The Role of Surface Chemistry in the Efficacy of Protein and DNA Microarrays for Label-Free Detection: An Overview
Abstract
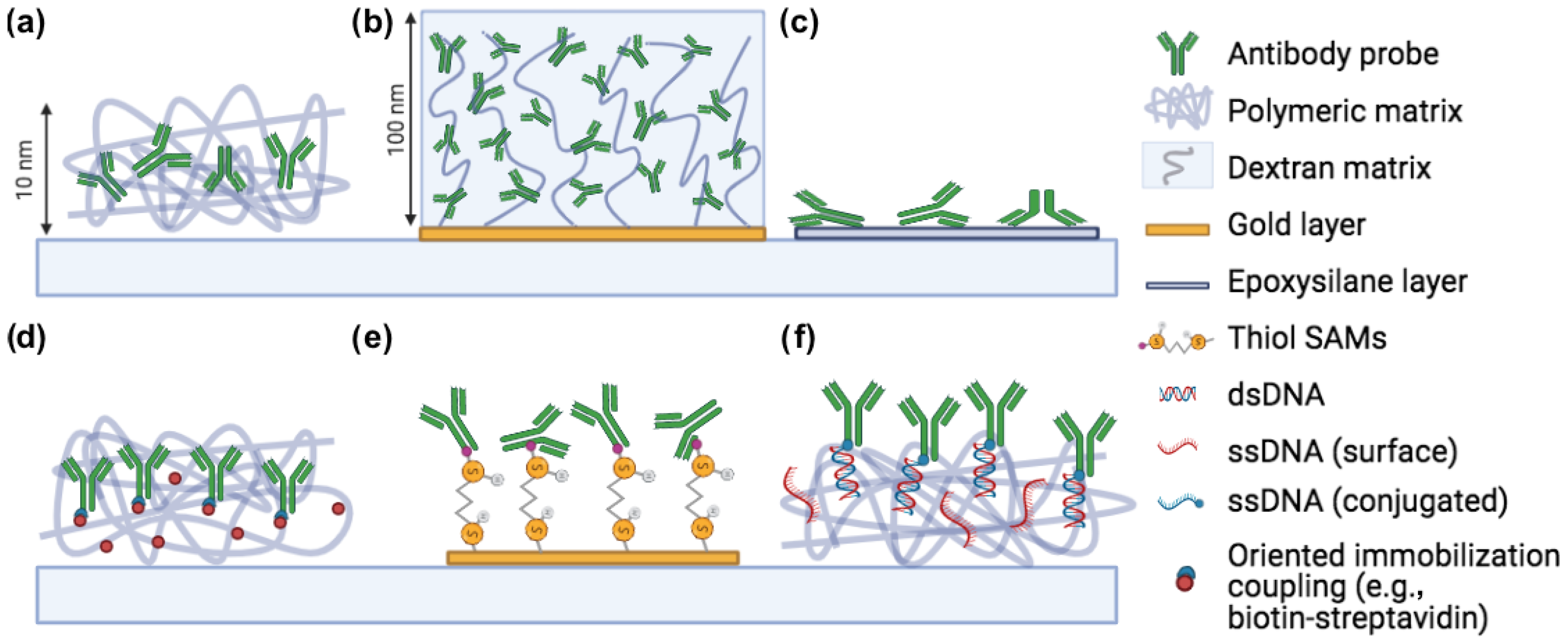
:1. Introduction
2. Popular Functionalization Methods
2.1. Types of Surfaces and Biorecognition Elements
2.2. Physisorption: Spontaneous Adsorption on the Surface
2.3. Polymeric Coatings
2.4. Nanostructure-Based Methods
2.5. Other Methods
3. Microarray Types and Specific Immobilization Strategies
3.1. Microarrays for Label-Free Single Particle Imaging
3.1.1. DNA-Directed Microarrays
3.1.2. Peptide Microarrays
3.2. Protein Microarrays for Binding Kinetics Assays
3.3. Surface Chemistry for Small Molecule Kinetics
4. Novel Technologies and Applications
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Govindarajan, R.; Duraiyan, J.; Kaliyappan, K.; Palanisamy, M. Microarray and its applications. J. Pharm. Bioallied Sci. 2012, 4, S310–S312. [Google Scholar] [CrossRef]
- Dufva, M. Fabrication of high quality microarrays. Biomol. Eng. 2005, 22, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Gergen, J.P.; Stern, R.H.; Wensink, P.C. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Res. 1979, 7, 2115–2136. [Google Scholar] [CrossRef] [Green Version]
- Bumgarner, R. Overview of DNA microarrays: Types, applications, and their future. Curr. Protoc. Mol. Biol. 2013, 101, 22.1.1–22.1.11. [Google Scholar] [CrossRef] [Green Version]
- Statnikov, A.; Aliferis, C.F.; Tsamardinos, I.; Hardin, D.; Levy, S. A comprehensive evaluation of multicategory classification methods for microarray gene expression cancer diagnosis. Bioinformatics 2004, 21, 631–643. [Google Scholar] [CrossRef] [Green Version]
- Statnikov, A.; Tsamardinos, I.; Dosbayev, Y.; Aliferis, C.F. GEMS: A system for automated cancer diagnosis and biomarker discovery from microarray gene expression data. Int. J. Med. Inform. 2005, 74, 491–503. [Google Scholar] [CrossRef]
- Perez-Diez, A.; Morgun, A.; Shulzhenko, N. Microarrays for Cancer Diagnosis and Classification; Springer: New York, NY, USA, 2007; pp. 74–85. [Google Scholar] [CrossRef]
- Cretich, M.; Monroe, M.R.; Reddington, A.; Zhang, X.; Daaboul, G.G.; Damin, F.; Sola, L.; Unlu, M.S.; Chiari, M. Interferometric silicon biochips for label and label-free DNA and protein microarrays. Proteomics 2012, 12, 2963–2977. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.S.; Nock, S. Functional protein microarrays. Curr. Opin. Chem. Biol. 2002, 6, 81–85. [Google Scholar] [CrossRef]
- LaBaer, J.; Ramachandran, N. Protein microarrays as tools for functional proteomics. Curr. Opin. Chem. Biol. 2005, 9, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer, B.; Kingsmore, S.F. Measuring proteins on microarrays. Curr. Opin. Biotechnol. 2002, 13, 14–19. [Google Scholar] [CrossRef]
- Zhu, H.; Snyder, M. Protein arrays and microarrays. Curr. Opin. Chem. Biol. 2001, 5, 40–45. [Google Scholar] [CrossRef]
- Jones, S.; Thornton, J.M. Principles of protein—Protein interactions. Proc. Natl. Acad. Sci. USA 1996, 93, 13–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nirschl, M.; Reuter, F.; Vörös, J. Review of Transducer Principles for Label-Free Biomolecular Interaction Analysis. Biosensors 2011, 1, 70–92. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Goldberg, A.F.; Stoltz, B.M. Label-free detection of single nanoparticles and biological molecules using microtoroid optical resonators. Light Sci. Appl. 2016, 5, e16001. [Google Scholar] [CrossRef] [Green Version]
- Han, A.; Creus, M.; Schürmann, G.; Linder, V.; Ward, T.R.; De Rooij, N.F.; Staufer, U. Label-Free Detection of Single Protein Molecules and Protein-Protein Interactions Using Synthetic Nanopores. Anal. Chem. 2008, 80, 4651–4658. [Google Scholar] [CrossRef] [Green Version]
- Mullard, A. 2019 FDA Drug Approvals. Nat. Rev. Drug Discov. 2020, 19, 79–84. [Google Scholar] [CrossRef]
- Piehler, J.; Brecht, A.; Gauglitz, G. Affinity Detection of Low Molecular Weight Analytes. Anal. Chem. 1996, 68, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Myszka, D.G. Analysis of small-molecule interactions using Biacore S51 technology. Anal. Biochem. 2004, 329, 316–323. [Google Scholar] [CrossRef]
- Chiodi, E.; Marn, A.M.; Geib, M.; Ekiz Kanik, F.; Rejman, J.; AnKrapp, D.; Unlu, M. Highly Multiplexed Label-Free Imaging Sensor for Accurate Quantification of Small-Molecule Binding Kinetics. ACS Omega 2020, 5, 25358–25364. [Google Scholar] [CrossRef]
- Mitchell, J. Small Molecule Immunosensing Using Surface Plasmon Resonance. Sensors 2010, 10, 7323–7346. [Google Scholar] [CrossRef] [Green Version]
- Papalia, G.A.; Leavitt, S.; Bynum, M.A.; Katsamba, P.S.; Wilton, R.; Qiu, H.; Steukers, M.; Wang, S.; Bindu, L.; Phogat, S.; et al. Comparative analysis of 10 small molecules binding to carbonic anhydrase II by different investigators using Biacore technology. Anal. Biochem. 2006, 359, 94–105. [Google Scholar] [CrossRef]
- Daaboul, G.; Vedula, R.; Ahn, S.; Lopez, C.; Reddington, A.; Ozkumur, E.; Ünlü, M. LED-based Interferometric Reflectance Imaging Sensor for quantitative dynamic monitoring of biomolecular interactions. Biosens. Bioelectron. 2011, 26, 2221–2227. [Google Scholar] [CrossRef]
- Bunroddith, K.; Viseshakul, N.; Chansiri, K.; Lieberzeit, P. QCM-based rapid detection of PCR amplification products of Ehrlichia canis. Anal. Chim. Acta 2018, 1001, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Soung, Y.; Ford, S.; Zhang, V.; Chung, J. Exosomes in Cancer Diagnostics. Cancers 2017, 9, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.; Rasool, H.I.; Palanisamy, V.; Mathisen, C.; Schmidt, M.; Wong, D.T.; Gimzewski, J.K. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano 2010, 4, 1921–1926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Deng, W.; Klinke, D.J.N. Exosomes: Improved methods to characterize their morphology, RNA content, and surface protein biomarkers. Analyst 2015, 140, 6631–6642. [Google Scholar] [CrossRef] [Green Version]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, C.A.; Daaboul, G.G.; Vedula, R.S.; Ozkumur, E.; Bergstein, D.A.; Geisbert, T.W.; Fawcett, H.E.; Goldberg, B.B.; Connor, J.H.; Unlü, M.S. Label-free multiplexed virus detection using spectral reflectance imaging. Biosens. Bioelectron. 2011, 26, 3432–3437. [Google Scholar] [CrossRef] [Green Version]
- Picciolini, S.; Gualerzi, A.; Vanna, R.; Sguassero, A.; Gramatica, F.; Bedoni, M.; Masserini, M.; Morasso, C. Detection and Characterization of Different Brain-Derived Subpopulations of Plasma Exosomes by Surface Plasmon Resonance Imaging. Anal. Chem. 2018, 90, 8873–8880. [Google Scholar] [CrossRef]
- Chiodi, E.; Marn, A.M.; Daaboul, G.; Unlu, M. Real-time measurements of extracellular vesicles binding kinetics in a multiplexed microarray modality. Chemrxiv 2021. [Google Scholar] [CrossRef]
- Pilot, R.; Signorini, R.; Durante, C.; Orian, L.; Bhamidipati, M.; Fabris, L. A Review on Surface-Enhanced Raman Scattering. Biosensors 2019, 9, 57. [Google Scholar] [CrossRef] [Green Version]
- Giavazzi, F.; Salina, M.; Cerbino, R.; Bassi, M.; Prosperi, D.; Ceccarello, E.; Damin, F.; Sola, L.; Rusnati, M.; Chiari, M.; et al. Multispot, label-free biodetection at a phantom plastic–water interface. Proc. Natl. Acad. Sci. USA 2013, 110, 9350–9355. [Google Scholar] [CrossRef] [Green Version]
- Zanchetta, G.; Lanfranco, R.; Giavazzi, F.; Bellini, T.; Buscaglia, M. Emerging applications of label-free optical biosensors. Nanophotonics 2017, 6, 627–645. [Google Scholar] [CrossRef]
- Tagliabue, G.; Faoro, V.; Rizzo, S.; Sblattero, D.; Saccani, A.; Riccio, G.; Bellini, T.; Salina, M.; Buscaglia, M.; Marcello, A. A label-free immunoassay for Flavivirus detection by the Reflective Phantom Interface technology. Biochem. Biophys. Res. Commun. 2017, 492, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Yaseen, T.; Pu, H.; Sun, D.W. Functionalization techniques for improving SERS substrates and their applications in food safety evaluation: A review of recent research trends. Trends Food Sci. Technol. 2018, 72, 162–174. [Google Scholar] [CrossRef]
- Trilling, A.K.; Harmsen, M.M.; Ruigrok, V.J.B.; Zuilhof, H.; Beekwilder, J. The effect of uniform capture molecule orientation on biosensor sensitivity: Dependence on analyte properties. Biosens. Bioelectron. 2013, 40, 219–226. [Google Scholar] [CrossRef]
- Nimse, S.; Song, K.; Sonawane, M.; Sayyed, D.; Kim, T. Immobilization Techniques for Microarray: Challenges and Applications. Sensors 2014, 14, 22208–22229. [Google Scholar] [CrossRef] [Green Version]
- Seurynck-Servoss, S.L.; White, A.M.; Baird, C.L.; Rodland, K.D.; Zangar, R.C. Evaluation of surface chemistries for antibody microarrays. Anal. Biochem. 2007, 371, 105–115. [Google Scholar] [CrossRef] [Green Version]
- Pirri, G.; Damin, F.; Chiari, M.; Bontempi, E.; Depero, L.E. Characterization of A Polymeric Adsorbed Coating for DNA Microarray Glass Slides. Anal. Chem. 2004, 76, 1352–1358. [Google Scholar] [CrossRef]
- Cretich, M.; Pirri, G.; Damin, F.; Solinas, I.; Chiari, M. A new polymeric coating for protein microarrays. Anal. Chem. 2004, 332, 67–74. [Google Scholar] [CrossRef]
- Erb, E.M.; Chen, X.; Allen, S.; Roberts, C.J.; Tendler, S.J.; Davies, M.C.; Forsén, S. Characterization of the surfaces generated by liposome binding to the modified dextran matrix of a surface plasmon resonance sensor chip. Anal. Biochem. 2000, 280, 29–35. [Google Scholar] [CrossRef]
- Kim, D.K.; Kerman, K.; Hiep, H.M.; Saito, M.; Yamamura, S.; Takamura, Y.; Kwon, Y.S.; Tamiya, E. Label-free optical detection of aptamer–protein interactions using gold-capped oxide nanostructures. Anal. Biochem. 2008, 379, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Khranovskyy, V.; Ekblad, T.; Yakimova, R.; Hultman, L. Surface morphology effects on the light-controlled wettability of ZnO nanostructures. Appl. Surf. Sci. 2012, 258, 8146–8152. [Google Scholar] [CrossRef] [Green Version]
- Tsougeni, K.; Ellinas, K.; Koukouvinos, G.; Petrou, P.S.; Tserepi, A.; Kakabakos, S.E.; Gogolides, E. Three-dimensional (3D) plasma micro-nanotextured slides for high performance biomolecule microarrays: Comparison with epoxy-silane coated glass slides. Colloids Surf. B Biointerfaces 2018, 165, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Odinolfi, M.T.; Romanato, A.; Bergamaschi, G.; Strada, A.; Sola, L.; Girella, A.; Milanese, C.; Chiari, M.; Gori, A.; Cretich, M. Clickable cellulosic surfaces for peptide-based bioassays. Talanta 2019, 205, 120152. [Google Scholar] [CrossRef] [PubMed]
- Cretich, M.; Sedini, V.; Damin, F.; Pelliccia, M.; Sola, L.; Chiari, M. Coating of nitrocellulose for colorimetric DNA microarrays. Anal. Biochem. 2010, 397, 84–88. [Google Scholar] [CrossRef]
- Yin, L.T.; Hu, C.Y.; Chang, C.H. A single layer nitrocellulose substrate for fabricating protein chips. Sens. Actuators B Chem. 2008, 130, 374–378. [Google Scholar] [CrossRef]
- Zhao, Z.; Peytavi, R.; Diaz-Quijada, G.A.; Picard, F.J.; Huletsky, A.; Leblanc, E.; Frenette, J.; Boivin, G.; Veres, T.; Dumoulin, M.M.; et al. Plastic polymers for efficient DNA microarray hybridization: Application to microbiological diagnostics. J. Clin. Microbiol. 2008, 46, 3752–3758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benters, R.; Niemeyer, C.M.; Wöhrle, D. Dendrimer-activated solid supports for nucleic acid and protein microarrays. Chembiochem 2001, 2, 686–694. [Google Scholar] [CrossRef]
- Guschin, D.; Yershov, G.; Zaslavsky, A.; Gemmell, A.; Shick, V.; Proudnikov, D.; Arenkov, P.; Mirzabekov, A. Manual Manufacturing of Oligonucleotide, DNA, and Protein Microchips. Anal. Biochem. 1997, 250, 203–211. [Google Scholar] [CrossRef] [Green Version]
- Arenkov, P.; Kukhtin, A.; Gemmell, A.; Voloshchuk, S.; Chupeeva, V.; Mirzabekov, A. Protein Microchips: Use for Immunoassay and Enzymatic Reactions. Anal. Biochem. 2000, 278, 123–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dufva, M.; Petronis, S.; Jensen, L.B.; Krag, C.; Christensen, C.B.V. Characterization of an inexpensive, nontoxic, and highly sensitive microarray substrate. Biotechniques 2004, 37, 286–292. [Google Scholar] [CrossRef]
- Sola, L.; Gori, A.; Cretich, M.; Finetti, C.; Zilio, C.; Chiari, M. Clickable Polymeric Coating for Oriented Peptide Immobilization. Methods Mol. Biol. 2016, 1352, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Zilio, C.; Bernardi, A.; Palmioli, A.; Salina, M.; Tagliabue, G.; Buscaglia, M.; Consonni, R.; Chiari, M. New “clickable” polymeric coating for glycan microarrays. Sens. Actuators B Chem. 2015, 215, 412–420. [Google Scholar] [CrossRef] [Green Version]
- Meyer, R.; Giselbrecht, S.; Rapp, B.E.; Hirtz, M.; Niemeyer, C.M. Advances in DNA-directed immobilization. Curr. Opin. Chem. Biol. 2014, 18, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, D.; Sola, L.; Chiari, M. Advantageous antibody microarray fabrication through DNA-directed immobilization: A step toward use of extracellular vesicles in diagnostics. Talanta 2021, 222, 121542. [Google Scholar] [CrossRef]
- Lee, J.M.; Park, H.K.; Jung, Y.; Kim, J.K.; Jung, S.O.; Chung, B.H. Direct Immobilization of Protein G Variants with Various Numbers of Cysteine Residues on a Gold Surface. Anal. Chem. 2007, 79, 2680–2687. [Google Scholar] [CrossRef]
- Lee, J.W.; Sim, S.J.; Cho, S.M.; Lee, J. Characterization of a self-assembled monolayer of thiol on a gold surface and the fabrication of a biosensor chip based on surface plasmon resonance for detecting anti-GAD antibody. Biosens. Bioelectron. 2005, 20, 1422–1427. [Google Scholar] [CrossRef]
- Brena, B.; González-Pombo, P.; Batista-Viera, F. Immobilization of Enzymes: A Literature Survey; Humana Press: Totowa, NJ, USA, 2013; pp. 15–31. [Google Scholar] [CrossRef]
- Spahn, C.; Minteer, S.D. Enzyme Immobilization in Biotechnology. Recent Patents Eng. 2008, 2, 195–200. [Google Scholar] [CrossRef]
- Hirsh, S.L.; Bilek, M.M.M.; Nosworthy, N.J.; Kondyurin, A.; Dos Remedios, C.G.; McKenzie, D.R. A Comparison of Covalent Immobilization and Physical Adsorption of a Cellulase Enzyme Mixture. Langmuir 2010, 26, 14380–14388. [Google Scholar] [CrossRef]
- Der Voort, P.V.; Vansant, E.F. Silylation of the Silica Surface A Review. J. Liq. Chromatogr. Relat. Technol. 1996, 19, 2723–2752. [Google Scholar] [CrossRef]
- Chiu, S.K.; Hsu, M.; Ku, W.C.; Tu, C.Y.; Tseng, Y.T.; Lau, W.K.; Yan, R.Y.; Ma, J.T.; Tzeng, C.M. Synergistic effects of epoxy- and amine-silanes on microarray DNA immobilization and hybridization. Biochem. J. 2003, 374, 625–632. [Google Scholar] [CrossRef]
- Cretich, M.; Reddington, A.; Monroe, M.; Bagnati, M.; Damin, F.; Sola, L.; Unlu, M.S.; Chiari, M. Silicon biochips for dual label-free and fluorescence detection: Application to protein microarray development. Biosens. Bioelectron. 2011, 26, 3938–3943. [Google Scholar] [CrossRef] [Green Version]
- Cretich, M.; Di Carlo, G.; Longhi, R.; Gotti, C.; Spinella, N.; Coffa, S.; Galati, C.; Renna, L.; Chiari, M. High Sensitivity Protein Assays on Microarray Silicon Slides. Anal. Chem. 2009, 81, 5197–5203. [Google Scholar] [CrossRef] [PubMed]
- Olle, E.W.; Messamore, J.; Deogracias, M.P.; McClintock, S.D.; Anderson, T.D.; Johnson, K.J. Comparison of antibody array substrates and the use of glycerol to normalize spot morphology. Exp. Mol. Pathol. 2005, 79, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Yalçın, A.; Damin, F.; Özkumur, E.; Di Carlo, G.; Goldberg, B.B.; Chiari, M.; Ünlü, M.S. Direct Observation of Conformation of a Polymeric Coating with Implications in Microarray Applications. Anal. Chem. 2009, 81, 625–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sola, L.; Damin, F.; Gagni, P.; Consonni, R.; Chiari, M. Synthesis of Clickable Coating Polymers by Postpolymerization Modification: Applications in Microarray Technology. Langmuir 2016, 32, 10284–10295. [Google Scholar] [CrossRef] [PubMed]
- Chikkaveeraiah, B.V.; Bhirde, A.A.; Morgan, N.Y.; Eden, H.S.; Chen, X. Electrochemical Immunosensors for Detection of Cancer Protein Biomarkers. ACS Nano 2012, 6, 6546–6561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Won, J.Y.; Seo, S.; Choi, J.W.; Min, J. The fabrication of protein nano arrays using 3-dimensional plastic nanopillar patterns. J. Nanosci. Nanotechnol. 2011, 11, 4231–4235. [Google Scholar] [CrossRef]
- Abdelhamid, H.N. Biointerface Between ZIF-8 and Biomolecules and their Applications. Biointerface Res. Appl. Chem. 2021, 11, 8283–8297. [Google Scholar]
- Neto, A.; Custodio, C.; Song, W.; Mano, J.F. High-throughput evaluation of interactions between biomaterials, proteins and cells using patterned superhydrophobic substrates. Soft Matter 2011, 7, 4147–4151. [Google Scholar] [CrossRef] [Green Version]
- Al-Rubaye, A.G.; Nabok, A.; Tsargorodska, A. LSPR Biosensor Based on Nanostructured Gold Films: Detection of Mycotoxins. Procedia Technol. 2017, 27, 131–132. [Google Scholar] [CrossRef]
- Ghanim Al-Rubaye, A.; Nabok, A.; Catanante, G.; Marty, J.L.; Takács, E.; Székács, A. Label-Free Optical Detection of Mycotoxins Using Specific Aptamers Immobilized on Gold Nanostructures. Toxins 2018, 10, 291. [Google Scholar] [CrossRef] [Green Version]
- Hwang, G.M.; Pang, L.; Mullen, E.H.; Fainman, Y. Plasmonic Sensing of Biological Analytes Through Nanoholes. IEEE Sens. J. 2008, 8, 2074–2079. [Google Scholar] [CrossRef]
- Lin, D.; Harris, K.D.; Chan, N.W.; Jemere, A.B. Nanostructured indium tin oxide electrodes immobilized with toll-like receptor proteins for label-free electrochemical detection of pathogen markers. Sens. Actuators B Chem. 2018, 257, 324–330. [Google Scholar] [CrossRef]
- Kusnezow, W.; Hoheisel, J.D. Solid supports for microarray immunoassays. J. Mol. Recognit. 2003, 16, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.; Chilkoti, A.; Moy, V.T. Direct force measurements of the streptavidin—biotin interaction. Biomol. Eng. 1999, 16, 45–55. [Google Scholar] [CrossRef]
- Chivers, C.E.; Koner, A.L.; Lowe, E.D.; Howarth, M. How the biotin-streptavidin interaction was made even stronger: Investigation via crystallography and a chimaeric tetramer. Biochem. J. 2011, 435, 55–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brambilla, D.; Sola, L.; Chiodi, E.; Zarovni, N.; Fortunato, D.; Criscuoli, M.; Dolo, V.; Giusti, I.; Murdica, V.; Vago, R.; et al. Separation of Extracellular Vesicles by DNA-Directed Immunocapturing Followed by Enzymatic Release. Chemrxiv 2020. [Google Scholar] [CrossRef]
- Zaraee, N.; Kanik, F.E.; Bhuiya, A.M.; Gong, E.S.; Geib, M.T.; Lortlar Ünlü, N.; Ozkumur, A.Y.; Dupuis, J.R.; Ünlü, M.S. Highly sensitive and label-free digital detection of whole cell E. coli with Interferometric Reflectance Imaging. Biosens. Bioelectron. 2020, 162, 112258. [Google Scholar] [CrossRef]
- Yurdakul, C.; Avci, O.; Matlock, A.; Devaux, A.J.; Quintero, M.V.; Ozbay, E.; Davey, R.A.; Connor, J.H.; Karl, W.C.; Tian, L.; et al. High-Throughput, High-Resolution Interferometric Light Microscopy of Biological Nanoparticles. ACS Nano 2020, 14, 2002–2013. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.W.; Sandoghdar, V. Interferometric Scattering Microscopy: Seeing Single Nanoparticles and Molecules via Rayleigh Scattering. Nano Lett. 2019, 19, 4827–4835. [Google Scholar] [CrossRef] [Green Version]
- Ekiz-Kanik, F.; Sevenler, D.D.; Ünlü, N.L.; Chiari, M.; Ünlü, M.S. Surface chemistry and morphology in single particle optical imaging. Nanophotonics 2017, 6, 713–730. [Google Scholar] [CrossRef]
- Niemeyer, C.M.; Sano, T.; Smith, C.L.; Cantor, C.R. Oligonucleotide-directed self-assembly of proteins: Semisynthetic DNA—Streptavidin hybrid molecules as connectors for the generation of macroscopic arrays and the construction of supramolecular bioconjugates. Nucleic Acids Res. 1994, 22, 5530–5539. [Google Scholar] [CrossRef] [Green Version]
- Niemeyer, C.M.; Boldt, L.; Ceyhan, B.; Blohm, D. DNA-Directed Immobilization: Efficient, Reversible, and Site-Selective Surface Binding of Proteins by Means of Covalent DNA—Streptavidin Conjugates. Anal. Biochem. 1999, 268, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Seymour, E.; Ünlü, N.L.; Carter, E.P.; Connor, J.H.; Ünlü, M.S. Configurable Digital Virus Counter on Robust Universal DNA Chips. ACS Sens. 2021. [Google Scholar] [CrossRef] [PubMed]
- Stoevesandt, O.; He, M.; Taussig, M.J. Protein microarrays printed from DNA microarrays. Methods Mol. Biol. 2011, 671, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Seymour, E.; Daaboul, G.G.; Zhang, X.; Scherr, S.M.; Ünlü, N.L.; Connor, J.H.; Ünlü, M.S. DNA-Directed Antibody Immobilization for Enhanced Detection of Single Viral Pathogens. Anal. Chem. 2015, 87, 10505–10512. [Google Scholar] [CrossRef]
- Wacker, R.; Niemeyer, C.M. DDI-FIA—A Readily Configurable Microarray-Fluorescence Immunoassay Based on DNA-Directed Immobilization of Proteins. ChemBioChem 2004, 5, 453–459. [Google Scholar] [CrossRef]
- Ladd, J.; Boozer, C.; Yu, Q.; Chen, S.; Homola, J.; Jiang, S. DNA-directed protein immobilization on mixed self-assembled monolayers via a streptavidin bridge. Langmuir 2004, 20, 8090–8095. [Google Scholar] [CrossRef]
- Douglas, E.S.; Hsiao, S.C.; Onoe, H.; Bertozzi, C.R.; Francis, M.B.; Mathies, R.A. DNA-barcode directed capture and electrochemical metabolic analysis of single mammalian cells on a microelectrode array. Lab Chip 2009, 9, 2010–2015. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, L.C.; Kuo, H.Y.; Mrksich, M. Peptide Arrays: Development and Application. Anal. Chem. 2018, 90, 266–282. [Google Scholar] [CrossRef]
- Gori, A.; Romanato, A.; Bergamaschi, G.; Strada, A.; Gagni, P.; Frigerio, R.; Brambilla, D.; Vago, R.; Galbiati, S.; Picciolini, S.; et al. Membrane-binding peptides for extracellular vesicles on-chip analysis. J. Extracell. Vesicles 2020, 9, 1751428. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.; Caciula, A.; Price, A.; Thakkar, R.; Ng, J.; Chauhan, L.V.; Jain, K.; Che, X.; Espinosa, D.A.; Montoya Cruz, M.; et al. Diagnosis of Zika Virus Infection by Peptide Array and Enzyme-Linked Immunosorbent Assay. mBio 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Fodor, S.P.; Read, J.L.; Pirrung, M.C.; Stryer, L.; Lu, A.T.; Solas, D. Light-directed, spatially addressable parallel chemical synthesis. Science 1991, 251, 767–773. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Chen, J.; Hu, G.; Wang, L.; Zheng, J.; Zhan, J.; Zhu, Y.; Zhong, C.; Shi, X.; Liu, S.; et al. Immobilization of an antimicrobial peptide on silicon surface with stable activity by click chemistry. J. Mater. Chem. B 2018, 6, 68–74. [Google Scholar] [CrossRef]
- Brambilla, D.; Chiari, M.; Gori, A.; Cretich, M. Towards precision medicine: The role and potential of protein and peptide microarrays. Analyst 2019, 144, 5353–5367. [Google Scholar] [CrossRef]
- Linman, M.J.; Abbas, A.; Cheng, Q. Interface design and multiplexed analysis with surface plasmon resonance (SPR) spectroscopy and SPR imaging. Analyst 2010, 135, 2759. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, S.; Pedrero, M.; Yáñez-Sedeño, P.; Pingarrón, J. Antifouling (Bio)materials for Electrochemical (Bio)sensing. Int. J. Mol. Sci. 2019, 20, 423. [Google Scholar] [CrossRef] [Green Version]
- Shumaker-Parry, J.S.; Zareie, M.H.; Aebersold, R.; Campbell, C.T. Microspotting streptavidin and double-stranded DNA arrays on gold for high-throughput studies of protein-DNA interactions by surface plasmon resonance microscopy. Anal. Chem. 2004, 76, 918–929. [Google Scholar] [CrossRef] [PubMed]
- Schasfoort, R.B.M. Handbook of Surface Plasmon Resonance; Royal Society of Chemistry (RSC): London, UK, 2017. [Google Scholar]
- Chiodi, E.; Sola, L.; Brambilla, D.; Cretich, M.; Marn, A.M.; Unlu, M.; Chiari, M. Simultaneous evaluation of multiple microarray surface chemistries through real-time interferometric imaging. Anal. Bioanal. Chem. 2020, 412, 3477–3487. [Google Scholar] [CrossRef] [Green Version]
- Nava, G.; Ceccarello, E.; Giavazzi, F.; Salina, M.; Damin, F.; Chiari, M.; Buscaglia, M.; Bellini, T.; Zanchetta, G. Label-free detection of DNA single-base mismatches using a simple reflectance-based optical technique. Phys. Chem. Chem. Phys. 2016, 18, 13395–13402. [Google Scholar] [CrossRef] [Green Version]
- Vanjur, L.; Carzaniga, T.; Casiraghi, L.; Chiari, M.; Zanchetta, G.; Buscaglia, M. Non-Langmuir Kinetics of DNA Surface Hybridization. Biophys. J. 2020, 119, 989–1001. [Google Scholar] [CrossRef]
- Sola, L.; Chiari, M. Modulation of electroosmotic flow in capillary electrophoresis using functional polymer coatings. J. Chromatogr. A 2012, 1270, 324–329. [Google Scholar] [CrossRef] [PubMed]
- Thierry, B.; Jasieniak, M.; De Smet, L.C.P.M.; Vasilev, K.; Griesser, H.J. Reactive Epoxy-Functionalized Thin Films by a Pulsed Plasma Polymerization Process. Langmuir 2008, 24, 10187–10195. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Grainger, D.W. Comparison of DNA immobilization efficiency on new and regenerated commercial amine-reactive polymer microarray surfaces. Surf. Sci. 2004, 570, 67–77. [Google Scholar] [CrossRef]
- Kodoyianni, V. Label-free analysis of biomolecular interactions using SPR imaging. BioTechniques 2011, 50, 32–40. [Google Scholar] [CrossRef]
- Linman, M.J.; Taylor, J.D.; Yu, H.; Chen, X.; Cheng, Q. Surface Plasmon Resonance Study of Protein-Carbohydrate Interactions Using Biotinylated Sialosides. Anal. Chem. 2008, 80, 4007–4013. [Google Scholar] [CrossRef] [Green Version]
- Needham, J.; Lortlar Ünlü, N.; Ünlü, M.S. Interferometric Reflectance Imaging Sensor (IRIS) for Molecular Kinetics with a Low-Cost, Disposable Fluidic Cartridge. In Methods in Molecular Biology; Springer Science and Business Media LLC: New York, NY, USA, 2019; Volume 2027. [Google Scholar]
- Fechner, P.; Bleher, O.; Ewald, M.; Freudenberger, K.; Furin, D.; Hilbig, U.; Kolarov, F.; Krieg, K.; Leidner, L.; Markovic, G.; et al. Size does matter! Label-free detection of small molecule–protein interaction. Anal. Bioanal. Chem. 2014, 406, 4033–4051. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Loo, J.; Chen, J.; Yam, Y.; Chen, S.C.; He, H.; Kong, S.; Ho, H. Recent Advances in Surface Plasmon Resonance Imaging Sensors. Sensors 2019, 19, 1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Yang, M.; Zhou, W.; Johnston, T.G.; Wang, R.; Zhu, J. Dextran hydrogel coated surface plasmon resonance imaging (SPRi) sensor for sensitive and label-free detection of small molecule drugs. Appl. Surf. Sci. 2015, 355, 570–576. [Google Scholar] [CrossRef]
- Fuller, M.A.; Koper, I. Biomedical applications of polyelectrolyte coated spherical gold nanoparticles. Nano Converg. 2019, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Haitami, A.E.E.; Martel, D.; Ball, V.; Nguyen, H.C.; Gonthier, E.; Labbe, P.; Voegel, J.C.; Schaaf, P.; Senger, B.; Boulmedais, F. Effect of the Supporting Electrolyte Anion on the Thickness of PSS/PAH Multilayer Films and on Their Permeability to an Electroactive Probe. Langmuir 2009, 25, 2282–2289. [Google Scholar] [CrossRef] [Green Version]
- Rubayea, A.A.; Naboka, A.; Catananteb, G.; Martyb, J.L.; Takacsc, E.; Szekacsca, A. Detection of ochratoxin A in aptamer assay using total internalreflection ellipsometry. Sens. Actuators B Chem. 2018, 263, 248–251. [Google Scholar] [CrossRef] [Green Version]
- Sola, L.; Romanato, A.; Siboni, M.; Damin, F.; Chiodi, E.; Brambilla, D.; Cretich, M.; Gori, A.; Chiari, M. Layer-by-layer deposition of functional click polymers for microarray applications. Express Polym. Lett. 2019, 13, 1004–1017. [Google Scholar] [CrossRef]
- Zhao, H.; Sha, J.; Wu, T.; Chen, T.; Chen, X.; Ji, H.; Wang, Y.; Zhu, H.; Xie, L.; Ma, Y. Spatial modulation of biomolecules immobilization by fabrication of hierarchically structured PEG-derived brush micropatterns: An versatile cellular microarray platform. Appl. Surf. Sci. 2020, 529, 147056. [Google Scholar] [CrossRef]
- Wu, S.; Sun, Z.; Peng, Y.; Han, Y.; Li, J.; Zhu, S.; Yin, Y.; Li, G. Peptide-functionalized metal-organic framework nanocomposite for ultrasensitive detection of secreted protein acidic and rich in cysteine with practical application. Biosens. Bioelectron. 2020, 169, 112613. [Google Scholar] [CrossRef]
- Celebi, I.; Geib, M.; Chiodi, E.; Lortlar-Unlu, N.; Ekiz Kanik, F.; Unlu, M. Instrument-Free Protein Microarray Fabrication for Accurate Affinity Measurements. Biosensors 2020, 10, 158. [Google Scholar] [CrossRef] [PubMed]






| Immobilization Strategy | Amide Coupling | Oriented | 3D-Structure | Kinetics | Single Step | Probe Modification |
|---|---|---|---|---|---|---|
| Physical adsorption [38] | no | no | no | no | yes | no |
| Epoxysilane [38,39] | yes | no | no | yes | yes | no |
| Copoly(DMA-NAS-MAPS) [40] | yes | no | yes | yes | yes | no |
| Dendrimers [50] | yes | no | yes | yes | no | no |
| Hydrogels [51,52,53] | no | no | yes | no | no | no |
| Click chemistry [54,55] | no | yes | yes | yes | yes | yes |
| DNA-directed [56,57] | yes | yes | yes | yes | no | yes |
| Biotin-SAV [38] | no | yes | no | yes | no | yes |
| Protein A/G [58] | no | no | no | yes | no | no |
| Carboxymethyl dextran [42] | no | no | yes | yes | yes | no |
| Thiol-gold coupling [59] | no | no | no | yes | yes | yes |
| Nanostructures [43] | no | no | no | yes | yes | no |
| Immobilization Strategy | Molecular Structure Preservation | Loading Capacity | Surface Morphology | Non-Specific Interactions | Accessibility |
|---|---|---|---|---|---|
| Physical adsorption [38] | ** | * | ** | * | **** |
| Epoxysilane [38,39] | * | ** | *** | *** | *** |
| Copoly(DMA-NAS-MAPS) [40] | **** | *** | **** | **** | ** |
| Dendrimers [50] | *** | *** | *** | **** | * |
| Hydrogels [51,52,53] | **** | **** | ** | *** | * |
| Click chemistry [54,55] | **** | *** | **** | **** | ** |
| DNA-directed [56,57] | **** | **** | ** | **** | * |
| Biotin-SAV [38] | *** | ** | ** | **** | *** |
| Protein A/G [58] | ** | ** | *** | ** | ** |
| Carboxymethyl dextran [42] | **** | **** | ** | *** | ** |
| Thiol-gold coupling [59] | * | ** | **** | ** | *** |
| Nanostructures [43] | ** | *** | ** | **** | * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiodi, E.; Marn, A.M.; Geib, M.T.; Ünlü, M.S. The Role of Surface Chemistry in the Efficacy of Protein and DNA Microarrays for Label-Free Detection: An Overview. Polymers 2021, 13, 1026. https://doi.org/10.3390/polym13071026
Chiodi E, Marn AM, Geib MT, Ünlü MS. The Role of Surface Chemistry in the Efficacy of Protein and DNA Microarrays for Label-Free Detection: An Overview. Polymers. 2021; 13(7):1026. https://doi.org/10.3390/polym13071026
Chicago/Turabian StyleChiodi, Elisa, Allison M. Marn, Matthew T. Geib, and M. Selim Ünlü. 2021. "The Role of Surface Chemistry in the Efficacy of Protein and DNA Microarrays for Label-Free Detection: An Overview" Polymers 13, no. 7: 1026. https://doi.org/10.3390/polym13071026
APA StyleChiodi, E., Marn, A. M., Geib, M. T., & Ünlü, M. S. (2021). The Role of Surface Chemistry in the Efficacy of Protein and DNA Microarrays for Label-Free Detection: An Overview. Polymers, 13(7), 1026. https://doi.org/10.3390/polym13071026









