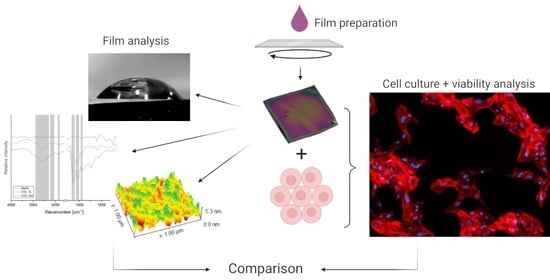
Investigating the Viability of Epithelial Cells on Polymer Based Thin-Films
Abstract
:1. Introduction
2. Results and Discussion
2.1. Thin-Film Preparation and Characterization
2.2. Attenuated Total Reflectance Fourier Transform Infrared Spectroscopy (FTIR-ATR)
2.3. Atomic Force Microscopy (AFM)
2.4. Contact Angle Measurement
2.5. Cell Culture and Viability Analysis
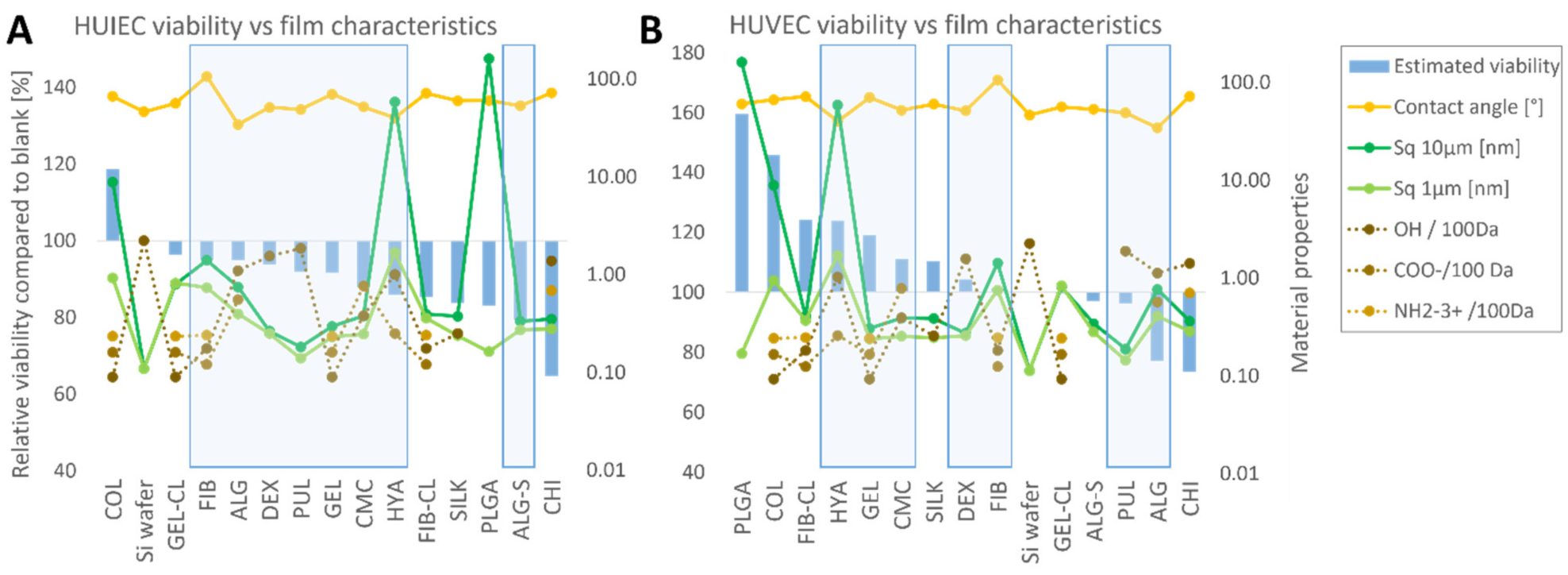
- Chemical interactions: The polymers may facilitate or prevent cell attachment, trigger or block membrane receptors, and act as chelators for nutrients or toxic compounds. For example, CHI is generally considered biodegradable and biocompatible with simultaneous antimicrobial activity, typically attributed to free amino groups [51,78].
- Structural interactions: Surface roughness and solubility of the polymers determine the attachment surface and its stability and change in viscosity of the culture medium near the thin-film surface.
- Depending on the solubility of the thin films in the nutrient medium under cell culture conditions (37 °C, 5% CO2), the cell-polymer interaction may vary. For soluble thin films, the effect on cell viability should be comparable for direct contact and exposure to extracts, with a correlation between metabolic activity and polymer concentration. For poorly soluble polymers, thin-film extracts are expected to have less effect on cell viability. It is important to note that surface properties such as roughness can only affect the cells if the polymer is poorly soluble and therefore sufficiently stable. In addition to the thin-film properties measured in this study, the available literature was analyzed to compare the influence of thin film on cell development with functional groups of the polymers with which they might interact. Specifically, the polymers were analyzed for hydroxy, carboxyl, or amino groups. The polysaccharides selected for this study consist of repeating hexoses with respective functional groups so that their number per molecular weight could be easily derived from the chemical structure of the individual polymers, as found in the literature [27,28,35,37,42,45,51]. The same method of assessing the functional groups per unit molecular weight was also used for PLGA [20]. With their complex amino acid sequences and folding and intertwining chains, the free functional groups of proteins are more difficult to estimate. Therefore, in the work presented here, the number of functional groups was estimated from the proportion of individual amino acids and their functional groups in the total composition alone. E.g., for silk fibroin, which is produced by the silk moth (Bombyx mori), the composition was estimated according to the genetic sequence, coding for 45.9% glycine, 30.3% alanine, 12.1% serine, 5.3% tyrosine, and 1.8% valine [72]. The same procedure was used to determine the amount of functional groups in COL [56], GEL [61], and FIB [66]. For the reasons mentioned above, the number of functional target groups per 100 kDa in proteins is only a rough estimate. It does not necessarily represent the actual value of the functional groups available for interaction with cultured cells.
2.5.1. HUVEC
2.5.2. HUIEC
2.6. Interpretation of the Results
3. Materials and Methods
3.1. Thin-Film Preparation
3.2. Attenuated Total Reflectance (ATR) FTIR
3.3. Atomic Force Microscopy (AFM)
3.4. Contact Angle Measurements
3.5. Cell Selection and Culturing
3.6. MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide) Test
3.7. Cell Culture with Thin-Film Extracts
3.8. Cell Culture on Thin Films
3.9. Cell Morphology Analysis
3.10. Statistical Analysis
3.11. Biocompatibility Score
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Gao, G.; Lee, J.H.; Jang, J.; Lee, D.H.; Kong, J.-S.; Kim, B.S.; Choi, Y.-J.; Jang, W.B.; Hong, Y.J.; Kwon, S.-M.; et al. Tissue Engineered Bio-Blood-Vessels Constructed Using a Tissue-Specific Bioink and 3D Coaxial Cell Printing Technique: A Novel Therapy for Ischemic Disease. Adv. Funct. Mater. 2017, 27. [Google Scholar] [CrossRef]
- Ott, H.C.; Matthiesen, T.S.; Goh, S.K.; Black, L.D.; Kren, S.M.; Netoff, T.I.; Taylor, D.A. Perfusion-decellularized matrix: Using nature’s platform to engineer a bioartificial heart. Nat. Med. 2008, 14, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, T.W.; Sellaro, T.L.; Badylak, S.F. Decellularization of tissues and organs. Biomaterials 2006, 27, 3675–3683. [Google Scholar] [CrossRef] [PubMed]
- Kundu, J.; Michaelson, A.; Talbot, K.; Baranov, P.; Young, M.J.; Carrier, R.L. Decellularized retinal matrix: Natural platforms for human retinal progenitor cell culture. Acta Biomater. 2016, 31, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.W.; Zhang, Y.; Luo, J.C.; Zhang, D.; Xiong, B.J.; Yang, J.Q.; Xie, H.Q.; Lv, Q. Hydrogel derived from decellularized porcine adipose tissue as a promising biomaterial for soft tissue augmentation. J. Biomed. Mater. Res. A 2017, 105, 1756–1764. [Google Scholar] [CrossRef]
- Noor, N.; Shapira, A.; Edri, R.; Gal, I.; Wertheim, L.; Dvir, T. 3D Printing of Personalized Thick and Perfusable Cardiac Patches and Hearts. Adv. Sci. 2019, 6, 1900344. [Google Scholar] [CrossRef] [Green Version]
- Pati, F.; Ha, D.H.; Jang, J.; Han, H.H.; Rhie, J.W.; Cho, D.W. Biomimetic 3D tissue printing for soft tissue regeneration. Biomaterials 2015, 62, 164–175. [Google Scholar] [CrossRef]
- Jang, J.; Park, H.J.; Kim, S.W.; Kim, H.; Park, J.Y.; Na, S.J.; Kim, H.J.; Park, M.N.; Choi, S.H.; Park, S.H.; et al. 3D printed complex tissue construct using stem cell-laden decellularized extracellular matrix bioinks for cardiac repair. Biomaterials 2017, 112, 264–274. [Google Scholar] [CrossRef]
- Pati, F.; Jang, J.; Ha, D.H.; Won Kim, S.; Rhie, J.W.; Shim, J.H.; Kim, D.H.; Cho, D.W. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat. Commun. 2014, 5, 3935. [Google Scholar] [CrossRef] [Green Version]
- Jang, J.; Kim, T.G.; Kim, B.S.; Kim, S.W.; Kwon, S.M.; Cho, D.W. Tailoring mechanical properties of decellularized extracellular matrix bioink by vitamin B2-induced photo-crosslinking. Acta Biomater. 2016, 33, 88–95. [Google Scholar] [CrossRef]
- Pati, F.; Song, T.H.; Rijal, G.; Jang, J.; Kim, S.W.; Cho, D.W. Ornamenting 3D printed scaffolds with cell-laid extracellular matrix for bone tissue regeneration. Biomaterials 2015, 37, 230–241. [Google Scholar] [CrossRef]
- Lee, H.; Han, W.; Kim, H.; Ha, D.H.; Jang, J.; Kim, B.S.; Cho, D.W. Development of Liver Decellularized Extracellular Matrix Bioink for Three-Dimensional Cell Printing-Based Liver Tissue Engineering. Biomacromolecules 2017, 18, 1229–1237. [Google Scholar] [CrossRef]
- Hospodiuk, M.; Dey, M.; Sosnoski, D.; Ozbolat, I.T. The bioink: A comprehensive review on bioprintable materials. Biotechnol. Adv. 2017, 35, 217–239. [Google Scholar] [CrossRef] [Green Version]
- Ng, W.L.; Chua, C.K.; Shen, Y.-F. Print Me An Organ! Why We Are Not There Yet. Prog. Polym. Sci. 2019, 97. [Google Scholar] [CrossRef]
- Naahidi, S.; Jafari, M.; Logan, M.; Wang, Y.; Yuan, Y.; Bae, H.; Dixon, B.; Chen, P. Biocompatibility of hydrogel-based scaffolds for tissue engineering applications. Biotechnol. Adv. 2017, 35, 530–544. [Google Scholar] [CrossRef]
- Paragkumar, N.T.; Edith, D.; Six, J.-L. Surface characteristics of PLA and PLGA films. Appl. Surf. Sci. 2006, 253, 2758–2764. [Google Scholar] [CrossRef]
- Okassa, L.N.; Marchais, H.; Douziech-Eyrolles, L.; Herve, K.; Cohen-Jonathan, S.; Munnier, E.; Souce, M.; Linassier, C.; Dubois, P.; Chourpa, I. Optimization of iron oxide nanoparticles encapsulation within poly(d,l-lactide-co-glycolide) sub-micron particles. Eur. J. Pharm. Biopharm. 2007, 67, 31–38. [Google Scholar] [CrossRef]
- Chen, G.; Sato, T.; Ohgushi, H.; Ushida, T.; Tateishi, T.; Tanaka, J. Culturing of skin fibroblasts in a thin PLGA-collagen hybrid mesh. Biomaterials 2005, 26, 2559–2566. [Google Scholar] [CrossRef]
- Zamani, F.; Amani-Tehran, M.; Latifi, M.; Shokrgozar, M.A. The influence of surface nanoroughness of electrospun PLGA nanofibrous scaffold on nerve cell adhesion and proliferation. J. Mater. Sci. Mater. Med. 2013, 24, 1551–1560. [Google Scholar] [CrossRef]
- Bukala, J.; Buszman, P.P.; Malachowski, J.; Mazurkiewicz, L.; Sybilski, K. Experimental Tests, FEM Constitutive Modeling and Validation of PLGA Bioresorbable Polymer for Stent Applications. Materials 2020, 13, 2003. [Google Scholar] [CrossRef]
- Papageorgiou, S.K.; Kouvelos, E.P.; Favvas, E.P.; Sapalidis, A.A.; Romanos, G.E.; Katsaros, F.K. Metal-carboxylate interactions in metal-alginate complexes studied with FTIR spectroscopy. Carbohydr. Res. 2010, 345, 469–473. [Google Scholar] [CrossRef]
- Maver, T.; Mohan, T.; Gradisnik, L.; Finsgar, M.; Stana Kleinschek, K.; Maver, U. Polysaccharide Thin Solid Films for Analgesic Drug Delivery and Growth of Human Skin Cells. Front. Chem 2019, 7, 217. [Google Scholar] [CrossRef] [Green Version]
- Hinton, T.J.; Jallerat, Q.; Palchesko, R.N.; Park, J.H.; Grodzicki, M.S.; Shue, H.J.; Ramadan, M.H.; Hudson, A.R.; Feinberg, A.W. Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci. Adv. 2015, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attalla, R.; Puersten, E.; Jain, N.; Selvaganapathy, P.R. 3D bioprinting of heterogeneous bi- and tri-layered hollow channels within gel scaffolds using scalable multi-axial microfluidic extrusion nozzle. Biofabrication 2018, 11, 015012. [Google Scholar] [CrossRef] [PubMed]
- Chansoria, P.; Narayanan, L.K.; Schuchard, K.; Shirwaiker, R. Ultrasound-assisted biofabrication and bioprinting of preferentially aligned three-dimensional cellular constructs. Biofabrication 2019, 11, 035015. [Google Scholar] [CrossRef] [PubMed]
- Colosi, C.; Shin, S.R.; Manoharan, V.; Massa, S.; Costantini, M.; Barbetta, A.; Dokmeci, M.R.; Dentini, M.; Khademhosseini, A. Microfluidic Bioprinting of Heterogeneous 3D Tissue Constructs Using Low-Viscosity Bioink. Adv. Mater. 2016, 28, 677–684. [Google Scholar] [CrossRef]
- Guo, X.; Wang, Y.; Qin, Y.; Shen, P.; Peng, Q. Structures, properties and application of alginic acid: A review. Int. J. Biol. Macromol. 2020, 162, 618–628. [Google Scholar] [CrossRef]
- Muller, M.; Ozturk, E.; Arlov, O.; Gatenholm, P.; Zenobi-Wong, M. Alginate Sulfate-Nanocellulose Bioinks for Cartilage Bioprinting Applications. Ann. Biomed. Eng. 2017, 45, 210–223. [Google Scholar] [CrossRef]
- Gershlak, J.R.; Hernandez, S.; Fontana, G.; Perreault, L.R.; Hansen, K.J.; Larson, S.A.; Binder, B.Y.; Dolivo, D.M.; Yang, T.; Dominko, T.; et al. Crossing kingdoms: Using decellularized plants as perfusable tissue engineering scaffolds. Biomaterials 2017, 125, 13–22. [Google Scholar] [CrossRef]
- Hickey, R.J.; Modulevsky, D.J.; Cuerrier, C.M.; Pelling, A.E. Customizing the Shape and Microenvironment Biochemistry of Biocompatible Macroscopic Plant-Derived Cellulose Scaffolds. ACS Biomater. Sci. Eng. 2018, 4, 3726–3736. [Google Scholar] [CrossRef] [Green Version]
- Modulevsky, D.J.; Lefebvre, C.; Haase, K.; Al-Rekabi, Z.; Pelling, A.E. Apple derived cellulose scaffolds for 3D mammalian cell culture. PLoS ONE 2014, 9, e97835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kono, H.; Onishi, K.; Nakamura, T. Characterization and bisphenol A adsorption capacity of beta-cyclodextrin-carboxymethylcellulose-based hydrogels. Carbohydr. Polym. 2013, 98, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Maver, T.; Maver, U.; Mostegel, F.; Griesser, T.; Spirk, S.; Smrke, D.M.; Stana-Kleinschek, K. Cellulose based thin films as a platform for drug release studies to mimick wound dressing materials. Cellulose 2015, 22, 749–761. [Google Scholar] [CrossRef]
- Milojevic, M.; Vihar, B.; Banovic, L.; Misko, M.; Gradisnik, L.; Zidaric, T.; Maver, U. Core/shell Printing Scaffolds For Tissue Engineering Of Tubular Structures. J. Vis. Exp. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Javanbakht, S.; Shaabani, A. Carboxymethyl cellulose-based oral delivery systems. Int. J. Biol. Macromol. 2019, 133, 21–29. [Google Scholar] [CrossRef]
- Shingel, K.I. Determination of structural peculiarities of dexran, pullulan and γ-irradiated pullulan by Fourier-transform IR spectroscopy. Carbohydr. Res. 2002, 337, 1445–1451. [Google Scholar] [CrossRef]
- Singh, R.S.; Kaur, N.; Rana, V.; Kennedy, J.F. Pullulan: A novel molecule for biomedical applications. Carbohydr. Polym. 2017, 171, 102–121. [Google Scholar] [CrossRef]
- Aschenbrenner, E.; Bley, K.; Koynov, K.; Makowski, M.; Kappl, M.; Landfester, K.; Weiss, C.K. Using the polymeric ouzo effect for the preparation of polysaccharide-based nanoparticles. Langmuir 2013, 29, 8845–8855. [Google Scholar] [CrossRef]
- Siddiqui, N.N.; Aman, A.; Silipo, A.; Qader, S.A.; Molinaro, A. Structural analysis and characterization of dextran produced by wild and mutant strains of Leuconostoc mesenteroides. Carbohydr. Polym. 2014, 99, 331–338. [Google Scholar] [CrossRef]
- Perez, J.M.; Asati, A.; Nath, S.; Kaittanis, C. Synthesis of biocompatible dextran-coated nanoceria with pH-dependent antioxidant properties. Small 2008, 4, 552–556. [Google Scholar] [CrossRef]
- Cadee, J.A.; van Luyn, M.J.; Brouwer, L.A.; Plantinga, J.A.; van Wachem, P.B.; de Groot, C.J.; den Otter, W.; Hennink, W.E. In vivo biocompatibility of dextran-based hydrogels. J. Biomed. Mater. Res. 2000, 50, 397–404. [Google Scholar] [CrossRef]
- Tuchilus, C.G.; Nichifor, M.; Mocanu, G.; Stanciu, M.C. Antimicrobial activity of chemically modified dextran derivatives. Carbohydr. Polym. 2017, 161, 181–186. [Google Scholar] [CrossRef]
- de Oliveira, S.A.; da Silva, B.C.; Riegel-Vidotti, I.C.; Urbano, A.; de Sousa Faria-Tischer, P.C.; Tischer, C.A. Production and characterization of bacterial cellulose membranes with hyaluronic acid from chicken comb. Int. J. Biol. Macromol. 2017, 97, 642–653. [Google Scholar] [CrossRef]
- Alkrad, J.A.; Mrestani, Y.; Stroehl, D.; Wartewig, S.; Neubert, R. Characterization of enzymatically digested hyaluronic acid using NMR, Raman, IR, and UV–Vis spectroscopies. J. Pharm. Biomed. Anal. 2003, 31, 545–550. [Google Scholar] [CrossRef]
- Collins, M.N.; Birkinshaw, C. Hyaluronic acid based scaffolds for tissue engineering—A review. Carbohydr. Polym. 2013, 92, 1262–1279. [Google Scholar] [CrossRef]
- Gruene, M.; Pflaum, M.; Hess, C.; Diamantouros, S.; Schlie, S.; Deiwick, A.; Koch, L.; Wilhelmi, M.; Jockenhoevel, S.; Haverich, A.; et al. Laser printing of three-dimensional multicellular arrays for studies of cell-cell and cell-environment interactions. Tissue Eng. Part. C Methods 2011, 17, 973–982. [Google Scholar] [CrossRef] [Green Version]
- Da Róz, A.L.; Leite, F.L.; Pereiro, L.V.; Nascente, P.A.P.; Zucolotto, V.; Oliveira, O.N.; Carvalho, A.J.F. Adsorption of chitosan on spin-coated cellulose films. Carbohydr. Polym. 2010, 80, 65–70. [Google Scholar] [CrossRef]
- Finsgar, M.; Uzunalic, A.P.; Stergar, J.; Gradisnik, L.; Maver, U. Novel chitosan/diclofenac coatings on medical grade stainless steel for hip replacement applications. Sci. Rep. 2016, 6, 26653. [Google Scholar] [CrossRef]
- Andrady, A.L.; Torikai, A.; Kobatake, T. Spectral sensitivity of chitosan photodegradation. J. Appl. Polym. Sci. 1996, 62, 1465–1471. [Google Scholar] [CrossRef]
- Kucharska, M.; Butruk, B.; Walenko, K.; Brynk, T.; Ciach, T. Fabrication of in-situ foamed chitosan/β-TCP scaffolds for bone tissue engineering application. Mater. Lett. 2012, 85, 124–127. [Google Scholar] [CrossRef]
- Croisier, F.; Jérôme, C. Chitosan-based biomaterials for tissue engineering. Eur. Polym. J. 2013, 49, 780–792. [Google Scholar] [CrossRef] [Green Version]
- Riaz, T.; Zeeshan, R.; Zarif, F.; Ilyas, K.; Muhammad, N.; Safi, S.Z.; Rahim, A.; Rizvi, S.A.A.; Rehman, I.U. FTIR analysis of natural and synthetic collagen. Appl. Spectrosc. Rev. 2018, 53, 703–746. [Google Scholar] [CrossRef]
- Veeruraj, A.; Arumugam, M.; Ajithkumar, T.; Balasubramanian, T. Isolation and characterization of collagen from the outer skin of squid (Doryteuthis singhalensis). Food Hydrocoll. 2015, 43, 708–716. [Google Scholar] [CrossRef]
- Belbachir, K.; Noreen, R.; Gouspillou, G.; Petibois, C. Collagen types analysis and differentiation by FTIR spectroscopy. Anal. Bioanal. Chem. 2009, 395, 829–837. [Google Scholar] [CrossRef]
- Chevallay, B.; Herbage, D. Collagen-based biomaterials as 3D scaffold for cell cultures: Applications for tissue engineering and gene therapy. Med. Biol. Eng. Comput. 2000, 38, 211–218. [Google Scholar] [CrossRef]
- Gauza-Wlodarczyk, M.; Kubisz, L.; Wlodarczyk, D. Amino acid composition in determination of collagen origin and assessment of physical factors effects. Int. J. Biol. Macromol. 2017, 104, 987–991. [Google Scholar] [CrossRef]
- Skardal, A.; Mack, D.; Kapetanovic, E.; Atala, A.; Jackson, J.D.; Yoo, J.; Soker, S. Bioprinted amniotic fluid-derived stem cells accelerate healing of large skin wounds. Stem Cells Transl. Med. 2012, 1, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.K.; Lanzi, A.M.; Ngo, H.; Yoo, S.S.; Vincent, P.A.; Dai, G.H. Generation of Multi-scale Vascular Network System Within 3D Hydrogel Using 3D Bio-printing Technology. Cell. Mol. Bioeng. 2014, 7, 460–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Liu, X.; Yu, L.; Shanks, R.; Petinaks, E.; Liu, H. Phase composition and interface of starch-gelatin blends studied by synchrotron FTIR micro-spectroscopy. Carbohydr. Polym. 2013, 95, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Hashim, D.M.; Man, Y.B.C.; Norakasha, R.; Shuhaimi, M.; Salmah, Y.; Syahariza, Z.A. Potential use of Fourier transform infrared spectroscopy for differentiation of bovine and porcine gelatins. Food Chem. 2010, 118, 856–860. [Google Scholar] [CrossRef]
- Van Vlierberghe, S.; Graulus, G.J.; Keshari Samal, S.; Van Nieuwenhove, I.; Dubruel, P. Porous hydrogel biomedical foam scaffolds for tissue repair. In Biomedical Foams for Tissue Engineering Applications; Woodhead Publishing: Cambridge, UK, 2014; pp. 335–390. [Google Scholar] [CrossRef]
- Wang, X.; Yan, Y.; Pan, Y.; Xiong, Z.; Liu, H.; Cheng, J.; Liu, F.; Lin, F.; Wu, R.; Zhang, R.; et al. Generation of three-dimensional hepatocyte/gelatin structures with rapid prototyping system. Tissue Eng. 2006, 12, 83–90. [Google Scholar] [CrossRef]
- Irvine, S.A.; Agrawal, A.; Lee, B.H.; Chua, H.Y.; Low, K.Y.; Lau, B.C.; Machluf, M.; Venkatraman, S. Printing cell-laden gelatin constructs by free-form fabrication and enzymatic protein crosslinking. Biomed. Microdevices 2015, 17, 16. [Google Scholar] [CrossRef] [Green Version]
- Laronda, M.M.; Rutz, A.L.; Xiao, S.; Whelan, K.A.; Duncan, F.E.; Roth, E.W.; Woodruff, T.K.; Shah, R.N. A bioprosthetic ovary created using 3D printed microporous scaffolds restores ovarian function in sterilized mice. Nat. Commun. 2017, 8, 15261. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, R.I.; Faizullin, D.A.; Zuev, Y.F.; Weisel, J.W. The alpha-helix to beta-sheet transition in stretched and compressed hydrated fibrin clots. Biophys. J. 2012, 103, 1020–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watt, K.W.; Takagi, T.; Doolittle, R.F. Amino acid sequence of the beta chain of human fibrinogen: Homology with the gamma chain. Proc. Natl. Acad. Sci. USA 1978, 75, 1731–1735. [Google Scholar] [CrossRef] [Green Version]
- Weisel, J.W.; Litvinov, R.I. Fibrin Formation, Structure and Properties. Subcell. Biochem. 2017, 82, 405–456. [Google Scholar] [CrossRef] [Green Version]
- Sionkowska, A.; Planecka, A. The influence of UV radiation on silk fibroin. Polym. Degrad. Stab. 2011, 96, 523–528. [Google Scholar] [CrossRef]
- Rockwood, D.N.; Preda, R.C.; Yucel, T.; Wang, X.; Lovett, M.L.; Kaplan, D.L. Materials fabrication from Bombyx mori silk fibroin. Nat. Protoc. 2011, 6, 1612–1631. [Google Scholar] [CrossRef]
- Voga, M.; Drnovsek, N.; Novak, S.; Majdic, G. Silk fibroin induces chondrogenic differentiation of canine adipose-derived multipotent mesenchymal stromal cells/mesenchymal stem cells. J. Tissue Eng. 2019. [Google Scholar] [CrossRef] [Green Version]
- Compaan, A.M.; Christensen, K.; Huang, Y. Inkjet Bioprinting of 3D Silk Fibroin Cellular Constructs Using Sacrificial Alginate. ACS Biomater. Sci. Eng. 2016, 3, 1519–1526. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.Z.; Confalonieri, F.; Jacquet, M.; Perasso, R.; Li, Z.G.; Janin, J. Silk fibroin: Structural implications of a remarkable amino acid sequence. Proteins 2001, 44, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Gradisnik, L.; Trapecar, M.; Rupnik, M.S.; Velnar, T. HUIEC, Human intestinal epithelial cell line with differentiated properties: Process of isolation and characterisation. Wien. Klin. Wochenschr. 2015, 127 (Suppl. S5), 204–209. [Google Scholar] [CrossRef] [PubMed]
- Stana, J.; Stergar, J.; Gradisnik, L.; Flis, V.; Kargl, R.; Frohlich, E.; Stana Kleinschek, K.; Mohan, T.; Maver, U. Multilayered Polysaccharide Nanofilms for Controlled Delivery of Pentoxifylline and Possible Treatment of Chronic Venous Ulceration. Biomacromolecules 2017, 18, 2732–2746. [Google Scholar] [CrossRef] [PubMed]
- Sirghi, L.; Kylián, O.; Gilliland, D.; Ceccone, G.; Rossi, F. Cleaning and Hydrophilization of Atomic Force Microscopy Silicon Probes. J. Phys. Chem. B 2006, 110, 25975–25981. [Google Scholar] [CrossRef]
- van Oss, C.J. Surface properties of fibrinogen and fibrin. J. Protein Chem. 1990, 9, 487–491. [Google Scholar] [CrossRef]
- Kubiak, K.J.; Wilson, M.C.T.; Mathia, T.G.; Carval, P. Wettability versus roughness of engineering surfaces. Wear 2011, 271, 523–528. [Google Scholar] [CrossRef] [Green Version]
- Peniche, C.; Argüelles-Monal, W.; Peniche, H.; Acosta, N. Chitosan: An Attractive Biocompatible Polymer for Microencapsulation. Macromol. Biosci. 2003, 3, 511–520. [Google Scholar] [CrossRef]
- Jabaji, Z.; Sears, C.M.; Brinkley, G.J.; Lei, N.Y.; Joshi, V.S.; Wang, J.; Lewis, M.; Stelzner, M.; Martín, M.G.; Dunn, J.C. Use of collagen gel as an alternative extracellular matrix for the in vitro and in vivo growth of murine small intestinal epithelium. Tissue Eng. Part. C Methods 2013, 19, 961–969. [Google Scholar] [CrossRef] [Green Version]
- Lovett, M.; Lee, K.; Edwards, A.; Kaplan, D.L. Vascularization Strategies for Tissue Engineering. Tissue Eng. Part. B Rev. 2009, 15, 353–370. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.; Poovaiah, N.; Forrester, M.; Cochran, E.; Wang, Q. Ex vivo culture of primary intestinal stem cells in collagen gels and foams. ACS Biomater. Sci. Eng. 2015, 1, 37–42. [Google Scholar] [CrossRef]
- Han, J.; Lazarovici, P.; Pomerantz, C.; Chen, X.; Wei, Y.; Lelkes, P.I. Co-electrospun blends of PLGA, gelatin, and elastin as potential nonthrombogenic scaffolds for vascular tissue engineering. Biomacromolecules 2011, 12, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Loh, J.W.; Yeoh, G.; Saunders, M.; Lim, L.Y. Uptake and cytotoxicity of chitosan nanoparticles in human liver cells. Toxicol Appl Pharm. 2010, 249, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Skardal, A. Perspective: “Universal” bioink technology for advancing extrusion bioprinting-based biomanufacturing. Bioprinting 2018, 10, e00026. [Google Scholar] [CrossRef]
- Aydin, L.; Kucuk, S.; Kenar, H. A universal self-eroding sacrificial bioink that enables bioprinting at room temperature. Polym. Adv. Technol. 2020, 31, 1634–1647. [Google Scholar] [CrossRef]
- Maver, U.; Gradišnik, L.; Smrke, D.M.; Stana Kleinschek, K.; Maver, T. Impact of growth factors on wound healing in polysaccharide blend thin films. Appl. Surf. Sci. 2019, 489, 485–493. [Google Scholar] [CrossRef]
- Maver, T.; Hribernik, S.; Mohan, T.; Smrke, D.M.; Maver, U.; Stana-Kleinschek, K. Functional wound dressing materials with highly tunable drug release properties. RSC Adv. 2015, 5, 77873–77884. [Google Scholar] [CrossRef] [Green Version]
- Milojević, M.; Gradišnik, L.; Stergar, J.; Skelin Klemen, M.; Stožer, A.; Vesenjak, M.; Dobnik Dubrovski, P.; Maver, T.; Mohan, T.; Stana Kleinschek, K.; et al. Development of multifunctional 3D printed bioscaffolds from polysaccharides and NiCu nanoparticles and their application. Appl. Surf. Sci. 2019, 488, 836–852. [Google Scholar] [CrossRef]
- Maver, U.; Xhanari, K.; Zizek, M.; Korte, D.; Gradisnik, L.; Franko, M.; Finsgar, M. A combination of interdisciplinary analytical tools for evaluation of multi-layered coatings on medical grade stainless steel for biomedical applications. Eur. J. Pharm. Biopharm. 2018, 128, 230–246. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Ferrari, M.; Fornasiero, M.C.; Isetta, A.M. MTT colorimetric assay for testing macrophage cytotoxic activity in vitro. J. Immunol. Methods 1990, 131, 165–172. [Google Scholar] [CrossRef]
- Van de Loosdrecht, A.; Beelen, R.; Ossenkoppele, g.; Broekhoven, M.; Langenhuijsen, M. A tetrazolium-based colorimetric MTT assay to quantitate human monocyte mediated cytotoxicity against leukemic cells from cell lines and patients with acute myeloid leukemia. J. Immunol. Methods 1994, 174, 311–320. [Google Scholar] [CrossRef]




| Vibration (cm−1) | ALG [21,22] | ALG-S [21,22] | CMC [32] | PUL [36] | DEX [36,39] | HYA [43,44] | CHI [47,48,49] |
|---|---|---|---|---|---|---|---|
| ν(OH) | 3300 | 3300 | 3300 | 3330 | 3220 | 3300 | 3300 |
| ν(CH)anomer | 2930 | 2930 | 2930 | 2920 | 2900 | 2925 | |
| ν(COO)asym | 1595 | 1595 | 1595 | ||||
| ν(COO)sym | 1425 | 1425 | 1415 | 1411 | |||
| δ(CCH)+δ(OCH) | 1300 | 1300 | 1310 | ||||
| ν(C-O) | 1024 | 1024 | |||||
| ν(NH)sym | 3300 | 3300 | |||||
| Amide I | 1650 | 1667 | |||||
| 1614 | |||||||
| ν(NH2) | 1560 | 1560 | |||||
| ν(C-N) | 1310 | 1309 | |||||
| ν(C-O-C) | 1043 | ||||||
| ν(C-O) | 1155 | 1155 | |||||
| 1107 | 1107 | ||||||
| Vibration (cm−1) | COL [52,53,54] | GEL [54,59,60] | GEL-CL [54,59,60] | FIB [65] | FIB-CL [65] | SILK [68] | PLGA [16,17] |
| ν(NH) | 3300 | 3290 | |||||
| ν(CH) | 3060 | 3000 | |||||
| Amide B band | 2928 | 2940 | 2940 | 2030 | |||
| ν(C=O) | 1750 | ||||||
| Amide I | 1600–1700 | ||||||
| Amide II | 1544 | 1525 | 1525 | 1520 | 1520 | 1520 | |
| δ(C-H2) | 1454 | 1450 | 1450 | 1450 | 1450 | 1410 | |
| δ(C-H3) | 1390 | 1400 | 1400 | 1390 | 1390 | 1380 | |
| Amide III | 1236 | 1235 | 1235 | 1240 | 1240 | 1230 | |
| ν(C–N) | 1300 | 1300 | |||||
| ω(C-H2) | 1330 | ||||||
| ν(C-H) methyl | 1450 | ||||||
| ν(C-O-C) | 1160 | 1085 | |||||
| ν(C–O) | 1035 | ||||||
| Polymer | Recipe |
|---|---|
| ALG | A total of 0.01 g was dissolved in 2 mL of high-purity dH2O with agitation on a magnetic stirrer at room temperature until completely dissolved. |
| ALG-S | A total of 0.01 g of dry material obtained from Zenobi Group (ETH Zürich, Switzerland) [28] was dissolved in 2 mL of high-purity dH2O with agitation on a magnetic stirrer at room temperature until completely dissolved. |
| CMC | A total of 0.01 g was dissolved in 2 mL of high-purity dH2O with agitation on a magnetic stirrer at room temperature until completely dissolved. |
| PUL | A total of 0.01 g was dissolved in 2 mL of high-purity dH2O with agitation on a magnetic stirrer at room temperature until completely dissolved. |
| DEX | A total of 0.01 g was dissolved in 2 mL of high-purity dH2O with agitation on a magnetic stirrer at room temperature until completely dissolved. |
| HYA | A total of 0.01 g was dissolved in 2 mL of phosphate buffered saline (PBS) with agitation on a magnetic stirrer at 90–95 °C until completely dissolved, followed by cooling to 37 °C. |
| CHI | A total of 0.01 g was dissolved in 2 mL of 17 mM solution of acetic acid with agitation on a magnetic stirrer at room temperature until completely dissolved. |
| GEL | A total of 0.01 g was dissolved in 2 mL of high-purity dH2O with agitation on a magnetic stirrer at 40 °C until completely dissolved and cooled to room temperature. |
| COL | A total of 0.01 g was dissolved in 2 mL of 0.2 M acetic acid with agitation on a magnetic stirrer at 45 °C overnight. |
| FIB | A total of 0.01 g was dissolved in 2 mL of 0.9% NaCl with agitation on a magnetic stirrer at room temperature until completely dissolved. |
| SILK | A prepared solution was obtained from the department of nanostructured materials (IJS, Ljubljana, Slovenia) and prepared as previously described [69,70]. |
| PLGA | A total of 0.01 g was dissolved in 2 mL of acetone with gentle manual agitation until completely dissolved. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vihar, B.; Rožanc, J.; Krajnc, B.; Gradišnik, L.; Milojević, M.; Činč Ćurić, L.; Maver, U. Investigating the Viability of Epithelial Cells on Polymer Based Thin-Films. Polymers 2021, 13, 2311. https://doi.org/10.3390/polym13142311
Vihar B, Rožanc J, Krajnc B, Gradišnik L, Milojević M, Činč Ćurić L, Maver U. Investigating the Viability of Epithelial Cells on Polymer Based Thin-Films. Polymers. 2021; 13(14):2311. https://doi.org/10.3390/polym13142311
Chicago/Turabian StyleVihar, Boštjan, Jan Rožanc, Boštjan Krajnc, Lidija Gradišnik, Marko Milojević, Laura Činč Ćurić, and Uroš Maver. 2021. "Investigating the Viability of Epithelial Cells on Polymer Based Thin-Films" Polymers 13, no. 14: 2311. https://doi.org/10.3390/polym13142311
APA StyleVihar, B., Rožanc, J., Krajnc, B., Gradišnik, L., Milojević, M., Činč Ćurić, L., & Maver, U. (2021). Investigating the Viability of Epithelial Cells on Polymer Based Thin-Films. Polymers, 13(14), 2311. https://doi.org/10.3390/polym13142311