Recent Advances in Antifouling Materials for Surface Plasmon Resonance Biosensing in Clinical Diagnostics and Food Safety
Abstract
:1. Introduction
2. Why Are the Antifouling Strategies So Essential for SPR Detection in Clinical Diagnostics and Food Safety?
3. Type of Antifouling Materials
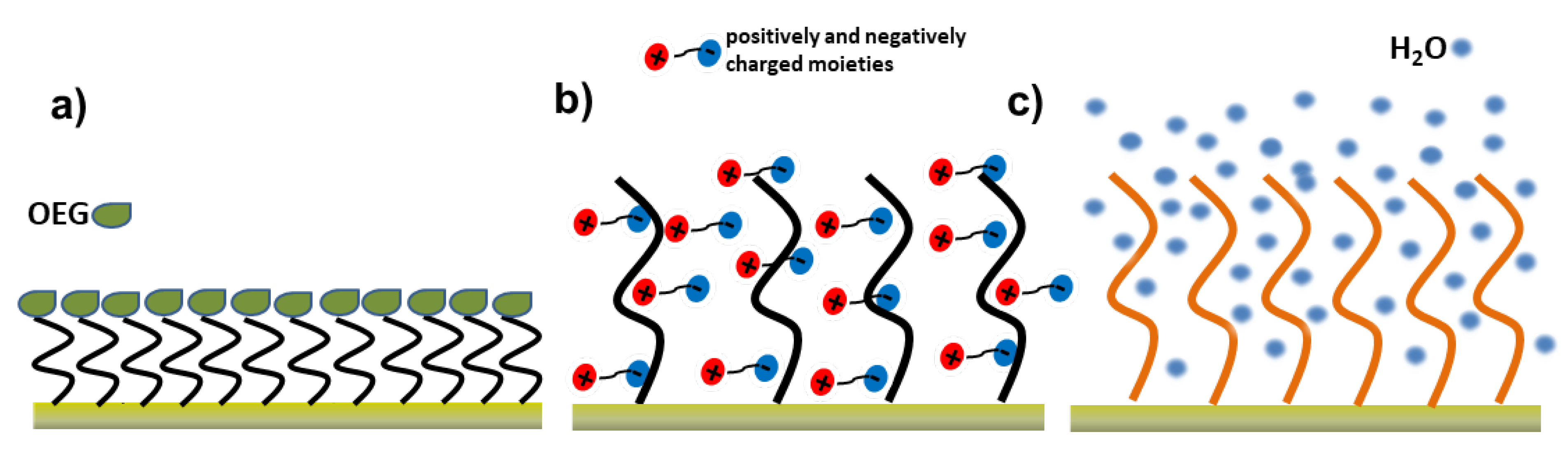
3.1. Poly (Ethylene Glycol)/Oligo (Ethylene Glycol)-Based Self-Assembled Monolayers
3.2. Zwitterionic Surface Layers
3.3. Natural Compounds and Biomimetic Materials
4. Antifouling Strategies for SPR Biosensing in Clinical Diagnostics
5. Antifouling Strategies for SPR Biosensing in Food Safety
6. Conclusions
7. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jiang, C.; Wang, G.; Hein, R.; Liu, N.; Luo, X.; Davis, J.J. Antifouling Strategies for Selective in Vitro and in Vivo Sensing. Chem. Rev. 2020, 120, 3852–3889. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, I.; Pangule, R.C.; Kane, R.S. Antifouling coatings: Recent developments in the design of surfaces that prevent fouling by proteins, bacteria, and marine organisms. Adv. Mater. 2011, 23, 690–718. [Google Scholar] [CrossRef]
- Blaszykowski, C.; Sheikh, S.; Thompson, M. Surface chemistry to minimize fouling from blood-based fluids. Chem. Soc. Rev. 2012, 41, 5599–5612. [Google Scholar] [CrossRef]
- Blaszykowski, C.; Sheikh, S.; Thompson, M. A survey of state-of-the-art surface chemistries to minimize fouling from human and animal biofluids. Biomater. Sci. 2015, 3, 1335–1370. [Google Scholar] [CrossRef] [PubMed]
- Lichtenberg, J.Y.; Ling, Y.; Kim, S. Non-specific adsorption reduction methods in biosensing. Sensors 2019, 19, 2488. [Google Scholar] [CrossRef] [Green Version]
- Bakker, E. So, You Have a Great New Sensor. How Will You Validate It? ACS Sens. 2018, 3, 1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaisocherová, H.; Brynda, E.; Homola, J. Functionalizable low-fouling coatings for label-free biosensing in complex biological media: Advances and applications. Anal. Bioanal. Chem. 2015, 407, 3927–3953. [Google Scholar] [CrossRef]
- Peláez, E.C.; Estevez, M.C.; Portela, A.; Salvador, J.P.; Marco, M.P.; Lechuga, L.M. Nanoplasmonic biosensor device for the monitoring of acenocoumarol therapeutic drug in plasma. Biosens. Bioelectron. 2018, 119, 149–155. [Google Scholar] [CrossRef]
- Ramirez-Priego, P.; Martens, D.; Elamin, A.A.; Soetaert, P.; Van Roy, W.; Vos, R.; Anton, B.; Bockstaele, R.; Becker, H.; Singh, M.; et al. Label-Free and Real-Time Detection of Tuberculosis in Human Urine Samples Using a Nanophotonic Point-of-Care Platform. ACS Sens. 2018, 3, 2079–2086. [Google Scholar] [CrossRef]
- Phillips, K.S.; Jong, H.H.; Cheng, Q. Development of a “membrane cloaking” method for amperometric enzyme immunoassay and surface plasmon resonance analysis of proteins in serum samples. Anal. Chem. 2007, 79, 899–907. [Google Scholar] [CrossRef]
- Masson, J.F.; Battaglia, T.M.; Khairallah, P.; Beaudoin, S.; Booksh, K.S. Quantitative measurement of cardiac markers in undiluted serum. Anal. Chem. 2007, 79, 612–619. [Google Scholar] [CrossRef]
- Trabucchi, A.; Guerra, L.L.; Faccinetti, N.I.; Iacono, R.F.; Poskus, E.; Valdez, S.N. Surface plasmon resonance reveals a different pattern of proinsulin autoantibodies concentration and affinity in diabetic patients. PLoS ONE 2012, 7, e33574. [Google Scholar] [CrossRef] [Green Version]
- Lewis, K.B.; Hughes, R.J.; Epstein, M.S.; Josephson, N.C.; Kempton, C.L.; Kessler, C.M.; Key, N.S.; Howard, T.E.; Kruse-Jarres, R.; Lusher, J.M.; et al. Phenotypes of Allo- and Autoimmune Antibody Responses to FVIII Characterized by Surface Plasmon Resonance. PLoS ONE 2013, 8, e61120. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Liu, X.; Shi, S.; Huang, R.; Su, R.; Qi, W.; He, Z. Design and mechanisms of antifouling materials for surface plasmon resonance sensors. Acta Biomater. 2016, 40, 100–118. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.H.; Dillen, A.; Saeys, W.; Lammertyn, J.; Spasic, D. Advancements in SPR biosensing technology: An overview of recent trends in smart layers design, multiplexing concepts, continuous monitoring and in vivo sensing. Anal. Chim. Acta 2020, 1104, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Gorodkiewicz, E.; Lukaszewski, Z. Recent progress in surface plasmon resonance biosensors (2016 to mid-2018). Biosensors 2018, 8, 132. [Google Scholar] [CrossRef] [Green Version]
- Piliarik, M.; Párová, L.; Homola, J. High-throughput SPR sensor for food safety. Biosens. Bioelectron. 2009, 24, 1399–1404. [Google Scholar] [CrossRef]
- Mauriz, E.; Dey, P.; Lechuga, L.M. Advances in nanoplasmonic biosensors for clinical applications. Analyst 2019, 144, 7105–7129. [Google Scholar] [CrossRef] [PubMed]
- Zanoli, L.M.; D’Agata, R.; Spoto, G. Functionalized gold nanoparticles for ultrasensitive DNA detection. Anal. Bioanal. Chem. 2012, 402, 1759–1771. [Google Scholar] [CrossRef]
- Mariani, S.; Scarano, S.; Ermini, M.L.; Bonini, M.; Minunni, M. Investigating nanoparticle properties in plasmonic nanoarchitectures with DNA by surface plasmon resonance imaging. Chem. Commun. 2015, 51, 6587–6590. [Google Scholar] [CrossRef]
- Giuffrida, M.C.; Cigliana, G.; Spoto, G. Ultrasensitive detection of lysozyme in droplet-based microfluidic devices. Biosens. Bioelectron. 2018, 104, 8–14. [Google Scholar] [CrossRef]
- Zeng, S.; Baillargeat, D.; Ho, H.P.; Yong, K.T. Nanomaterials enhanced surface plasmon resonance for biological and chemical sensing applications. Chem. Soc. Rev. 2014, 43, 3426–3452. [Google Scholar] [CrossRef]
- Guo, L.; Jackman, J.A.; Yang, H.-H.; Chen, P.; Cho, N.-J.; Kim, D.-H. Strategies for enhancing the sensitivity of plasmonic nanosensors. Nano Today 2015, 10, 213–239. [Google Scholar] [CrossRef] [Green Version]
- Bellassai, N.; D’Agata, R.; Jungbluth, V.; Spoto, G. Surface Plasmon Resonance for Biomarker Detection: Advances in Non-invasive Cancer Diagnosis. Front. Chem. 2019, 7, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Jalali, M.; Mahshid, S.; Wachsmann-Hogiu, S. Are plasmonic optical biosensors ready for use in point-of-need applications? Analyst 2020, 145, 364–384. [Google Scholar] [CrossRef] [Green Version]
- Bocková, M.; Slabý, J.; Špringer, T.; Homola, J. Advances in Surface Plasmon Resonance Imaging and Microscopy and Their Biological Applications. Ann. Rev. Anal. Chem. 2019, 12, 151–176. [Google Scholar] [CrossRef] [PubMed]
- D’Agata, R.; Bellassai, N.; Allegretti, M.; Rozzi, A.; Korom, S.; Manicardi, A.; Melucci, E.; Pescarmona, E.; Corradini, R.; Giacomini, P.; et al. Direct plasmonic detection of circulating RAS mutated DNA in colorectal cancer patients. Biosens. Bioelectron. 2020, 170, 112648. [Google Scholar] [CrossRef]
- Bellassai, N.; Spoto, G. Biosensors for liquid biopsy: Circulating nucleic acids to diagnose and treat cancer. Anal. Bioanal. Chem. 2016, 408, 7255–7264. [Google Scholar] [CrossRef] [PubMed]
- Bellassai, N.; Marti, A.; Spoto, G.; Huskens, J. Low-fouling, mixed-charge poly-l-lysine polymers with anionic oligopeptide side-chains. J. Mater. Chem. B 2018, 6, 7662–7673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.-S.; Yau, S.; Chau, L.-K.; Mohamed, A.; Huang, C.-J. Functional Biointerfaces Based on Mixed Zwitterionic Self-Assembled Monolayers for Biosensing Applications. Langmuir 2019, 35, 1652–1661. [Google Scholar] [CrossRef]
- D’Agata, R.; Bellassai, N.; Giuffrida, M.C.; Aura, A.M.; Petri, C.; Kögler, P.; Vecchio, G.; Jonas, U.; Spoto, G. A new ultralow fouling surface for the analysis of human plasma samples with surface plasmon resonance. Talanta 2021, 221, 121483. [Google Scholar] [CrossRef]
- Jung, L.S.; Campbell, C.T.; Chinowsky, T.M.; Mar, M.N.; Yee, S.S. Quantitative interpretation of the response of surface plasmon resonance sensors to adsorbed films. Langmuir 1998, 14, 5636–5648. [Google Scholar] [CrossRef]
- Bahadir, E.B.; Sezgintürk, M.K. Applications of commercial biosensors in clinical, food, environmental, and biothreat/biowarfare analyses. Anal. Biochem. 2015, 478, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Soler, M.; Huertas, C.S.; Lechuga, L.M. Label-free plasmonic biosensors for point-of-care diagnostics: A review. Expert Rev. Mol. Diagn. 2019, 19, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Unsworth, L.D.; Tun, Z.; Sheardown, H.; Brash, J.L. Chemisorption of thiolated poly(ethylene oxide) to gold: Surface chain densities measured by ellipsometry and neutron reflectometry. J. Colloid Interface Sci. 2005, 281, 112–121. [Google Scholar] [CrossRef]
- Cescon, D.W.; Bratman, S.V.; Chan, S.M.; Siu, L.L. Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer 2020, 1, 276–290. [Google Scholar] [CrossRef]
- Masson, J.F. Surface Plasmon Resonance Clinical Biosensors for Medical Diagnostics. ACS Sens. 2017, 2, 16–30. [Google Scholar] [CrossRef]
- Mauriz, E. Low-fouling substrates for plasmonic sensing of circulating biomarkers in biological fluids. Biosensors 2020, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Qin, Y.; Peng, Z.; Chen, S.; Chen, S.; Deng, C.; Xiang, J. The simultaneous detection of free and total prostate antigen in serum samples with high sensitivity and specificity by using the dual-channel surface plasmon resonance. Biosens. Bioelectron. 2014, 62, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Vega, B.; Calle, A.; Sánchez, A.; Lechuga, L.M.; Ortiz, A.M.; Armelles, G.; Rodríguez-Frade, J.M.; Mellado, M. Real-time detection of the chemokine CXCL12 in urine samples by surface plasmon resonance. Talanta 2013, 109, 209–215. [Google Scholar] [CrossRef]
- Pisitkun, T.; Shen, R.F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef] [Green Version]
- Nolen, B.M.; Lokshin, A.E. The advancement of biomarker-based diagnostic tools for ovarian, breast, and pancreatic cancer through the use of urine as an analytical biofluid. Int. J. Biol. Markers 2011, 26, 141–152. [Google Scholar] [CrossRef]
- Aro, K.; Wei, F.; Wong, D.T.; Tu, M. Saliva liquid biopsy for point-of-care applications. Front. Public Health 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emmenegger, C.R.; Brynda, E.; Riedel, T.; Sedlakova, Z.; Houska, M.; Alles, A.B. Interaction of blood plasma with antifouling surfaces. Langmuir 2009, 25, 6328–6333. [Google Scholar] [CrossRef]
- Cornelius, R.M.; Archambault, J.G.; Berry, L.; Chan, A.K.C.; Brash, J.L. Adsorption of proteins from infant and adult plasma to biomaterial surfaces. J. Biomed. Mater. Res. 2002, 60, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.D.L.S.; Rodriguez-Emmenegger, C.; Surman, F.; Riedel, T.; Alles, A.B.; Brynda, E. Use of pooled blood plasmas in the assessment of fouling resistance. RSC Adv. 2014, 4, 2318–2321. [Google Scholar] [CrossRef]
- Aubé, A.; Charbonneau, D.M.; Pelletier, J.N.; Masson, J.F. Response Monitoring of Acute Lymphoblastic Leukemia Patients Undergoing l -Asparaginase Therapy: Successes and Challenges Associated with Clinical Sample Analysis in Plasmonic Sensing. ACS Sens. 2016, 1, 1358–1365. [Google Scholar] [CrossRef]
- Carlsson, J.; Gullstrand, C.; Westermark, G.T.; Ludvigsson, J.; Enander, K.; Liedberg, B. An indirect competitive immunoassay for insulin autoantibodies based on surface plasmon resonance. Biosens. Bioelectron. 2008, 24, 876–881. [Google Scholar] [CrossRef]
- Špringer, T.; Bocková, M.; Homola, J. Label-free biosensing in complex media: A referencing approach. Anal. Chem. 2013, 85, 5637–5640. [Google Scholar] [CrossRef]
- Kirsch, J.; Siltanen, C.; Zhou, Q.; Revzin, A.; Simonian, A. Biosensor technology: Recent advances in threat agent detection and medicine. Chem. Soc. Rev. 2013, 42, 8733–8768. [Google Scholar] [CrossRef]
- Dwivedi, H.P.; Jaykus, L.A. Detection of pathogens in foods: The current state-of-the-art and future directions. Crit. Rev. Microbiol. 2011, 37, 40–63. [Google Scholar] [CrossRef]
- Wang, B.; Park, B. Immunoassay Biosensing of Foodborne Pathogens with Surface Plasmon Resonance Imaging: A Review. J. Agric. Food Chem. 2020, 68, 12927–12939. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Salazar, J.K. Culture-Independent Rapid Detection Methods for Bacterial Pathogens and Toxins in Food Matrices. Compr. Rev. Food Sci. Food Saf. 2016, 15, 183–205. [Google Scholar] [CrossRef]
- López-Campos, G.; Martínez-Suárez, J.V.; Aguado-Urda, M.; López-Alonso, V. Detection, identification, and analysis of foodborne pathogens. In Microarray Detection and Characterization of Bacterial Foodborne Pathogens; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–126. ISBN 9781461432500. [Google Scholar]
- Taylor, A.D.; Ladd, J.; Yu, Q.; Chen, S.; Homola, J.; Jiang, S. Quantitative and simultaneous detection of four foodborne bacterial pathogens with a multi-channel SPR sensor. Biosens. Bioelectron. 2006, 22, 752–758. [Google Scholar] [CrossRef]
- Wei, D.; Oyarzabal, O.A.; Huang, T.S.; Balasubramanian, S.; Sista, S.; Simonian, A.L. Development of a surface plasmon resonance biosensor for the identification of Campylobacter jejuni. J. Microbiol. Methods 2007, 69, 78–85. [Google Scholar] [CrossRef]
- Wang, Y.; Ye, Z.; Si, C.; Ying, Y. Monitoring of Escherichia coli O157:H7 in food samples using lectin based surface plasmon resonance biosensor. Food Chem. 2013, 136, 1303–1308. [Google Scholar] [CrossRef]
- Ferracci, G.; Marconi, S.; Mazuet, C.; Jover, E.; Blanchard, M.P.; Seagar, M.; Popoff, M.; Lévêque, C. A label-free biosensor assay for botulinum neurotoxin B in food and human serum. Anal. Biochem. 2011, 410, 281–288. [Google Scholar] [CrossRef]
- Hossain, M.Z.; McCormick, S.P.; Maragos, C.M. An imaging surface plasmon resonance biosensor assay for the detection of t-2 toxin and masked t-2 toxin-3-glucoside in wheat. Toxins 2018, 10, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prime, K.L.; Whitesides, G.M. Adsorption of Proteins onto Surfaces Containing End-Attached Oligo(ethylene oxide): A Model System Using Self-Assembled Monolayers. J. Am. Chem. Soc. 1993, 115, 10714–10721. [Google Scholar] [CrossRef]
- Whitesides, G.M. Poly(Ethylene Glycol) Chemistry: Biotechnical and Biomedical Applications; Harris, J.M., Ed.; Springer: Berlin/Heidelberg, Germany, 1993; Volume 41, ISBN 9781489907059. [Google Scholar]
- Blackman, L.D.; Gunatillake, P.A.; Cass, P.; Locock, K.E.S. An introduction to zwitterionic polymer behavior and applications in solution and at surfaces. Chem. Soc. Rev. 2019, 48, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, R.; Su, R.; Qi, W.; Wang, L.; He, Z. Grafting hyaluronic acid onto gold surface to achieve low protein fouling in surface plasmon resonance biosensors. ACS Appl. Mater. Interfaces 2014, 6, 13034–13042. [Google Scholar] [CrossRef] [PubMed]
- Ederth, T.; Lerm, M.; Orihuela, B.; Rittschof, D. Resistance of Zwitterionic Peptide Monolayers to Biofouling. Langmuir 2019, 35, 1818–1827. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Coyle, R.C.; Richards, D.J.; Berry, C.L.; Barrs, R.W.; Biggs, J.; Chou, C.J.; Trusk, T.C.; Mei, Y. Development of peptide-functionalized synthetic hydrogel microarrays for stem cell and tissue engineering applications. Acta Biomater. 2016, 45, 110–120. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Huang, H.; Hung, H.C.; Leng, C.; Wei, S.; Crisci, R.; Jiang, S.; Chen, Z. Strong Hydration at the Poly(ethylene glycol) Brush/Albumin Solution Interface. Langmuir 2020, 36, 2030–2036. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Ren, B.; Zhang, D.; Xie, S.; Chang, Y.; Yang, J.; Wu, J.; Xu, L.; Zheng, J. Fundamentals and applications of zwitterionic antifouling polymers. J. Phys. D. Appl. Phys. 2019, 52, 403001. [Google Scholar] [CrossRef]
- Ostuni, E.; Chapman, R.G.; Holmlin, R.E.; Takayama, S.; Whitesides, G.M. A survey of structure-property relationships of surfaces that resist the adsorption of protein. Langmuir 2001, 17, 5605–5620. [Google Scholar] [CrossRef]
- Gasteier, P.; Reska, A.; Schulte, P.; Salber, J.; Offenhäusser, A.; Moeller, M.; Groll, J. Surface grafting of PEO-based star-shaped molecules for bioanalytical and biomedical applications. Macromol. Biosci. 2007, 7, 1010–1023. [Google Scholar] [CrossRef]
- Riedel, T.; Riedelová-Reicheltová, Z.; Májek, P.; Rodriguez-Emmenegger, C.; Houska, M.; Dyr, J.E.; Brynda, E. Complete identification of proteins responsible for human blood plasma fouling on poly(ethylene glycol)-based surfaces. Langmuir 2013, 29, 3388–3397. [Google Scholar] [CrossRef] [PubMed]
- Chapman, P.; Ducker, R.E.; Hurley, C.R.; Hobbs, J.K.; Leggett, G.J. Fabrication of two-component, brush-on-brush topographical microstructures by combination of atom-transfer radical polymerization with polymer end-functionalization and photopatterning. Langmuir 2015, 31, 5935–5944. [Google Scholar] [CrossRef] [Green Version]
- Wei, Q.; Becherer, T.; Angioletti-Uberti, S.; Dzubiella, J.; Wischke, C.; Neffe, A.T.; Lendlein, A.; Ballauff, M.; Haag, R. Protein interactions with polymer coatings and biomaterials. Angew. Chem. Int. Ed. 2014, 53, 8004–8031. [Google Scholar] [CrossRef] [PubMed]
- Konradi, R.; Acikgoz, C.; Textor, M. Polyoxazolines for nonfouling surface coatings—A direct comparison to the gold standard PEG. Macromol. Rapid Commun. 2012, 33, 1663–1676. [Google Scholar] [CrossRef]
- Pop-Georgievski, O.; Verreault, D.; Diesner, M.O.; Proks, V.; Heissler, S.; Rypáček, F.; Koelsch, P. Nonfouling poly(ethylene oxide) layers end-tethered to polydopamine. Langmuir 2012, 28, 14273–14283. [Google Scholar] [CrossRef] [Green Version]
- Baggerman, J.; Smulders, M.M.J.; Zuilhof, H. Romantic Surfaces: A Systematic Overview of Stable, Biospecific, and Antifouling Zwitterionic Surfaces. Langmuir 2019, 35, 1072–1084. [Google Scholar] [CrossRef] [Green Version]
- Tai, F.I.; Sterner, O.; Andersson, O.; Ekblad, T.; Ederth, T. PH-control of the protein resistance of thin hydrogel gradient films. Soft Matter 2014, 10, 5955–5964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaisocherová-Lísalová, H.; Víšová, I.; Ermini, M.L.; Špringer, T.; Song, X.C.; Mrázek, J.; Lamačová, J.; Lynn, N.S.; Šedivák, P.; Homola, J. Low-fouling surface plasmon resonance biosensor for multi-step detection of foodborne bacterial pathogens in complex food samples. Biosens. Bioelectron. 2016, 80, 84–90. [Google Scholar] [CrossRef]
- Riedel, T.; Surman, F.; Hageneder, S.; Pop-Georgievski, O.; Noehammer, C.; Hofner, M.; Brynda, E.; Rodriguez-Emmenegger, C.; Dostálek, J. Hepatitis B plasmonic biosensor for the analysis of clinical serum samples. Biosens. Bioelectron. 2016, 85, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Du, M.; Wu, Z.; Song, Y.; Zheng, Q. Strategy to construct polyzwitterionic hydrogel coating with antifouling, drag-reducing and weak swelling performance. RSC Adv. 2019, 9, 2081–2091. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Li, M.; Zhu, C.; Nurumbetov, G.; Li, Z.; Wilson, P.; Kempe, K.; Haddleton, D.M. Well-Defined Protein/Peptide-Polymer Conjugates by Aqueous Cu-LRP: Synthesis and Controlled Self-Assembly. J. Am. Chem. Soc. 2015, 137, 9344–9353. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Zhang, M.; Yang, J.; Zhao, C.; Hu, R.; Chen, Q.; Chang, Y.; Zheng, J. Synthesis and characterization of antifouling poly(N-acryloylaminoethoxyethanol) with ultralow protein adsorption and cell attachment. Langmuir 2014, 30, 10398–10409. [Google Scholar] [CrossRef]
- Dang, Y.; Quan, M.; Xing, C.M.; Wang, Y.B.; Gong, Y.K. Biocompatible and antifouling coating of cell membrane phosphorylcholine and mussel catechol modified multi-arm PEGs. J. Mater. Chem. B 2015, 3, 2350–2361. [Google Scholar] [CrossRef] [PubMed]
- Knight, A.S.; Zhou, E.Y.; Francis, M.B.; Zuckermann, R.N. Sequence Programmable Peptoid Polymers for Diverse Materials Applications. Adv. Mater. 2015, 27, 5665–5691. [Google Scholar] [CrossRef] [PubMed]
- Chou, Y.N.; Sun, F.; Hung, H.C.; Jain, P.; Sinclair, A.; Zhang, P.; Bai, T.; Chang, Y.; Wen, T.C.; Yu, Q.; et al. Ultra-low fouling and high antibody loading zwitterionic hydrogel coatings for sensing and detection in complex media. Acta Biomater. 2016, 40, 31–37. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Li, X.; Li, L.; Cheng, G.; Gong, X.; Zheng, J. Dual functionality of antimicrobial and antifouling of poly(n -hydroxyethylacrylamide)/salicylate hydrogels. Langmuir 2013, 29, 1517–1524. [Google Scholar] [CrossRef]
- Damodaran, V.B.; Murthy, N.S. Bio-inspired strategies for designing antifouling biomaterials. Biomater. Res. 2016, 20, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Yang, Y.; Zhang, A. Polymer-Engineered Nanostructures for Advanced Energy; Springer: Berlin/Heidelberg, Germany, 2017; ISBN 9783319570020. [Google Scholar]
- Ye, H.; Wang, L.; Huang, R.; Su, R.; Liu, B.; Qi, W.; He, Z. Superior Antifouling Performance of a Zwitterionic Peptide Compared to an Amphiphilic, Non-Ionic Peptide. ACS Appl. Mater. Interfaces 2015, 7, 22448–22457. [Google Scholar] [CrossRef]
- Leng, C.; Buss, H.G.; Segalman, R.A.; Chen, Z. Surface Structure and Hydration of Sequence-Specific Amphiphilic Polypeptoids for Antifouling/Fouling Release Applications. Langmuir 2015, 31, 9306–9311. [Google Scholar] [CrossRef]
- Kruis, I.C.; Löwik, D.W.P.M.; Boelens, W.C.; Van Hest, J.C.M.; Pruijn, G.J.M. An integrated, peptide-based approach to site-specific protein immobilization for detection of biomolecular interactions. Analyst 2016, 141, 5321–5328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Han, R.; Su, X.; Li, Y.; Xu, G.; Luo, X. Zwitterionic peptide anchored to conducting polymer PEDOT for the development of antifouling and ultrasensitive electrochemical DNA sensor. Biosens. Bioelectron. 2017, 92, 396–401. [Google Scholar] [CrossRef]
- Peungthum, P.; Sudprasert, K.; Amarit, R.; Somboonkaew, A.; Sutapun, B.; Vongsakulyanon, A.; Seedacoon, W.; Kitpoka, P.; Kunakorn, M.; Srikhirin, T. Surface plasmon resonance imaging for ABH antigen detection on red blood cells and in saliva: Secretor status-related ABO subgroup identification. Analyst 2017, 142, 1471–1481. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Mu, L.; Sawtelle, S.D.; Rajan, N.K.; Han, Z.; Wang, Y.; Qu, H.; Reed, M.A. Functionalized polyelectrolytes assembling on nano-BioFETs for biosensing applications. Adv. Funct. Mater. 2015, 25, 2279–2286. [Google Scholar] [CrossRef]
- Di Iorio, D.; Marti, A.; Koeman, S.; Huskens, J. Clickable poly-l-lysine for the formation of biorecognition surfaces. RSC Adv. 2019, 9, 35608–35613. [Google Scholar] [CrossRef] [Green Version]
- Movilli, J.; Di Iorio, D.; Rozzi, A.; Hiltunen, J.; Corradini, R.; Huskens, J. “Plug-n-Play” Polymer Substrates: Surface Patterning with Reactive-Group-Appended Poly- l -lysine for Biomolecule Adhesion. ACS Appl. Polym. Mater. 2019, 1, 3165–3173. [Google Scholar] [CrossRef] [Green Version]
- Bellassai, N.; D’Agata, R.; Marti, A.; Rozzi, A.; Volpi, S.; Allegretti, M.; Corradini, R.; Giacomini, P.; Huskens, J.; Spoto, G. Detection of tumor DNA in human plasma with a functional PLL based surface layer and plasmonic biosensing. ACS Sens. 2021. [Google Scholar] [CrossRef]
- Lowe, S.; O’Brien-Simpson, N.M.; Connal, L.A. Antibiofouling polymer interfaces: Poly(ethylene glycol) and other promising candidates. Polym. Chem. 2015, 6, 198–212. [Google Scholar] [CrossRef] [Green Version]
- Löfås, S.; Johnsson, B. A novel hydrogel matrix on gold surfaces in surface plasmon resonance sensors for fast and efficient covalent immobilization of ligands. J. Chem. Soc. Chem. Commun. 1990, 1526–1528. [Google Scholar] [CrossRef]
- Mohseni, S.; Moghadam, T.T.; Dabirmanesh, B.; Jabbari, S.; Khajeh, K. Development of a label-free SPR sensor for detection of matrixmetalloproteinase-9 by antibody immobilization on carboxymethyldextran chip. Biosens. Bioelectron. 2016, 81, 510–516. [Google Scholar] [CrossRef]
- Rath, D.; Kumar, S.; Panda, S. pH-Based Detection of Target Analytes in Diluted Serum Samples Using Surface Plasmon Resonance Immunosensor. Appl. Biochem. Biotechnol. 2019, 187, 1272–1284. [Google Scholar] [CrossRef] [PubMed]
- Castiello, F.R.; Tabrizian, M. Multiplex Surface Plasmon Resonance Imaging-Based Biosensor for Human Pancreatic Islets Hormones Quantification. Anal. Chem. 2018, 90, 3132–3139. [Google Scholar] [CrossRef] [PubMed]
- Love, J.C.; Estroff, L.A.; Kriebel, J.K.; Nuzzo, R.G.; Whitesides, G.M. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem. Rev. 2005, 105, 1103–1170. [Google Scholar] [CrossRef]
- Peláez, E.C.; Estevez, M.C.; Domínguez, R.; Sousa, C.; Cebolla, A.; Lechuga, L.M. A compact SPR biosensor device for the rapid and efficient monitoring of gluten-free diet directly in human urine. Anal. Bioanal. Chem. 2020, 412, 6407–6417. [Google Scholar] [CrossRef]
- Nie, W.; Wang, Q.; Zou, L.; Zheng, Y.; Liu, X.; Yang, X.; Wang, K. Low-Fouling Surface Plasmon Resonance Sensor for Highly Sensitive Detection of MicroRNA in a Complex Matrix Based on the DNA Tetrahedron. Anal. Chem. 2018, 90, 12584–12591. [Google Scholar] [CrossRef] [PubMed]
- Daems, D.; Pfeifer, W.; Rutten, I.; Saccà, B.; Spasic, D.; Lammertyn, J. Three-Dimensional DNA Origami as Programmable Anchoring Points for Bioreceptors in Fiber Optic Surface Plasmon Resonance Biosensing. ACS Appl. Mater. Interfaces 2018, 10, 23539–23547. [Google Scholar] [CrossRef]
- Bellassai, N.; D’Agata, R.; Spoto, G. Novel nucleic acid origami structures and conventional molecular beacon–based platforms: A comparison in biosensing applications. Anal. Bioanal. Chem. 2021, 1–15. [Google Scholar] [CrossRef]
- Wu, W.; Yu, X.; Wu, J.; Wu, T.; Fan, Y.; Chen, W.; Zhao, M.; Wu, H.; Li, X.; Ding, S. Surface plasmon resonance imaging-based biosensor for multiplex and ultrasensitive detection of NSCLC-associated exosomal miRNAs using DNA programmed heterostructure of Au-on-Ag. Biosens. Bioelectron. 2021, 175, 112835. [Google Scholar] [CrossRef]
- Mckeating, K.S.; Hinman, S.S.; Rais, N.A.; Zhou, Z.; Cheng, Q. Antifouling lipid membranes over protein a for orientation-controlled immunosensing in undiluted serum and plasma. ACS Sens. 2019, 4, 1774–1782. [Google Scholar] [CrossRef]
- Cui, M.; Ma, Y.; Wang, L.; Wang, Y.; Wang, S.; Luo, X. Antifouling sensors based on peptides for biomarker detection. TrAC Trends Anal. Chem. 2020, 127, 115903. [Google Scholar] [CrossRef]
- Qian, H.; Huang, Y.; Duan, X.; Wei, X.; Fan, Y.; Gan, D.; Yue, S.; Cheng, W.; Chen, T. Fiber optic surface plasmon resonance biosensor for detection of PDGF-BB in serum based on self-assembled aptamer and antifouling peptide monolayer. Biosens. Bioelectron. 2019, 140, 111350. [Google Scholar] [CrossRef]
- Nowinski, A.K.; Sun, F.; White, A.D.; Keefe, A.J.; Jiang, S. Sequence, structure, and function of peptide self-assembled monolayers. J. Am. Chem. Soc. 2012, 134, 6000–6005. [Google Scholar] [CrossRef] [Green Version]
- Jia, X.C.; Yi, W.; Shwu, J.C.; Ching-Jung, C.; Jen-Tsai, L. Peptide-based antifouling aptasensor for cardiac troponin I detection by surface plasmon resonance applied in medium sized Myocardial Infarction. Ann. Biomed. Sci. Eng. 2020, 4, 001–008. [Google Scholar] [CrossRef] [Green Version]
- Vaisocherová, H.; Yang, W.; Zhang, Z.; Cao, Z.; Cheng, G.; Piliarik, M.; Homola, J.; Jiang, S. Ultralow fouling and functionalizable surface chemistry based on a zwitterionic polymer enabling sensitive and specific protein detection in undiluted blood plasma. Anal. Chem. 2008, 80, 7894–7901. [Google Scholar] [CrossRef] [PubMed]
- Vaisocherová, H.; Zhang, Z.; Yang, W.; Cao, Z.; Cheng, G.; Taylor, A.D.; Piliarik, M.; Homola, J.; Jiang, S. Functionalizable surface platform with reduced non-specific protein adsorption from full blood plasma-Material selection and protein immobilization optimization. Biosens. Bioelectron. 2009, 24, 1924–1930. [Google Scholar] [CrossRef]
- Vaisocherová, H.; Šípová, H.; Víšová, I.; Bocková, M.; Špringer, T.; Ermini, M.L.; Song, X.; Krejčík, Z.; Chrastinová, L.; Pastva, O.; et al. Rapid and sensitive detection of multiple microRNAs in cell lysate by low-fouling surface plasmon resonance biosensor. Biosens. Bioelectron. 2015, 70, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Riedel, T.; Hageneder, S.; Surman, F.; Pop-Georgievski, O.; Noehammer, C.; Hofner, M.; Brynda, E.; Rodriguez-Emmenegger, C.; Dostálek, J. Plasmonic Hepatitis B Biosensor for the Analysis of Clinical Saliva. Anal. Chem. 2017, 89, 2972–2977. [Google Scholar] [CrossRef]
- Vaisocherová, H.; Ševců, V.; Adam, P.; Špačková, B.; Hegnerová, K.; Pereira, A.D.L.S.; Rodriguez-Emmenegger, C.; Riedel, T.; Houska, M.; Brynda, E.; et al. Functionalized ultra-low fouling carboxy- and hydroxy-functional surface platforms: Functionalization capacity, biorecognition capability and resistance to fouling from undiluted biological media. Biosens. Bioelectron. 2014, 51, 150–157. [Google Scholar] [CrossRef]
- Kotlarek, D.; Curti, F.; Vorobii, M.; Corradini, R.; Careri, M.; Knoll, W.; Rodriguez-Emmenegger, C.; Dostálek, J. Surface plasmon resonance-based aptasensor for direct monitoring of thrombin in a minimally processed human blood. Sens. Actuators B Chem. 2020, 320, 128380. [Google Scholar] [CrossRef]
- Aura, A.M.; D’Agata, R.; Spoto, G. Ultrasensitive Detection of Staphylococcus aureus and Listeria monocytogenes Genomic DNA by Nanoparticle-Enhanced Surface Plasmon Resonance Imaging. ChemistrySelect 2017, 2, 7024–7030. [Google Scholar] [CrossRef]
- Chen, J.; Park, B. Label-free screening of foodborne Salmonella using surface plasmon resonance imaging. Anal. Bioanal. Chem. 2018, 410, 5455–5464. [Google Scholar] [CrossRef]
- Tran, D.T.; Knez, K.; Janssen, K.P.; Pollet, J.; Spasic, D.; Lammertyn, J. Selection of aptamers against Ara h 1 protein for FO-SPR biosensing of peanut allergens in food matrices. Biosens. Bioelectron. 2013, 43, 245–251. [Google Scholar] [CrossRef]
- Mihai, I.; Vezeanu, A.; Polonschii, C.; Albu, C.; Radu, G.L.; Vasilescu, A. Label-free detection of lysozyme in wines using an aptamer based biosensor and SPR detection. Sens. Actuators B Chem. 2015, 206, 198–204. [Google Scholar] [CrossRef]
- Joshi, S.; Annida, R.M.; Zuilhof, H.; Van Beek, T.A.; Nielen, M.W.F. Analysis of Mycotoxins in Beer Using a Portable Nanostructured Imaging Surface Plasmon Resonance Biosensor. J. Agric. Food Chem. 2016, 64, 8263–8271. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Zuilhof, H.; Van Beek, T.A.; Nielen, M.W.F. Biochip Spray: Simplified Coupling of Surface Plasmon Resonance Biosensing and Mass Spectrometry. Anal. Chem. 2017, 89, 1427–1432. [Google Scholar] [CrossRef]
- Joshi, S.; Segarra-Fas, A.; Peters, J.; Zuilhof, H.; Van Beek, T.A.; Nielen, M.W.F. Multiplex surface plasmon resonance biosensing and its transferability towards imaging nanoplasmonics for detection of mycotoxins in barley. Analyst 2016, 141, 1307–1318. [Google Scholar] [CrossRef]
- Rodriguez-Emmenegger, C.; Avramenko, O.A.; Brynda, E.; Skvor, J.; Alles, A.B. Poly(HEMA) brushes emerging as a new platform for direct detection of food pathogen in milk samples. Biosens. Bioelectron. 2011, 26, 4545–4551. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Li, L.; Wang, Q.; Yu, Q.; Zheng, J. Effect of film thickness on the antifouling performance of poly(hydroxy-functional methacrylates) grafted surfaces. Langmuir 2011, 27, 4906–4913. [Google Scholar] [CrossRef] [PubMed]
- Vaisocherova-Lisalova, H.; Surman, F.; Víšová, I.; Vala, M.; Špringer, T.; Ermini, M.L.; Šípová, H.; Šedivák, P.; Houska, M.; Riedel, T.; et al. Copolymer Brush-Based Ultralow-Fouling Biorecognition Surface Platform for Food Safety. Anal. Chem. 2016, 88, 10533–10539. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Gupta, B.D. Detection of heavy metal ions in contaminated water by surface plasmon resonance based optical fibre sensor using conducting polymer and chitosan. Food Chem. 2015, 166, 568–575. [Google Scholar] [CrossRef]
- Wei, M.; Gao, Y.; Li, X.; Serpe, M.J. Stimuli-responsive polymers and their applications. Polym. Chem. 2017, 8, 127–143. [Google Scholar] [CrossRef] [Green Version]
- Ijäs, H.; Nummelin, S.; Shen, B.; Kostiainen, M.A.; Linko, V. Dynamic DNA origami devices: From strand-displacement reactions to external-stimuli responsive systems. Int. J. Mol. Sci. 2018, 19, 2114. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.S.; In, I.; Park, S.Y. PH and redox responsive polymer for antifouling surface coating. Appl. Surf. Sci. 2014, 313, 532–536. [Google Scholar] [CrossRef]





| Antifouling Layer | Target | Sensing Performance | Ref. | ||
|---|---|---|---|---|---|
| Operational Range | LOD | Matrix | |||
| carboxymethyldextran (CMD) | DON OTA mycotoxines | 60–2000 ng mL−1 10–120 ng mL−1 | 17 ng mL−1 7 ng mL−1 | beer | [123] |
| CMD | DON OTA ZEA T-2 FB1 AFB1 | 26–3200 µg kg−1 13–320 µg kg−1 16–160 µg kg−1 0.6–290 µg kg−1 10–1200 µg kg−1 3–260 µg kg−1 | 26 µg kg−1 3 µg kg−1 6 µg kg−1 0.6 µg kg−1 2 µg kg−1 0.6 µg kg−1 | barley | [125] |
| mixed SAM of PEG and 11-mercaptohexadecanoic acid | Peanut allergen Ara h1 | 0, 211, 423, 634, 846, 1058 nM | 75 nM (in buffer) | candy bar | [121] |
| mixed SAM of PEG and 11-mercaptohexadecanoic acid | LYS | 0.05-80 μg mL−1 | 0.035 μg mL−1 (2.44 nM) | red and white wines | [122] |
| mixed SAM of PEG and 16-mercaptohexadecanoic acid | insulin glucagon somatostatin | 34–633 ng mL−1 85–1592 ng mL−1 719–4000 ng mL−1 | 1 nM (8 ng/mL) 4 nM (14 ng/mL) 246 nM (403 ng/mL) | pancreatic islet secretome | [101] |
| ethylene glycol layer and an extra-blocking step comprising BSA | α2-gliadin (33mer) GIP | Using G12 mAB: 3.6 ± 0.2–56.2 ± 13.3 (33mer) 3.4 ± 0.1–35.4 ± 3.0 (GIP) Using A1 mAB: 14.7 ± 3.2–702.3 ± 110 (33mer) 9.1 ± 1.2–172.1 ± 43.3 (GIP) | 1.6 ng mL−1 (33mer) 1.7 ng mL−1 (GIP) 4.7 ng mL−1 (33mer) 4.0 ng mL−1 (GIP) | 100% urine | [103] |
| DNA tetrahedron probes (DTPs) | miR let-7a | 0–2 pM | 0.8 fM | 100% serum, 100% plasma, 9.85 × 108 red blood cells/mL, 5% whole blood and cell lysate | [104] |
| DNA tetrahedral framework (DTF) | NSCLC-associated exosomal miRNA-21, miRNA-378, miRNA-200, and miRNA-139 | 2 fM–20 nM | 1.68 fM | 10% plasma exosomes in clinical samples | [107] |
| positively charged lipid bilayer mimic, 2-dioleoyl-sn-glycero-3-ethylphosphocholine (EPC+) | cholera toxin | 0, 10, 20, 30 μg mL−1 | 0.05 μg mL−1 | 100% serum or plasma | [108] |
| mixed SAMs of CPPPP-EKEKEKE peptide | platelet-derived growth factor PDGF-BB | 1–1000 pM | 0.35 pM | 10% human serum | [110] |
| N’-biotin-EKEKEKE-PPPPC | cardiac troponin I (cTnI) | 20−600 ng mL−1 | ~20 ng mL−1 | 10% FBS | [112] |
| poly(HEMA) brushes | Cronobacter | 108–106 cells mL−1 | 106 cells mL−1 | fresh milk powdered milk and PIF samples | [126] |
| polycarboxybetaine acrylamide (pCBAA) | protein cancer biomarker (ALCAM) | 7.8–1000 ng mL−1 | ~10 ng mL−1 (unblocked polyCBAA surface) ~100 ng mL−1 (blocked COOH/OH OEG) | 100% human blood plasma | [113] |
| pCBAA brushes | miR-16 miR-181 miR-34a miR-125b | 0.1–100 pM | 0.35 pM (miR-16) 0.39 pM (miR-181) 0.50 pM (miR-34a) 0.95 pM (miR-125b) | crude erythrocyte lysates | [115] |
| pCBAA brushes | E. coli O157:H7 Salmonella sp. | 1.5 × 101–1.5 × 107 CFU mL−1 2.5 × 102–2.5 × 107 CFU mL−1 | 57 CFU mL−1 (E. coli in cucumber) 17 CFU mL−1 (E. coli in hamburger) 7.4 × 10³ CFU mL−1 (Salmonella sp. in cucumber) 11.7 × 10³ CFU mL−1 (Salmonella sp. in hamburger) | crude cucumber and hamburger | [77] |
| copolymer brushes of poly[N-(2-hydroxypropyl) methacrylamide] (poly(HPMAA)) and poly(carboxy betaine methacrylamide) (poly(CBMA)) | E. coli O157:H7 | 104−107 CFU mL−1 (direct E.coli detection) 102−106 CFU mL−1 (SA-AuNP-enhanced E. coli detection) | 2.1 × 104 CFU mL−1 poly (CBMAA 15 mol %-ran-HPMAA) 81 CFU mL−1 poly (CBMAA 15 mol %-ran-HPMAA) in cucumber samples | cucumber, hamburger, sprouts, and lettuce | [128] |
| poly[HPMA-co-CBMAA] | hepatitis B surface antigen | 0.01 and 1 IU·mL−1 | Not available | 10% saliva | [116] |
| poly[HPMA-co-CBMAA] | thrombin | 0–20 nM | 0.7 nM (aptamer HD1) 1 nM (aptamer HD22) | 10% blood | [118] |
| triethylene glycol-PEG(3)-pentrimer carboxybetaine (PPCB) | Human Arginase I | 0, 12.5, 50 nM | Not available | 10% pooled human plasma | [31] |
| poly-l-lysine (PLL)-mal(26%)-PNA-CEEEEE oligopeptide | KRAS G13D mutated ctDNA | 0.5–20 pg μL−1 | MDC = 0.58 pg μL−1 RDL = 1.45 pg μL−1 | 10% plasma from cancer patient | [96] |
| Surface Layer | Clinical Samples | Food Matrices | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Blood | Serum | Plasma | Urine | Cell Lysate | Saliva | Exosome and Secretome | Beer | Barley | Milk | Wine and Candy Bar | Cucumber and Hamburger | Ref. | |
| CMD | ✔ mycotoxins | ✔ mycotoxins | [123] [125] | ||||||||||
| OEG/SAM or PEG- zwitterionic | ✔ lung, gastrointestinal and bladder cancers | ✔ allergens | ✔ hormones | ✔ Allergens and additives | [121] [122] [101] [103] [31] | ||||||||
| DNA-based structures | ✔ | ✔ breast, lung cancer and hepatocellular carcinoma | ✔ | ✔ breast, lung cancer and hepatocellular carcinoma | ✔ colon adenocarcinoma, breast cancer, and lung cancer | [104] [107] | |||||||
| Peptide | ✔ sarcomas and glioblastomas, acute myocardial infarction | ✔ colorectal cancer | [110] [112] [96] | ||||||||||
| Zwitterionic brushes | ✔ | ✔ various carcinomas | ✔ myelodysplastic syndrome | ✔ bacteria | ✔ bacteria | [113] [115] [77] [126] | |||||||
| Copolymer brushes | ✔ thrombosis | ✔ hepatitis B | ✔ bacteria | [128] [116] [118] | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Agata, R.; Bellassai, N.; Jungbluth, V.; Spoto, G. Recent Advances in Antifouling Materials for Surface Plasmon Resonance Biosensing in Clinical Diagnostics and Food Safety. Polymers 2021, 13, 1929. https://doi.org/10.3390/polym13121929
D’Agata R, Bellassai N, Jungbluth V, Spoto G. Recent Advances in Antifouling Materials for Surface Plasmon Resonance Biosensing in Clinical Diagnostics and Food Safety. Polymers. 2021; 13(12):1929. https://doi.org/10.3390/polym13121929
Chicago/Turabian StyleD’Agata, Roberta, Noemi Bellassai, Vanessa Jungbluth, and Giuseppe Spoto. 2021. "Recent Advances in Antifouling Materials for Surface Plasmon Resonance Biosensing in Clinical Diagnostics and Food Safety" Polymers 13, no. 12: 1929. https://doi.org/10.3390/polym13121929








