Curr. Med. Chem. 2009, 16, 2338–2344. [Google Scholar] [CrossRef] [PubMed]
- She, Y.M.; Li, X.; Cyr, T.D. Remarkable structural diversity of N-glycan sulfation on influenza vaccines. Anal. Chem. 2019, 91, 5083–5090. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Shah, P.; Eshghi, S.T.; Yang, W.; Trikannad, N.; Yang, S.; Chen, L.; Aiyetan, P.; Höti, N.; Zhang, Z.; et al. Comprehensive analysis of protein glycosylation by solid-phase extraction of N-linked glycans and glycosite-containing peptides. Nat. Biotechnol. 2016, 34, 84–88. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Veillon, L.; Dong, X.; Huang, Y.; Mechref, Y. Direct comparison of derivatization strategies for LC-MS/MS analysis of N-glycans. Physiol. Behav. 2017, 142, 4446–4455. [Google Scholar] [CrossRef]
- Xiao, K.; Han, Y.; Yang, H.; Lu, H.; Tian, Z. Mass spectrometry-based qualitative and quantitative N-glycomics: An update of 2017–2018. Anal. Chim. Acta 2019, 1091, 1–22. [Google Scholar] [CrossRef]
- Keser, T.; Pavić, T.; Lauc, G.; Gornik, O. Comparison of 2-aminobenzamide, procainamide and RapiFluor-MS as derivatizing agents for high-throughput HILIC-UPLC-FLR-MS N-glycan analysis. Front. Chem. 2018, 6, 324. [Google Scholar] [CrossRef]
- Klapoetke, S.; Zhang, J.; Becht, S.; Gu, X.; Ding, X. The evaluation of a novel approach for the profiling and identification of N-linked glycan with a procainamide tag by HPLC with fluorescent and mass spectrometric detection. J. Pharm. Biomed. Anal. 2010, 53, 315–324. [Google Scholar] [CrossRef]
- Mechref, Y.; Hu, Y.; Desantos-Garcia, J.L.; Hussein, A.; Tang, H. Quantitative glycomics strategies. Mol. Cell. Proteom. 2013, 12, 874–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrou, G.; Crouzier, T. Mucins as multifunctional building blocks of biomaterials. Biomater. Sci. 2018, 6, 2282–2297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strous, G.J.; Dekker, J. Mucin-type glycoproteins. Crit. Rev. Biochem. Mol. Biol. 1992, 27, 57–92. [Google Scholar] [CrossRef] [PubMed]
- Marczynski, M.; Balzer, B.N.; Jiang, K.; Lutz, T.M.; Crouzier, T.; Lieleg, O. Charged glycan residues critically contribute to the adsorption and lubricity of mucins. Colloids Surf. B 2020, 187, 110614. [Google Scholar] [CrossRef] [PubMed]
- Authimoolam, S.P.; Dziubla, T.D. Biopolymeric mucin and synthetic polymer analogs: Their structure, function and role in biomedical applications. Polymer 2016, 8, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergstrom, K.S.B.; Xia, L. Mucin-type O-glycans and their roles in intestinal homeostasis. Glycobiology 2013, 23, 1026–1037. [Google Scholar] [CrossRef]
- Taniguchi, T.; Woodward, A.M.; Magnelli, P.; McColgan, N.M.; Lehoux, S.; Jacobo, S.M.P.; Mauris, J.; Argüeso, P. N-glycosylation affects the stability and barrier function of the MUC16 mucin. J. Biol. Chem. 2017, 292, 11079–11090. [Google Scholar] [CrossRef] [Green Version]
- Ho, J.J.L.; Jaituni, R.S.; Crawley, S.C.; Yang, S.C.; Gum, J.R.; Kim, Y.S. N-glycosylation is required for the surface localization of MUC17 mucin. Int. J. Oncol. 2003, 23, 585–592. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.; Jang, Y.; Ha, J.; Kim, D.; Ji, M.; Lee, Y.K.; Kim, W.; You, S.; Do, J.; et al. N-glycans of bovine submaxillary mucin contain core-fucosylated and sulfated glycans but not sialylated glycans. Int. J. Biol. Macromol. 2019, 138, 1072–1078. [Google Scholar] [CrossRef]
- Bettelheim, F.A.; Hashimoto, Y.; Pigman, W. Light-scattering studies of bovine submaxillary mucin. Biochim. Biophys. Acta 1962, 63, 235–242. [Google Scholar] [CrossRef]
- Polak, R.; Crouzier, T.; Lim, R.M.; Ribbeck, K.; Beppu, M.M.; Pitombo, R.N.M.; Cohen, R.E.; Rubner, M.F. Sugar-mediated disassembly of mucin/lectin multilayers and their use as ph-tolerant, on-demand sacrificial layers. Biomacromolecules 2014, 15, 3093–3098. [Google Scholar] [CrossRef] [PubMed]
- Bansil, R.; Turner, B.S. Mucin structure, aggregation, physiological functions and biomedical applications. Curr. Opin. Colloid Interface Sci. 2006, 11, 164–170. [Google Scholar] [CrossRef]
- Yan, H.; Hjorth, M.; Winkeljann, B.; Dobryden, I.; Lieleg, O.; Crouzier, T. Glyco-modification of mucin hydrogels to investigate their immune activity. Acs Appl. Mater. Interfaces 2020, 12, 19324–19336. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Seignez, C.; Hjorth, M.; Winkeljann, B.; Blakeley, M.; Lieleg, O.; Phillipson, M.; Crouzier, T. Immune-informed mucin hydrogels evade fibrotic foreign body response in vivo. Adv. Funct. Mater. 2019, 29, 1–11. [Google Scholar] [CrossRef]
- Shi, L.; Ardehali, R.; Caldwell, K.D.; Valint, P. Mucin coating on polymeric material surfaces to suppress bacterial adhesion. Colloids Surf. B 2000, 17, 229–239. [Google Scholar] [CrossRef]
- Sandberg, T.; Carlsson, J.; Karlsson, O.M. Mucin coatings suppress neutrophil adhesion to a polymeric model biomaterial. Microsc. Res. Tech. 2007, 70, 864–868. [Google Scholar] [CrossRef]
- Wang, C.; Yuan, J.; Wang, Z.; Huang, L. Separation of one-pot procedure released O-glycans as 1-phenyl-3-methyl-5-pyrazolone derivatives by hydrophilic interaction and reversed-phase liquid chromatography followed by identification using electrospray mass spectrometry and tandem mass spectromet. J. Chromatogr. A 2013, 1274, 107–117. [Google Scholar] [CrossRef]
- Kameyama, A.; Thet Tin, W.W.; Toyoda, M.; Sakaguchi, M. A practical method of liberating O-linked glycans from glycoproteins using hydroxylamine and an organic superbase. Biochem. Biophys. Res. Commun. 2019, 513, 186–192. [Google Scholar] [CrossRef]
- Kim, J.; Ryu, C.; Ha, J.; Lee, J.; Kim, D.; Ji, M.; Park, C.S.; Lee, J.; Kim, D.K.; Kim, H.H. Structural and quantitative characterization of mucin-type O-glycans and the identification of O-glycosylation sites in bovine submaxillary mucin. Biomolecules 2020, 10, 636. [Google Scholar] [CrossRef] [Green Version]
- Packer, N.H.; Lawson, M.A.; Jardine, D.R.; Redmond, J.W. A general approach to desalting oligosaccharides released from glycoproteins. Glycoconj. J. 1998, 15, 737–747. [Google Scholar] [CrossRef]
- Nwosu, C.; Yau, H.K.; Becht, S. Assignment of core versus antenna fucosylation types in protein N-glycosylation via procainamide labeling and tandem mass spectrometry. Anal. Chem. 2015, 87, 5905–5913. [Google Scholar] [CrossRef] [PubMed]
- Bansil, R.; Stanley, E.; LaMont, J.T. Mucin biophysics. Annu. Rev. Physiol. 1995, 57, 635–657. [Google Scholar] [CrossRef] [PubMed]
- Wedepohl, S.; Kaup, M.; Riese, S.B.; Berger, M.; Dernedde, J.; Tauber, R.; Blanchard, V. N-glycan analysis of recombinant L-selectin reveals sulfated GalNAc and GalNAc-GalNAc motifs. J. Proteome Res. 2010, 9, 3403–3411. [Google Scholar] [CrossRef] [PubMed]
- Mendicino, J.; Sangadala, S. Synthesis of sulfated oligosaccharides by cystic fibrosis trachea epithelial cells. Mol. Cell. Biochem. 1999, 201, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, E.; Moriwaki, K.; Terao, N.; Tan, C.C.; Terao, M.; Nakagawa, T.; Matsumoto, H.; Shinzaki, S.; Kamada, Y. Fucosylation is a promising target for cancer diagnosis and therapy. Biomolecules 2012, 2, 34–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moran, A.P.; Gupta, A.; Joshi, L. Sweet-talk: Role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut 2011, 60, 1412–1425. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, A.; Xu, F.; Lee, S. Human saliva and model saliva at bulk to adsorbed phases—Similarities and differences. Adv. Colloid Interface Sci. 2019, 273, 102034. [Google Scholar] [CrossRef]
- Ruhaak, L.R.; Zauner, G.; Huhn, C.; Bruggink, C.; Deelder, A.M.; Wuhrer, M. Glycan labeling strategies and their use in identification and quantification. Anal. Bioanal. Chem. 2010, 397, 3457–3481. [Google Scholar] [CrossRef] [Green Version]
- Lei, M.; Novotny, M.V.; Mechref, Y. Sequential enrichment of sulfated glycans by strong anion-exchange chromatography prior to mass spectrometric measurements. J. Am. Soc. Mass Spectrom. 2010, 21, 348–357. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.Y.; Khoo, K.H.; Yang, Z.; Herp, A.; Wu, A.M. Glycomic mapping of O- and N-linked glycans from major rat sublingual mucin. Glycoconj. J. 2008, 25, 199–212. [Google Scholar] [CrossRef]
- Parry, S.; Hanisch, F.G.; Leir, S.H.; Sutton-Smith, M.; Morris, H.R.; Dell, A.; Harris, A. N-Glycosylation of the MUC1 mucin in epithelial cells and secretions. Glycobiology 2006, 16, 623–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandberg, T.; Blom, H.; Caldwell, K.D. Potential use of mucins as biomaterial coatings. I. Fractionation, characterization, and model adsorption of bovine, porcine, and human mucins. J. Biomed. Mater. Res. A 2009, 91, 762–772. [Google Scholar] [CrossRef] [PubMed]

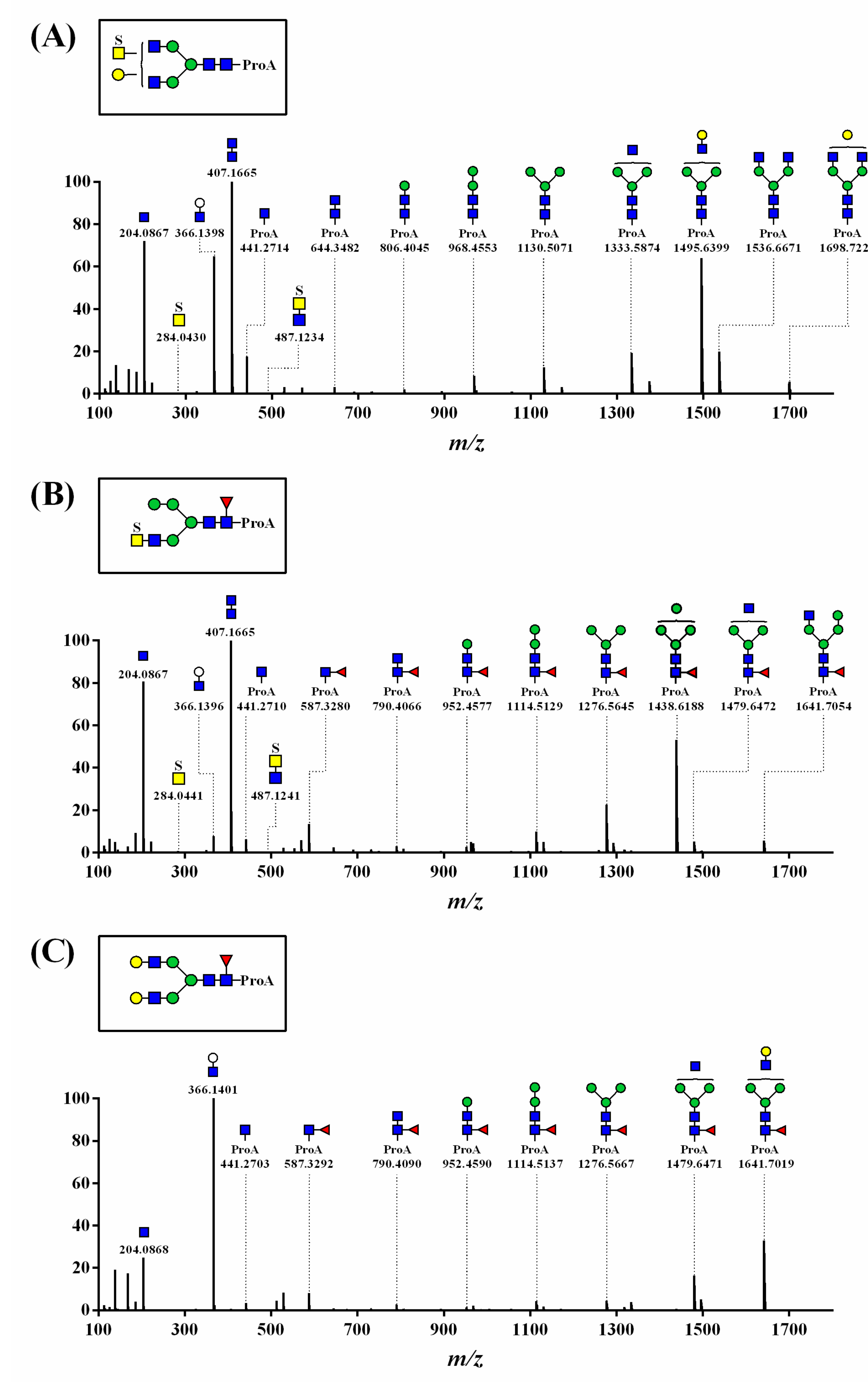


| Group | Sub-Group (%) a | Proposed Structure b | Mass (m/z) | Relative Quantity (%) e | Peak No f | ||
|---|---|---|---|---|---|---|---|
| Calculated [M + H]+ | Observed [M + H]+ c | Error d (ppm) | |||||
| 12 sulfated N-glycans (27.9%) | 5 sulfated N-glycans (6.2%) |  | 910.3554 | 910.3557 g | 0.3 | 1.9 | 7 |
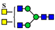 | 1011.8951 | 1011.8954 g | 0.3 | 1.6 | 10 | ||
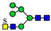 | 970.8685 | 970.8686 g | 0.1 | 1.2 | 23 | ||
 | 1051.8735 | 1051.8738 g | 0.3 | 0.8 | 9 | ||
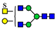 | 991.3818 | 991.3793 g | −2.6 | 0.7 | 18 | ||
| 7 sulfatedand core-fucosylated N-glycans (21.7%) |  | 983.3844 | 983.3817 g | −2.8 | 10.2 | 13 | |
 | 1064.4108 | 1064.4102 g | −0.6 | 2.6 | 24 | ||
 | 881.8447 | 881.8420 g | −3.1 | 2.1 | 5 | ||
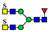 | 1124.9025 | 1124.9021 g | −0.3 | 2.1 | 8 | ||
 | 2273.8722 | 2273.8745 | 1.0 | 2.0 | 27 | ||
 | 1084.9241 | 1084.9227 g | −1.2 | 1.9 | 20 | ||
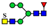 | 962.8711 | 962.8702 g | −0.9 | 0.8 | 16 | ||
| 20 non-sulfated N-glycans (72.1%) | 11 core-fucosylated N-glycans (50.9%) |  | 841.8663 | 841.8639 g | −2.8 | 16.3 | 12 |
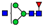 | 740.3266 | 740.3243 g | −3.0 | 8.9 | 4 | ||
 | 943.4060 | 943.4033 g | −2.9 | 6.0 | 19 | ||
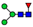 | 638.7869 | 638.7852 g | −2.6 | 4.9 | 2 | ||
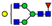 | 922.8927 | 922.8900 g | −2.9 | 4.1 | 22 | ||
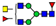 | 2031.8625 | 2031.8624 | −0.1 | 3.0 | 30 | ||
 | 2193.9154 | 2193.9148 | −0.3 | 2.1 | 32 | ||
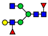 | 995.9216 | 995.9197 g | −1.9 | 1.7 | 26 | ||
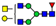 | 1024.4324 | 1024.4302 g | −2.3 | 1.6 | 29 | ||
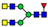 | 1044.9456 | 1044.9454 g | −2.1 | 1.5 | 25 | ||
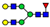 | 1003.9190 | 1003.9183 g | −0.8 | 0.8 | 31 | ||
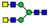
| 9 non-core-fucosylated N-glycans (21.2%) | 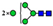 | 727.8108 | 727.8090 g | −2.5 | 6.5 | 15 | |
 | 768.8373 | 768.8350 g | −3.0 | 3.3 | 6 | ||
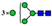 | 808.8372 | 808.8351 g | −2.6 | 3.0 | 28 | ||
 | 1333.5880 | 1333.5873 | −0.5 | 2.2 | 3 | ||
 | 870.3770 | 870.3747 g | −2.7 | 2.1 | 14 | ||
 | 849.8637 | 849.8613 g | −2.9 | 1.4 | 17 | ||
 | 1130.5086 | 1130.5090 | 0.3 | 1.0 | 1 | ||
 | 971.9167 | 971.9145 g | −2.3 | 1.0 | 21 | ||
 | 1495.6408 | 1495.6390 | −1.3 | 0.7 | 11 | ||
| Sum | - | 100.0 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Lee, B.; Lee, J.; Ji, M.; Park, C.S.; Lee, J.; Kang, M.; Kim, J.; Jin, M.; Kim, H.H. N-Glycan Modifications with Negative Charge in a Natural Polymer Mucin from Bovine Submaxillary Glands, and Their Structural Role. Polymers 2021, 13, 103. https://doi.org/10.3390/polym13010103
Kim J, Lee B, Lee J, Ji M, Park CS, Lee J, Kang M, Kim J, Jin M, Kim HH. N-Glycan Modifications with Negative Charge in a Natural Polymer Mucin from Bovine Submaxillary Glands, and Their Structural Role. Polymers. 2021; 13(1):103. https://doi.org/10.3390/polym13010103
Chicago/Turabian StyleKim, Jihye, Byoungju Lee, Junmyoung Lee, Minkyoo Ji, Chi Soo Park, Jaeryong Lee, Minju Kang, Jeongeun Kim, Mijung Jin, and Ha Hyung Kim. 2021. "N-Glycan Modifications with Negative Charge in a Natural Polymer Mucin from Bovine Submaxillary Glands, and Their Structural Role" Polymers 13, no. 1: 103. https://doi.org/10.3390/polym13010103





