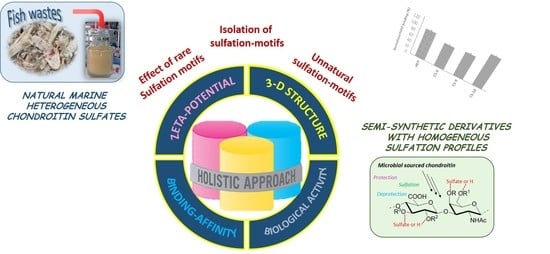
Deciphering Structural Determinants in Chondroitin Sulfate Binding to FGF-2: Paving the Way to Enhanced Predictability of Their Biological Functions
Abstract
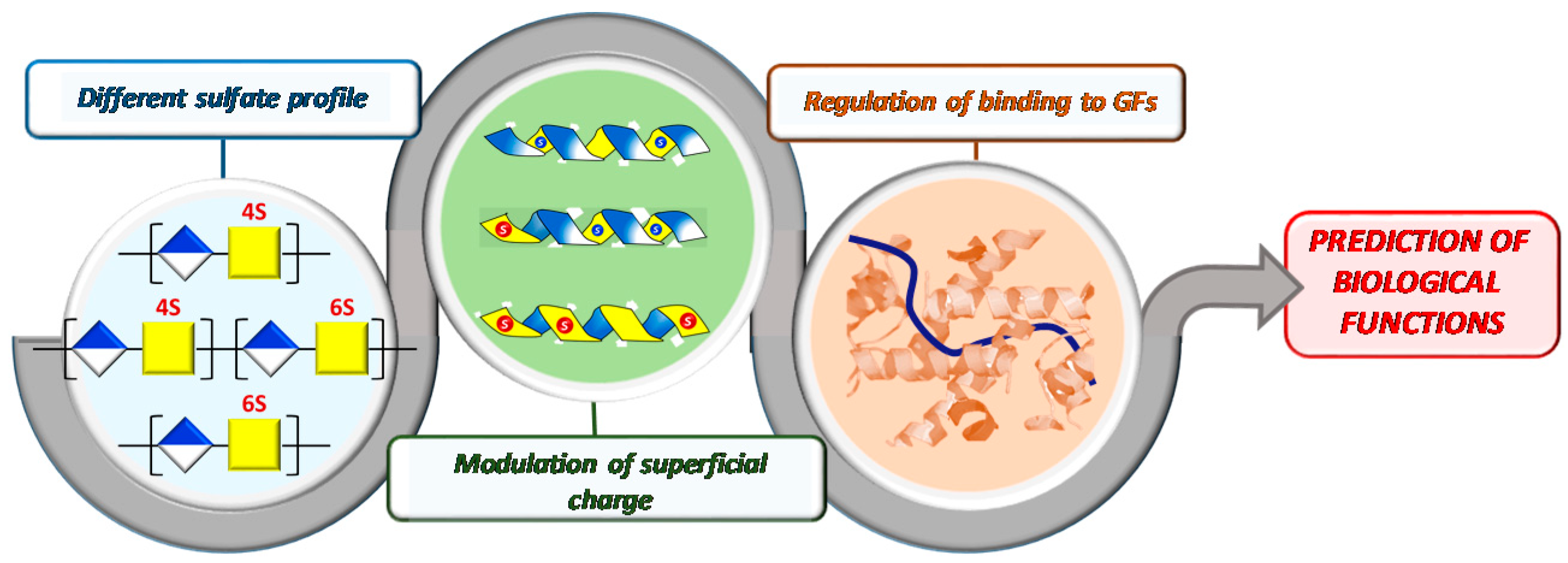
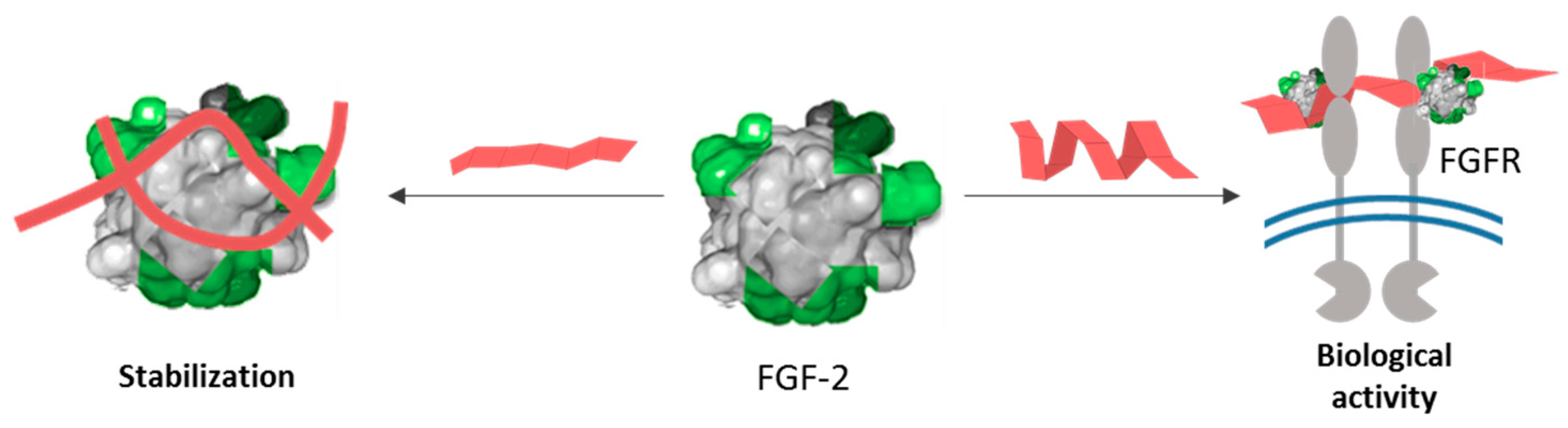
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Production of CSs from Marine Sources
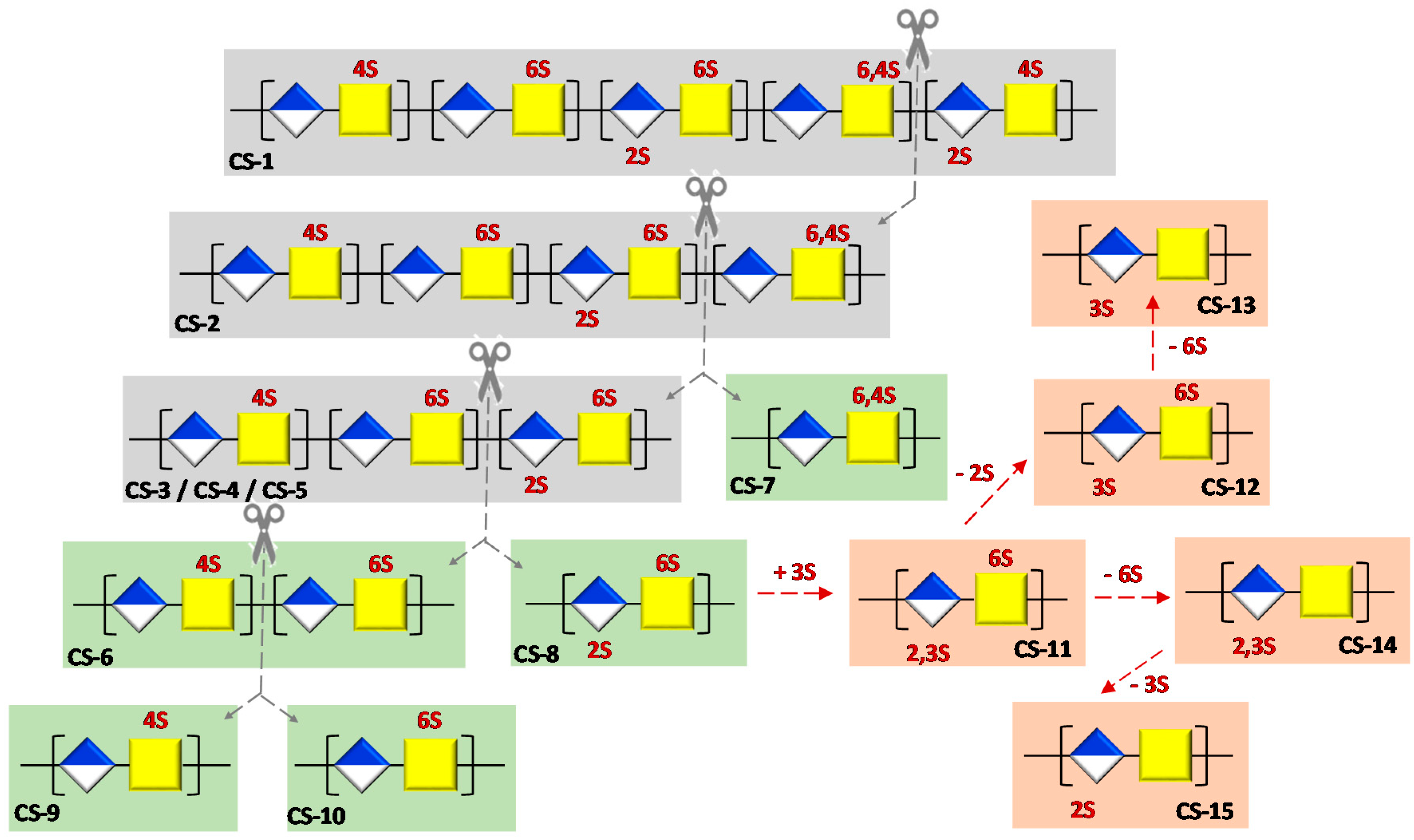
2.3. Preparation of Semi-Synthetic CSs
2.3.1. Semi-Synthesis of CS-7
2.3.2. Semi-Synthesis of CS-9 and CS-10
2.3.3. Semi-Synthesis of CS-13, CS-14 and CS-15
2.3.4. Semi-Synthesis of CS-8, CS-11 and CS-12
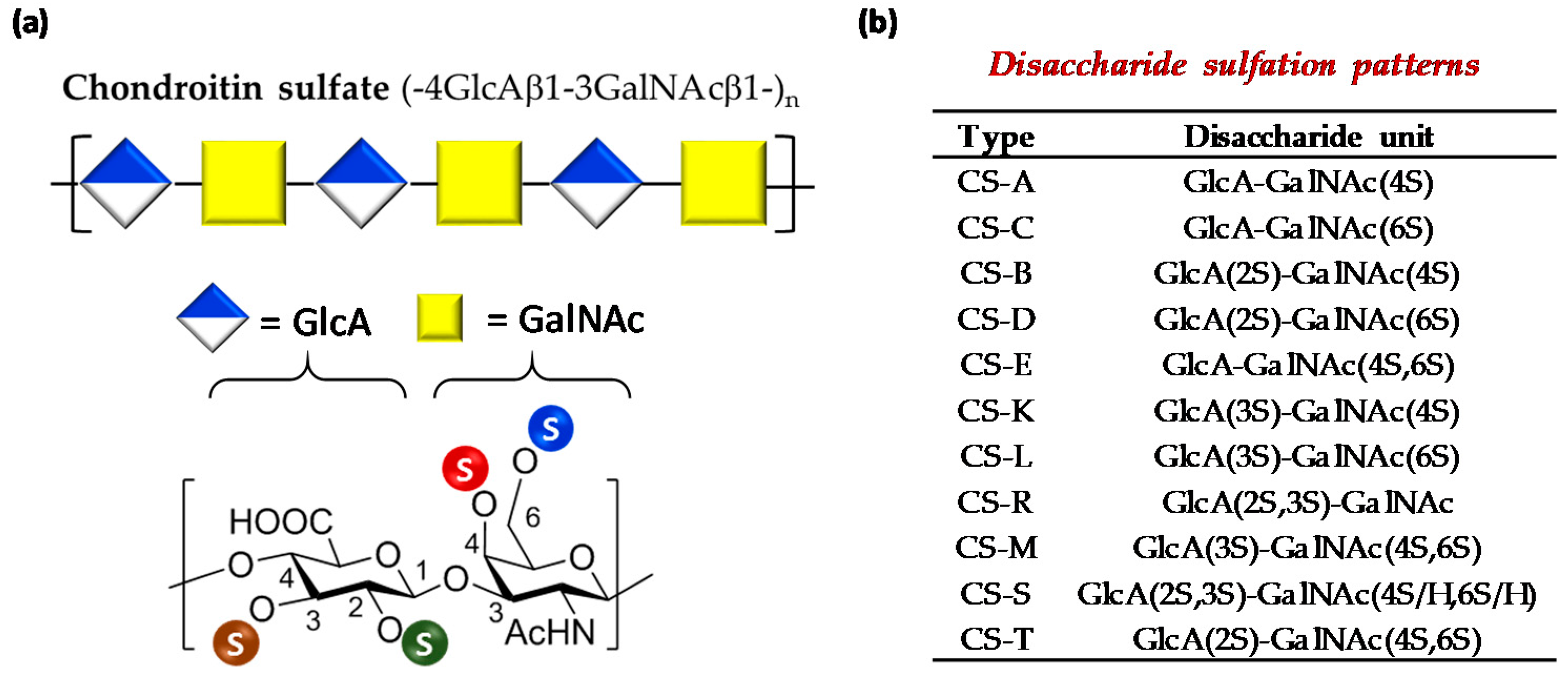
2.4. Characterization of Sulfated Polysaccharides
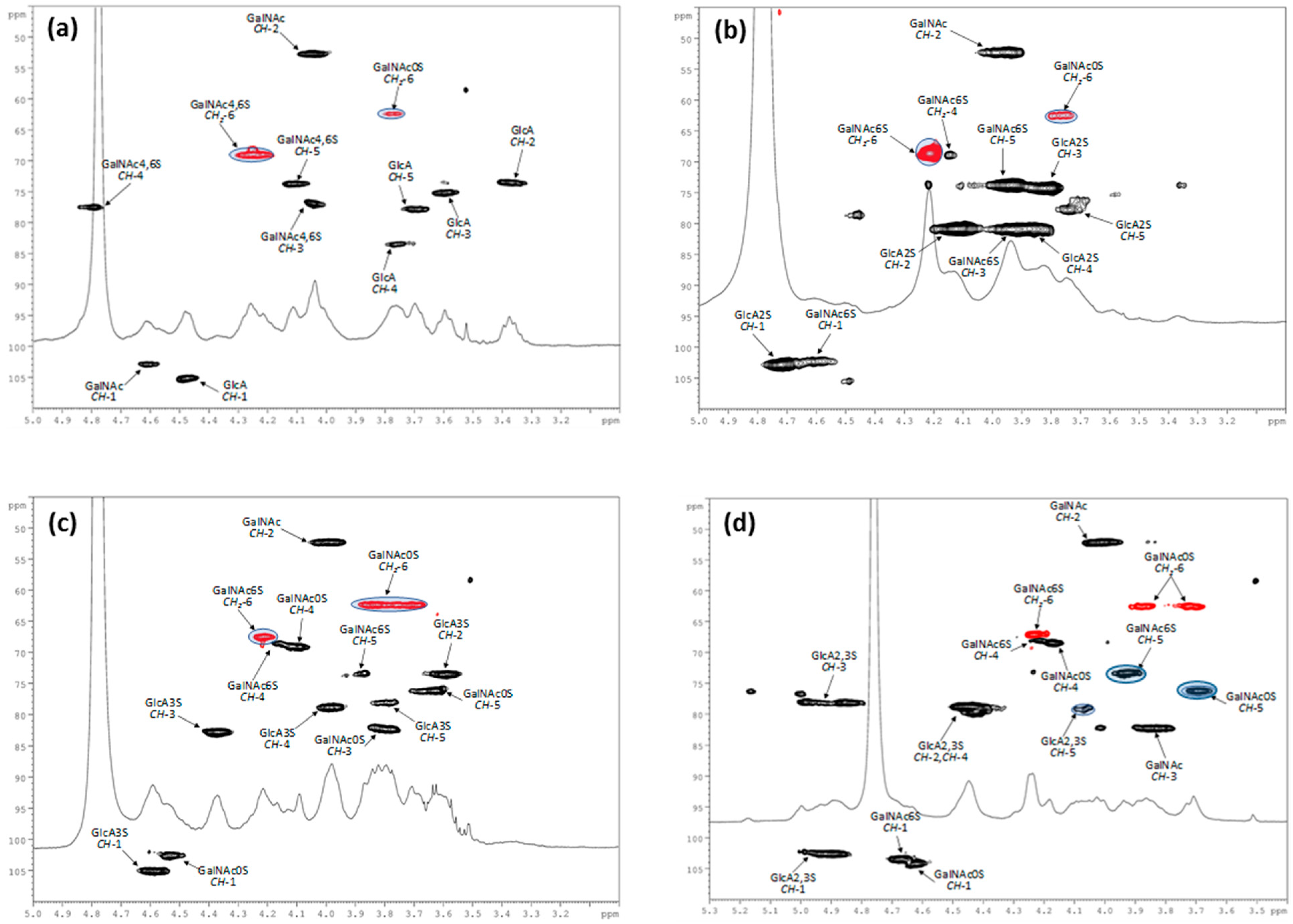
2.4.1. Disaccharide Composition
2.4.2. Nuclear Magnetic Resonance
2.4.3. Molecular Weight Determination
2.4.4. Zeta Potential Analysis
2.4.5. Circular Dichroism
2.5. Determination of Binding Affinity of Polysaccharides with FGF-2 Using Surface Plasmon Resonance (SPR)
3. Results and Discussion
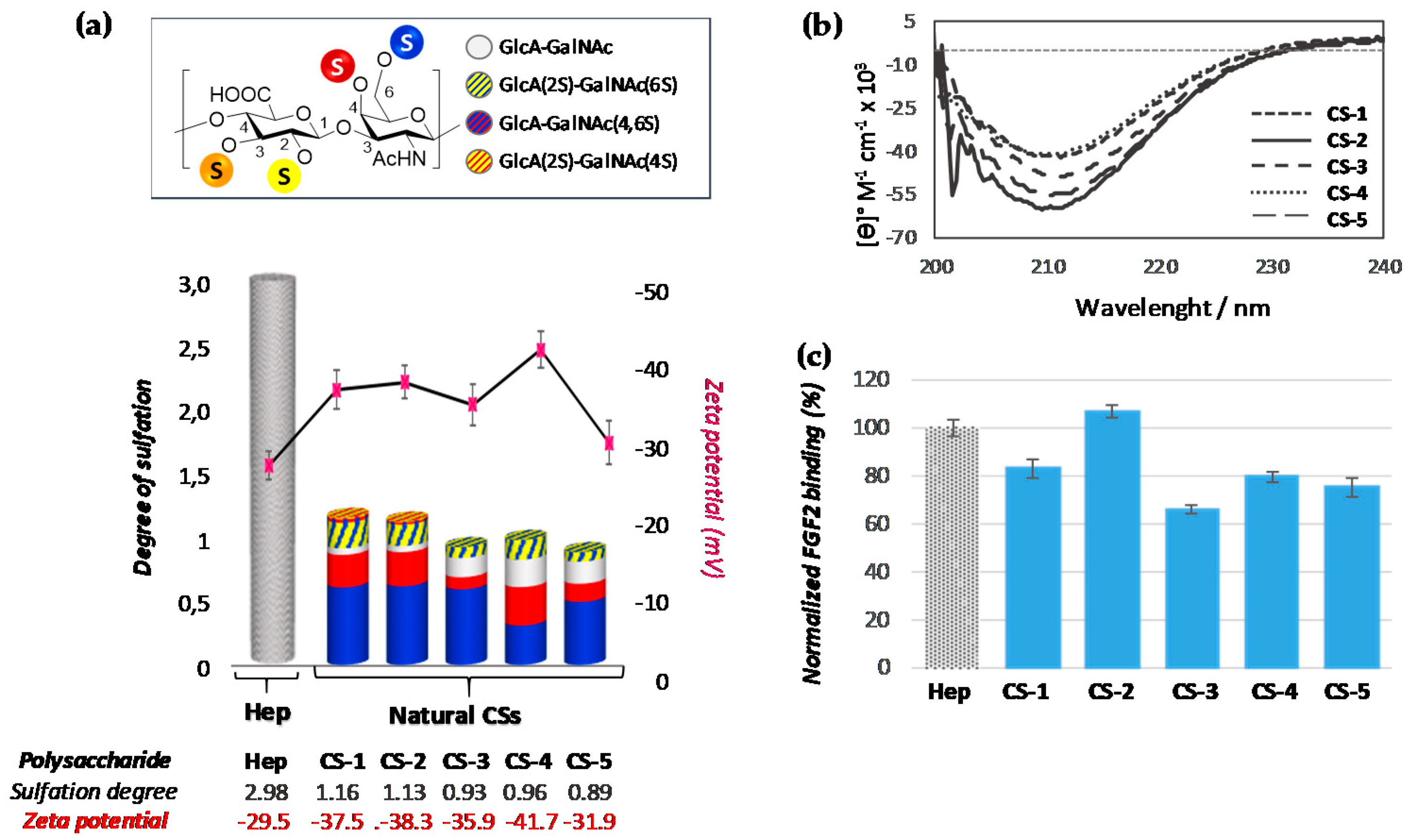
3.1. Characterization of Polysaccharides
3.2. Influence of Polysaccharide 3D-Sulfate Distribution on Their Binding Capacities with FGF-2
3.2.1. Analysis of Natural Chondroitin Sulfates
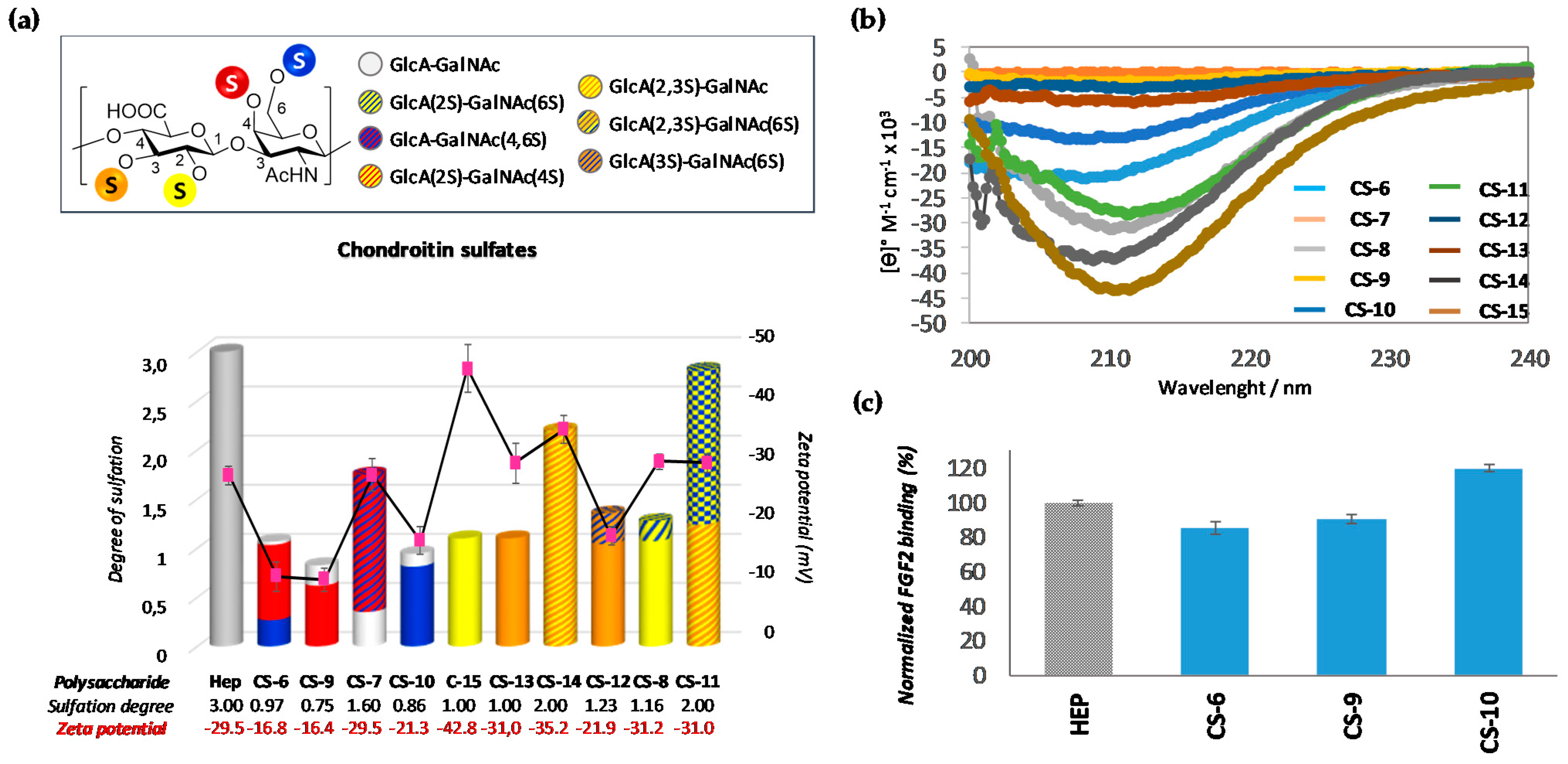
3.2.2. Analysis of Semi-Synthetic Chondroitin Sulfates with Defined Sulfation Profile
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bedini, E.; Laezza, A.; Iadonisi, A. Chemical Derivatization of Sulfated Glycosaminoglycans. Eur. J. Org. Chem. 2016, 18, 3018–3043. [Google Scholar] [CrossRef]
- López-Álvarez, M.; López-Senra, E.; Valcárcel, J.; Vázquez, J.A.; Serra, J.; González, P. Quantitative Evaluation of Sulfation Position Prevalence in Chondroitin Sulphate by Raman Spectroscopy. J. Raman Spectrosc. 2019, 50, 656–664. [Google Scholar] [CrossRef]
- Bedini, E.; Corsaro, M.M.; Fernández-Mayoralas, A.; Iadonisi, A. Chondroitin, Dermatan, Heparan, and Keratan Sulfate: Structure and Functions. In Extracellular Sugar-Based Biopolymers Matrices. Biologically-Inspired Systems, 1st ed.; Cohen, E., Merzendorfer, H., Eds.; Springer: Cham, Switzerland, 2019; Volume 12, pp. 187–233. ISBN 978-3-030-12919-4. [Google Scholar]
- Kjellén, L.; Lindahl, U. Specificity of Glycosaminoglycan-protein Interactions. Curr. Opin. Struct. Biol. 2018, 50, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Gama, C.I.; Tully, S.E.; Sotogaku, N.; Clark, P.M.; Rawat, M.; Vaidehi, N.; Goddard, W.A.; Nishi, A.; Hsieh-Wilson, L.C. Sulfation Patterns of Glycosaminoglycans Encode Molecular Recognition and Activity. Nat. Chem. Biol. 2006, 2, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Swarup, V.P.; Hsiao, T.W.; Zhang, J.; Prestwich, G.D.; Kuberan, B.; Hlady, V. Exploiting Differential Surface Display of Chondroitin Sulfate Variants for Directing Neuronal Outgrowth. J. Am. Chem. Soc. 2013, 135, 13488–13494. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Zhanga, S.; Ruoyu, J.; Qiao, M.; Jiao, R.; Li, J.; Song, P.; Zhang, X.; Huang, H. Synthesis of Structurally Defined Chondroitin Sulfate: Paving the Way to the Structure-activity Relationship Studies. Carb. Polym. 2020, 248, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Revuelta, J.; Fuentes, R.; Lagartera, L.; Hernáiz, M.J.; Bastida, A.; García-Junceda, E.; Fernández-Mayoralas, A. Assembly of Glycoamino Acid Building Blocks: A New Strategy for the Straightforward Synthesis of Heparan Sulfate Mimics. Chem. Commun. 2018, 54, 13455–13458. [Google Scholar] [CrossRef] [PubMed]
- Valcarcel, J.; Novoa-Carballal, R.; Pérez-Martín, R.I.; Reis, R.L.; Vázquez, J.A. Glycosaminoglycans from Marine Sources as Therapeutic Agents. Biotechnol. Adv. 2017, 35, 711–725. [Google Scholar] [CrossRef]
- Bedini, E.; Laezza, A.; Parrilli, M.; Iadonisi, A. A Review of Chemical Methods for the Selective Sulfation and Desulfation of Polysaccharides. Carbohydr. Polym. 2017, 174, 1224–1239. [Google Scholar] [CrossRef]
- Bedini, E.; De Castro, C.; De Rosa, M.; Di Nola, A.; Iadonisi, A.; Restaino, O.F.; Schiraldi, C.; Parrilli, M. A Microbiological-chemical Strategy to Produce Chondroitin Sulfate, A, C. Angew. Chem. Int. Ed. 2011, 50, 6160–6163. [Google Scholar] [CrossRef]
- Bedini, E.; De Castro, C.; De Rosa, M.; Di Nola, A.; Restaino, O.F.; Schiardi, C.; Parrilli, M. Semi-synthesis of Unusual Chondroitin Sulfate Polysaccharides Containing GlcA(3-O-sulfate) or GlcA(2,3-di-O-sulfate) Units. Chem. Eur. J. 2012, 18, 2123–2130. [Google Scholar] [CrossRef] [PubMed]
- Vessella, G.; Traboni, S.; Cimini, D.; Iadonisi, A.; Schiraldi, C.; Bedini, E. Development of Semisynthetic, Regioselective Pathways for Accessing the Missing Sulfation Patterns of Chondroitin Sulfate. Biomacromolecules 2019, 20, 3021–3030. [Google Scholar] [CrossRef] [PubMed]
- Joffrin, A.M.; Hsieh-Wilson, L.C. Photoaffinity Probes for the Identification of Sequence-Specific Glycosaminoglycan-Binding Proteins. J. Am. Chem. Soc. 2020, 142, 13672–13676. [Google Scholar] [CrossRef] [PubMed]
- Benito-Arenas, R.; Doncel-Pérez, E.; Fernández-Gutiérrez, M.; Garrido, L.; García-Junceda, E.; Revuelta, J.; Bastida, A.; Fernández-Mayoralas, A. A Holistic Approach to Unravelling Chondroitin Sulfation: Correlations Between Surface Charge, Structure and Binding to Growth Factors. Carbohydr. Polym. 2018, 202, 211–218. [Google Scholar] [CrossRef]
- Turnbull, J.; Powell, A.; Guimond, S. Heparan Sulfate: Decoding a Dynamic Multifunctional Cell Regulator. Trends Cell Biol. 2001, 11, 75–82. [Google Scholar] [CrossRef]
- Xu, D.; Esko, J.D. Demystifying Heparan Sulfate-Protein Interactions. Annu. Rev. Biochem. 2014, 83, 129–157. [Google Scholar] [CrossRef]
- Smock, R.G.; Meijers, R. Roles of Glycosaminoglycans as Regulators of Ligand/receptor Complexes. Open Biol. 2018, 8, 180026. [Google Scholar] [CrossRef]
- Blanco, M.; Fraguas, J.; Sotelo, C.G.; Pérez-Martín, R.I.; Vázquez, J.A. Production of Chondroitin Sulphate from Head, Skeleton and Fins of Scyliorhinus canicula By-products by Combination of Enzymatic, Chemical precipitation and Ultrafiltration Methodologies. Mar. Drugs 2015, 13, 3287–3308. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Blanco, M.; Fraguas, J.; Pastrana, L.; Pérez-Martín, R. Optimisation of the Extraction and Purification of Chondroitin Sulphate from Head By-products of Prionace glauca by Environmental Friendly Processes. Food Chem. 2016, 198, 28–35. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Fraguas, J.; Novoa-Carvallal, R.; Reis, R.; Antelo, L.; Pérez-Martín, R.; Valcarcel, J. Isolation and Chemical Characterization of Chondroitin Sulfate from Cartilage by-products of Blackmouth Catshark (Galeus melastomus). Mar. Drugs 2018, 16, 344. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Fraguas, J.; Novoa-Carballal, R.; Reis, R.L.; Pérez-Martín, R.I.; Valcarcel, J. Optimal Isolation and Characterisation of Chondroitin Sulfate from Rabbit Fish (Chimaera monstrosa). Carbohydr. Polym. 2019, 210, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Volpi, N. Hyaluronic Acid and Chondroitin Sulfate Unsaturated Disaccharides Analysis by High-Performance Liquid Chromatography and Fluorimetric Detection with Dansylhydrazine. Anal. Biochem. 2000, 277, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Vessella, G.; Traboni, S.; Pirozzi, A.V.A.; Laezza, A.; Iadonisi, A.; Schiraldi, C.; Bedini, E. A Study for the Access to a Semi-synthetic Regioisomer of Natural Fucosylated Chondroitin Sulfate with Fucosyl Branches on N-acetyl-Galactosamine Units. Mar. Drugs 2019, 17, 655. [Google Scholar] [CrossRef] [PubMed]
- Laezza, A.; Iadonisi, A.; Pirozzi, A.V.A.; Diana, P.; De Rosa, M.; Schiraldi, C.; Parrilli, M.; Bedini, E. A Modular Approach to a Library of Semi-Synthetic Fucosylated Chondroitin Sulfate Polysaccharides with Different Sulfation and Fucosylation Patterns. Chem. Eur. J. 2016, 22, 18215–18226. [Google Scholar] [CrossRef] [PubMed]
- Cimini, D.; Restaino, O.F.; Catapano, A.; De Rosa, M.; Schiraldi, C. Production of Capsular Polysaccharide from Escherichia coli K4 for Biotechnological Applications. Appl. Microbiol. Biotechnol. 2010, 85, 1779–1787. [Google Scholar] [CrossRef]
- Valoti, E.; Miraglia, N.; Bianchi, D.; Valetti, M.; Bazza, P. Shark-Like Chondroitin Sulphate and Process for the Preparation Thereof. US Patent 8,664,196 B2, 22 November 2012. [Google Scholar]
- Restaino, O.F.; Finamore, R.; Diana, P.; Marseglia, M.; Vitiello, M.; Casillo, A.; Bedini, E.; Parrilli, M.; Corsaro, M.M.; Trifuoggi, M.; et al. A Multi-analytical Approach to Better Assess the Keratan Sulfate Contamination in Animal Origin Chondroitin Sulfate. Anal. Chim. Acta 2017, 958, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Higashi, K.; Okamoto, Y.; Mukuno, A.; Wakai, J.; Hosoyama, S.; Linhardt, R.J.; Toida, T. Functional Chondroitin Sulfate from Enteroctopus dofleini Containing 3-O-Sulfo Glucuronic Acid Residue. Carbohydr. Polym. 2015, 134, 557–565. [Google Scholar] [CrossRef]
- Novoa-Carballal, R.; Pérez-Martín, R.; Blanco, M.; Sotelo, C.G.; Fassini, D.; Nunes, C.; Coimbra, M.A.; Silva, T.H.; Reis, R.L.; Vázquez, J.A. By-products of Scyliorhinus canícula, Prionace glauca and Raja clavata: A Valuable Source of Predominantly 6S Sulfated Chondroitin Sulfate. Carbohydr. Polym. 2017, 157, 31–37. [Google Scholar] [CrossRef]
- Guerrini, M.; Naggi, A.; Guglieri, S.; Santarsiero, R.; Torri, G. Complex Glycosaminoglycans: Profiling Substitution Patterns by Two-dimensional Nuclear Magnetic Resonance Spectroscopy. Anal. Biochem. 2005, 337, 35–47. [Google Scholar] [CrossRef]
- Lim, T.C.; Cai, S.; Huber, R.G.; Bond, P.J.; Chia, P.X.S.; Khou, S.L.; Gao, S.; Lee, S.S.; Song-Gil, L. Facile Saccharide-free Mimetics that Recapitulate Key Features of Glycosaminoglycan Sulfation Patterns. Chem. Sci. 2018, 9, 7940–7947. [Google Scholar] [CrossRef]
- Zong, C.; Venot, A.; Li, X.; Lu, W.; Xiao, W.; Wilkes, J.S.L.; Salanga, C.L.; Handel, T.M.; Wang, L.; Wolfert, M.A.; et al. Binding Exhibits Different Ligand Requirements Heparan Sulfate Microarray Reveals That Heparan Sulfate-Protein. J. Am. Chem. Soc. 2017, 139, 9534–9543. [Google Scholar] [CrossRef] [PubMed]
- Pudełko, A.; Wisowski, G.; Olczyk, K.; Koźma, E.M. The Dual Role of the Glycosaminoglycan Chondroitin-6-sulfate in the Development, Progression and Metastasis of Cancer. FEBS J. 2019, 286, 1815–1837. [Google Scholar] [CrossRef] [PubMed]
- Nagel, Y.A.; Raschle, P.S.; Wennemers, H. Effect of Preorganized Charge-Display on the Cell-Penetrating Properties of Cationic Peptides. Angew. Chem. Int. Ed. 2017, 56, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Raman, R.; Venkataraman, G.; Ernst, S.; Sasisekharan, V.; Sasisekharan, R. Structural Specificity of Heparin Binding in the Fibroblast Growth Factor Family of Proteins. PNAS 2003, 100, 2357–2362. [Google Scholar] [CrossRef] [PubMed]
- Deepa, S.S.; Umehara, Y.; Higashiyama, S.; Itoh, N.; Sugahara, K. Specific Molecular Interactions of Oversulfated Chondroitin Sulfate E with Various Heparin-binding Growth Factors. J. Biol. Chem. 2002, 277, 43707–43716. [Google Scholar] [CrossRef]
- Liu, Z.; Masuko, S.; Solakyildirim, K.; Pu, D.; Linhardt, R.J.; Zhang, F. Glycosaminoglycans of the Porcine Central Nervous System. Biochemistry 2010, 49, 9839–9847. [Google Scholar] [CrossRef][Green Version]
- Doncel-Pérez, E.; Aranaz, I.; Bastida, A.; Revuelta, J.; Camacho, C.; Acosta, N.; Garrido, L.; Civera, C.; García-Junceda, E.; Heras, A.; et al. Synthesis, Physicochemical Characterization and Biological Evaluation of Chitosan Sulfate as Heparan Sulfate Mimics. Carbohydr. Polym. 2018, 191, 225–233. [Google Scholar]
- Revuelta, J.; Aranaz, I.; Acosta, N.; Civera, C.; Bastida, A.; Peña, N.; Monterrey, D.T.; Doncel-Pérez, E.; Garrido, L.; Heras, A.; et al. Unraveling the Structural Landscape of Chitosan-Based Heparan Sulfate Mimics Binding to Growth Factors: Deciphering Structural Determinants for Optimal Activity. ACS Appl. Mater. Interfaces 2020, 12, 25534–25545. [Google Scholar] [CrossRef]
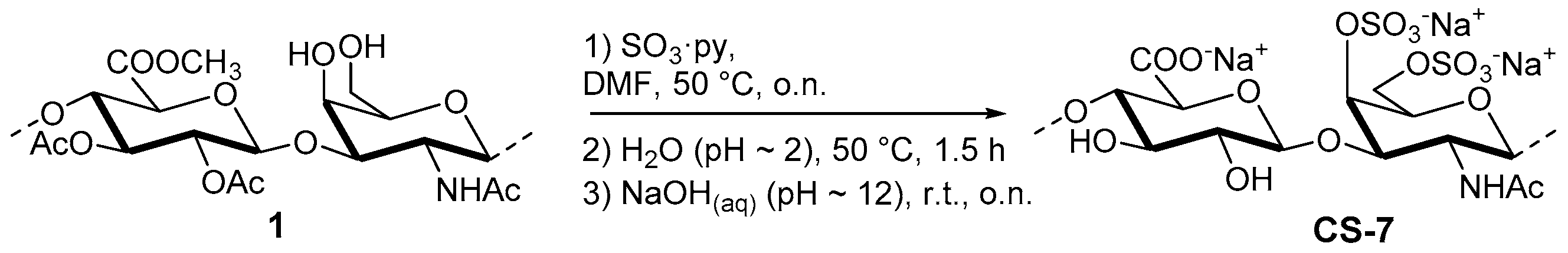


| C. monstrosa (CS-1) | G. melastomus (CS-2) | P. glauca (CS-3) | S. canicula (CS-4) | R. clavata (CS-5) | |
|---|---|---|---|---|---|
| GlcA-GalNAc(4S)[a] | 22.1 ± 0.3 | 23.8 ± 0.1 | 10.1 ± 0.1 | 31.5 ± 0.6 | 16.0 ± 0.1 |
| GlcA-GalNAc(6S)[a] | 52.7 ± 0.1 | 54.9 ± 0.4 | 64.2 ± 0.4 | 32.4 ± 0.1 | 55.9 ± 0.1 |
| GlcA-GalNAc[a] | 4.4 ± 0.2 | 4.2 ± 0.5 | 16.3 ± 0.6 | 22.5 ± 1.4 | 19.5 ± 0.1 |
| GlcA(2S)-GalNAc(6S)[a] | 17.4 ± 0.1 | 15.0 ± 0.1 | 9.3 ± 0.2 | 16.3 ± 0.2 | 8.6 ± 0.1 |
| GlcA-GalNAc(4,6S)[a] | 2.4 ± 0.1 | 1.5 ± 0.0 | n.o[b] | n.o[b] | n.o[b] |
| GlcA(2S)-GalNAc(4S)[a] | 1.0 ± 0.0 | 0.6 | n.o[b] | n.o[b] | n.o[b] |
| Sulfation degree | 1.16 | 1.13 | 0.93 | 0.96 | 0.89 |
| 4S/6S ratio | 0.32 | 0.34 | 0.15 | 0.64 | 0.26 |
| Polysaccharide | Disaccharide Units | Ratio | Sulfation Degree |
|---|---|---|---|
| CS-6[a] | GlcA-GalNAc(6S)/GlcA-GalNAc(4S)/GlcA-GalNAc | 25:72:3 | 0.97 |
| CS-7 | GlcA-GalNAc(4,6S)/GlcA-GalNAc | 80:20 | 1.60 |
| CS-8 | GlcA(2S)-GalNAc(6S)/GlcA(2S)-GalNAc | 16:84 | 1.16 |
| CS-9 | GlcA-GalNAc(4S)/GlcA-GalNAc | 75:25 | 0.75 |
| CS-10 | GlcA-GalNAc(6S)/GlcA-GalNAc | 86:14 | 0.86 |
| CS-11 | GlcA(2S,3S)-GalNAc/GlcA(2,3S)-GalNAc(6S) | 44:56 | 2.56 |
| CS-12 | GlcA(3S)-GalNAc/GlcA(3S)-GalNAc(6S) | 77:23 | 1.23 |
| CS-13 | GlcA(3S)-GalNAc | 100 | 1.00 |
| CS-14 | GlcA(2,3S)-GalNAc | 100 | 2.00 |
| CS-15 | GlcA(2S)-GalNAc | 100 | 1.00 |
| Polysaccharide | kD1 (M) | kD2 (M) |
|---|---|---|
| CS-1[a] | 1.8 × 10−5 | 8.6 × 10−8 |
| CS-2 | 7.4 × 10−6 | 2.1 × 10−8 |
| CS-4 | 3.4 × 10−6 | 6.8 × 10−9 |
| Entry | Polysaccharide | kD1 (M) | kD2 (M) |
|---|---|---|---|
| 1 | CS-6 | 1.3 × 10−4 | 1.08 × 10−6 |
| 2 | CS-7[a] | 1.31 × 10−6 | 3.88 × 10−9 |
| 3 | CS-15[a] | 3.6 × 10−6 | 7.77 × 10−9 |
| 4 | CS-13 | 6.6 × 10−6 | - |
| 5 | CS-14 | 8.6 × 10−6 | 3.37 × 10−8 |
| 6 | CS-12 | 4.0 × 10−8 | 2.9 × 10−6 |
| 7 | CS-8 | 1.75 × 10−8 | 2.56 × 10−6 |
| 8 | CS-11 | 1.33 × 10−6 | 4.75 × 10−9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vessella, G.; Vázquez, J.A.; Valcárcel, J.; Lagartera, L.; Monterrey, D.T.; Bastida, A.; García-Junceda, E.; Bedini, E.; Fernández-Mayoralas, A.; Revuelta, J. Deciphering Structural Determinants in Chondroitin Sulfate Binding to FGF-2: Paving the Way to Enhanced Predictability of Their Biological Functions. Polymers 2021, 13, 313. https://doi.org/10.3390/polym13020313
Vessella G, Vázquez JA, Valcárcel J, Lagartera L, Monterrey DT, Bastida A, García-Junceda E, Bedini E, Fernández-Mayoralas A, Revuelta J. Deciphering Structural Determinants in Chondroitin Sulfate Binding to FGF-2: Paving the Way to Enhanced Predictability of Their Biological Functions. Polymers. 2021; 13(2):313. https://doi.org/10.3390/polym13020313
Chicago/Turabian StyleVessella, Giulia, José Antonio Vázquez, Jesús Valcárcel, Laura Lagartera, Dianélis T. Monterrey, Agatha Bastida, Eduardo García-Junceda, Emiliano Bedini, Alfonso Fernández-Mayoralas, and Julia Revuelta. 2021. "Deciphering Structural Determinants in Chondroitin Sulfate Binding to FGF-2: Paving the Way to Enhanced Predictability of Their Biological Functions" Polymers 13, no. 2: 313. https://doi.org/10.3390/polym13020313
APA StyleVessella, G., Vázquez, J. A., Valcárcel, J., Lagartera, L., Monterrey, D. T., Bastida, A., García-Junceda, E., Bedini, E., Fernández-Mayoralas, A., & Revuelta, J. (2021). Deciphering Structural Determinants in Chondroitin Sulfate Binding to FGF-2: Paving the Way to Enhanced Predictability of Their Biological Functions. Polymers, 13(2), 313. https://doi.org/10.3390/polym13020313