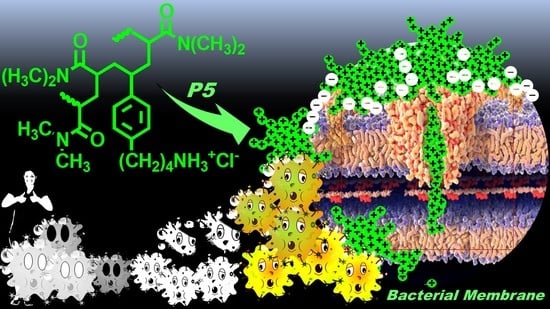
Synthesis, Characterization, and Bactericidal Activity of a 4-Ammoniumbuthylstyrene-Based Random Copolymer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Instruments
2.2. Synthesis of 4-(4-Amminobuthyl)styrene Hydrochloride M5 (5)
2.2.1. p-Vinylphenylmagnesium Chloride (1)
2.2.2. 4-(4-Bromobuthyl)styrene (2)
2.2.3. N-[(4-Vinylphenyl)buthyl]phthalimide (3)
2.2.4. 4-(4-Amminobuthyl)styrene (4)
2.2.5. 4-(4-Amminobuthyl)styrene Hydrochloride (5)
2.3. Preparation of Copolymer P5 by Radical Copolymerization in Solution
Fractioning of P5
2.4. Determination of the Average Molecular Mass (Mn) of P5
2.4.1. Calibration
2.4.2. Measurements
2.5. Determination of NH2 Equivalents Contained in P5
2.6. Dynamic Light Scattering (DLS) Analysis
2.7. Microbiology
2.7.1. Microorganisms
2.7.2. Determination of the MIC and MBC
2.7.3. Killing Curves
2.7.4. Evaluation of the Antimicrobial Effect of P5 by Turbidimetric Studies
3. Results and Discussion
3.1. Synthesis and Spectrophotometric Characterization of 4-(4-Amminobuthyl)styrene Hydrochloride M5 (5)
3.2. Preparation of Copolymer P5 by Radical Copolymerization in Solution and Its Spectroscopic Characterization
3.3. Determination of the Average Molecular Mass (Mn) of Copolymer P5
3.3.1. The Technique
3.3.2. Calibration and Measurements
3.4. Determinations of NH2 Content of P5
3.5. Particle Size, ζ-p and PDI of P5
3.6. Antibacterial Properties
3.6.1. Design of the Structure of the Cationic Monomer M5 and the Choice of the Not Charged Comonomer (DMAA)
3.6.2. Antimicrobial Activity of P5
3.6.3. Time-Killing Curves
3.6.4. Effect of P5 on the Growth Curve of P. aeruginosa, K. pneumoniae, and S. aureus
4. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alfei, S.; Schito, A.M. From Nanobiotechnology, Positively Charged Biomimetic Dendrimers as Novel Antibacterial Agents: A Review. Nanomaterials 2020, 10, 2022. [Google Scholar] [CrossRef] [PubMed]
- Alfei, S.; Schito, A.M. Positively Charged Polymers as Promising Devices against Multidrug Resistant Gram-Negative Bacteria: A Review. Polymers 2020, 12, 1195. [Google Scholar] [CrossRef] [PubMed]
- Gelman, M.A.; Weisblum, B.; Lynn, D.M.; Gellman, S.H. Biocidal activity of polystyrenes that are cationic by virtue of protonation. Org. Lett. 2004, 6, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Palermo, E.; Kuroda, K. Chemical structure of cationic groups in amphiphilic polymethacrylates modulates the antimicrobial and hemolytic activities. Biomacromolecules 2009, 10, 1416–1428. [Google Scholar] [CrossRef]
- Schito, A.M.; Alfei, S. Antibacterial Activity of Non-Cytotoxic, Amino Acid-Modified Polycationic Dendrimers against Pseudomonas aeruginosa and Other Non-Fermenting Gram-Negative Bacteria. Polymers 2020, 12, 1818. [Google Scholar] [CrossRef]
- Schito, A.M.; Schito, G.C.; Alfei, S. Synthesis and Antibacterial Activity of Cationic Amino Acid-Conjugated Dendrimers Loaded with a Mixture of Two Triterpenoid Acids. Polymers 2021, 13, 521. [Google Scholar] [CrossRef]
- Ergene, C.; Yasuharab, K.; Palermo, E.F. Biomimetic antimicrobial polymers: Recent advances in molecular design. Polym. Chem. 2018, 9, 2047. [Google Scholar] [CrossRef] [Green Version]
- Ganewatta, M.S.; Tang, C. Controlling macromolecular structures towards effective antimicrobial polymers. Polymer 2015, 63, A1–A29. [Google Scholar] [CrossRef]
- Matsuzaki, K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta 2009, 1788, 1687–1692. [Google Scholar] [CrossRef] [Green Version]
- Gabriel, G.J.; Som, A.; Madkour, A.E.; Eren, T.; Tew, G.N. Infectious disease: Connecting innate immunity to biocidal polymers. Mater. Sci. Eng. R Rep. 2007, 57, 28–64. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, K.; Sugishita, K.; Fujii, N.; Miyajima, K. Molecular Basis for Membrane Selectivity of an Antimicrobial Peptide, Magainin 2. Biochemistry 1995, 34, 3423–3429. [Google Scholar] [CrossRef]
- Bertini, V.; Alfei, S.; Pocci, M.; Lucchesini, F.; Picci, N.; Iemma, F. Monomers containing substrate or inhibitor residues for copper amine oxidases and their hydrophilic beaded resins designed for enzyme interaction studies. Tetrahedron 2004, 60, 11407–11414. [Google Scholar] [CrossRef]
- Alfei, S.; Marengo, B.; Domenicotti, C. Polyester-Based Dendrimer Nanoparticles Combined with Etoposide Have an Improved Cytotoxic and Pro-Oxidant Effect on Human Neuroblastoma Cells. Antioxidants 2020, 9, 50. [Google Scholar] [CrossRef] [Green Version]
- Hirao, A.; Hayashi, M.; Nakahama, S. Synthesis of End-Functionalized Polymer by Means of Anionic Living Polymerization. 6. Synthesis of Well-Defined Polystyrenes and Polyisoprenes with 4-Vinylphenyl End Group. Macromolecules 1996, 29, 3353–3358. [Google Scholar] [CrossRef]
- Alfei, S.; Castellaro, S. Synthesis and Characterization of Polyester-Based Dendrimers Containing Peripheral Arginine or Mixed Amino Acids as Potential Vectors for Gene and Drug Delivery. Macromol. Res. 2017, 25, 1172–1186. [Google Scholar] [CrossRef]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. Available online: https://www.eucast.org/ast_of_bacteria/ (accessed on 24 November 2020).
- Pearson, R.D.; Steigbigel, R.T.; Davis, H.T.; Chapman, S.W. Method for reliable determination of minimal lethal antibiotic concentrations. Antimicrob. Agents Chemother. 1980, 18, 699–708. [Google Scholar] [CrossRef] [Green Version]
- Schito, A.M.; Piatti, G.; Stauder, M.; Bisio, A.; Giacomelli, E.; Romussi, G.; Pruzzo, C. Effects of demethylfruticuline A and fruticuline A from Salvia corrugata Vahl. on biofilm production in vitro by multiresistant strains of Staphylococcus aureus, Staphylococcus epidermidis and Enterococcus faecalis. Int. J. Antimicrob. Agents 2011, 37, 129–134. [Google Scholar] [CrossRef] [Green Version]
- Dalgaard, P.; Ross, T.; Kamperman, L.; Neumeyer, K.; McMeekin, T.A. Estimation of bacterial growth rates from turbidimetric and viable count data. Int. J. Food Microbiol. 1994, 23, 391–404. [Google Scholar] [CrossRef]
- Andringa, H.; Hanekamp, J.; Brandsma, L. The Copper Halide-Catalyzed Mono-Substitution of Bromine in α,-dibromoalkanes by Grignard reagents. A Reinvestigation. Synth. Commun. 1990, 20, 2349–2351. [Google Scholar] [CrossRef]
- Arshady, R.; Atherton, E.; Clive, D.L.J.; Sheppard, R.C. Peptide synthesis. Part 1. Preparation and use of polar supports based on poly(dimethylacrylamide). J. Chem. Soc. Perkin Trans. 1 1981, 529–537. [Google Scholar] [CrossRef]
- Bersted, B.H. Molecular Weight Determination of High Polymers by Means of Vapor Pressure Osmometry and the Solute Dependence of the Constant of Calibration. J. Appl. Polym. Sci. 1973, 17, 1415–1430. [Google Scholar] [CrossRef]
- Chalmers, J.M.; Meier, R.J. Molecular Characterization and Analysis of Polymers. In Wilson & Wilson’s Comprehensive Analytical Chemistry, Barcelo, D., Ed.; Elsevier Science: Oxford, UK, 2008; Volume 53, pp. 1–763. [Google Scholar]
- Wen, Q.; Xu, L.; Xiao, X.; Wang, Z. Preparation, characterization, and antibacterial activity of cationic nanopolystyrenes. J. Appl. Polym. Sci. 2019, 137, 48405. [Google Scholar] [CrossRef]
- Hu, F.X.; Neoh, K.G.; Cen, L.; Kang, E.T. Antibacterial and antifungal efficacy of surface functionalized polymeric beads in repeated applications. Biotechnol. Bioeng. 2005, 89, 474–484. [Google Scholar] [CrossRef]
- Sellenet, P.H.; Allison, B.; Applegate, B.M.; Youngblood, J.P. Synergistic activity of hydrophilic modification in antibiotic polymers. Biomacromolecules 2007, 8, 19–23. [Google Scholar] [CrossRef]
- Tiller, J.C.; Liao, C.J.; Lewis, K.; Klibanov, A.M. Designing surfaces that kill bacteria on contact. Proc. Natl. Acad. Sci. USA 2001, 98, 5981–5985. [Google Scholar] [CrossRef] [Green Version]
- Timofeeva, L.M.; Kleshcheva, N.A.; Moroz, A.F.; Didenko, L.V. Secondary and tertiary polydiallylammonium salts: Novel polymers with high antimicrobial activity. Biomacromolecules 2009, 10, 2976–2986. [Google Scholar] [CrossRef]
- Gilbert, P.; Mc Bain, A.J. Potential impact of increased use of biocides in consumer products on prevalence of antibiotic resistance. Clin. Microbiol. Rev. 2003, 16, 189–208. [Google Scholar] [CrossRef] [Green Version]
- Kanazawa, A.; Ikeda, T.; Endo, T. Novel polycationic biocides: Synthesis and antibacterial activity of polymeric phosphonium salts. J. Polym. Sci. A Polym. Chem. 1993, 31, 335–343. [Google Scholar] [CrossRef]
- Santos, M.R.E.; Mendonça, P.V.; Almeida, M.C.; Branco, R.; Serra, A.C.; Morais, P.V.; Coelho, J.F.J. Increasing the Antimicrobial Activity of Amphiphilic Cationic Copolymers by the Facile Synthesis of High Molecular Weight Stars by Supplemental Activator and Reducing Agent Atom Transfer Radical Polymerization. Biomacromolecules 2019, 20, 1146–1156. [Google Scholar] [CrossRef]
- Locock, K.E.S.; Michl, T.D.; Stevens, N.; Hayball, J.D.; Vasilev, K.; Postma, A.; Griesser, H.J.; Meagher, L.; Haeussler, M. Antimicrobial Polymethacrylates Synthesized as Mimics of Tryptophan-Rich Cationic Peptides. ACS Macro Lett. 2014, 3, 319–323. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Park, Y.K.; Chung, E.S.; Na, I.Y.; Ko, K.S. Evolved Resistance to Colistin and Its Loss Due to Genetic Reversion in Pseudomonas Aeruginosa. Sci. Rep. 2016, 6, 25543. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, T.; Bustos, R.-H.; González, D.; Garzón, V.; García, J.-C.; Ramírez, D. An Approach to Measuring Colistin Plasma Levels Regarding the Treatment of Multidrug-Resistant Bacterial Infection. Antibiotics 2019, 8, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Zhu, J.; Hu, Q.; Rao, X. Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 2016, 50, 10–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Band, V.I.; Weiss, D.S. Mechanisms of Antimicrobial Peptide Resistance in Gram-Negative Bacteria. Antibiotics 2015, 4, 18–41. [Google Scholar] [CrossRef] [Green Version]
- Ostrowska, K.; Kamysz, W.; Dawgul, M.; Różalski, A. Synthetic amphibian peptides and short amino-acids derivatives against planktonic cells and mature biofilm of Providencia stuartii clinical strains. Pol. J. Microbiol. 2014, 63, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, M.; Palermo, E.F.; Thoma, L.M.; Satoh, K.; Kamigaito, M.; Kuroda, K. Design and Synthesis of Self-Degradable Antibacterial Polymers by Simultaneous Chain- and Step-Growth Radical Copolymerization. Biomacromolecules 2012, 13, 1554–1563. [Google Scholar] [CrossRef]
- Weiyang, L.; Shrinivas, V.; Guansheng, Z.; Bisha, D.; Tan, P.K.J.; Xu, L.; Weimin, F.; Yang, Y.Y. Antimicrobial polymers as therapeutics for treatment of multidrug-resistant Klebsiella pneumoniae lung infection. Acta Biomater. 2018, 78, 78–88. [Google Scholar] [CrossRef]
- Landman, D.; Georgescu, C.; Martin, D.A.; Quale, J. Polymyxins revisited. Clin. Microbiol. Rev. 2008, 21, 449–465. [Google Scholar] [CrossRef] [Green Version]
- Sudip, M.; Swagatam, B.; Riya, M.; Jayanta, H. Amphiphilic Cationic Macromolecules Highly Effective against Multi-Drug Resistant Gram-Positive Bacteria and Fungi with No Detectable Resistance. Front. Bioeng. Biotechnol. 2020, 8, e55. [Google Scholar] [CrossRef]
- Gough, N.R. Stressing bacteria to death. Sci. Signal. 2011, 4, e164. [Google Scholar] [CrossRef]








| M5 | DMAA | DMF | AIBN | Time | P5 |
|---|---|---|---|---|---|
| (mg, mmol, % 1) | (mg, mmol) | (mL) | (mg, % 2) | (h) | (g, % 3) |
| 697.7, 3.30, 42.8 | 765.9, 7.7 | 6.5 | 15.3, 1.0 | 7 | 0.6677, 46 |
| Calibration | Measurements | ||
|---|---|---|---|
| c (mol/Kg)PEO | MV/c (Kg/mol)PEO | c (g/Kg)P5 | Mn (g/mol) |
| 0.0048410 | 856 | 2.1949 | 5100 |
| 0.0058304 | 930 | 5.6195 | |
| 0.0068966 | 1007 | 7.7859 | |
| Kcal (Kg/mol) = 501 | Kmeas (Kg/g) = 0.0982 | ||
| P5 (5100) 1 | HClO4 0.1612 N | NH2 | µequiv.NH2/gP5 | µequiv.NH2/µmolP5 |
|---|---|---|---|---|
| mg (mmol) | (mL) | (mmol) | ||
| 300.5 (0.0589) | 1.67 | 0.2686 | 894 | 4.6 |
| mg/mL | Z-AVE Size (nm) | ζ-p (mV) | PDI | |
| 3 | 334 ± 27 | +57.6 ± 1.7 | 1.012 ± 0.007 | |
| P5 (5100) 2 | M5 (212) 2 | |||
|---|---|---|---|---|
| Strains | MIC µM (µg/mL) | MBC µM (µg/mL) | MIC µM (µg/mL) | MBC µM (µg/mL) |
| Gram-positive species of genus Enterococcus | ||||
| E. faecalis 1 * | 6.3 (32) | 12.5 (64) | 1208 (256) | 2416 (512) |
| E. faecalis 18 * | 6.3 (32) | 12.5 (64) | 604 (128) | 1208 (256) |
| E. faecalis 51 * | 6.3 (32) | 12.5 (64) | 1208 (256) | 2416 (512) |
| E. faecalis 365 * | 6.3 (32) | 12.5 (64) | 1208(256) | 2416 (512) |
| E. faecium 325 * | 3.15 (16) | 6.3 (32) | 604 (128) | 604 (128) |
| E. faecium 341 * | 3.15 (16) | 6.3 (32) | 1208 (256) | 2416 (512) |
| E. faecium 364 * | 3.15 (16) | 6.3 (32) | 302 (64) | 302 (64) |
| Minor strains | ||||
| E. casseliflavus 184 ° | 3.15 (16) | 3.15 (16) | 1208 (256) | 2416 (512) |
| E. durans 103 ° | 3.15 (16) | 6.3 (32) | 604 (128) | 1208 (256) |
| E. gallinarum 150 * | 3.15 (16) | 6.3 (32) | 1208 (256) | 2416 (512) |
| Gram-positive species of genus Staphylococcus | ||||
| S. aureus 18 ** | 12.6 (64) | 12.5 (64) | 2415 (512) | 4832 (1024) |
| S. aureus 195 ** | 12.6 (64) | 25 (128) | 2415 (512) | 4830 (1024) |
| S. aureus 189 | 12.6 (64) | 12.5 (64) | 1208 (256) | 2415 (512) |
| S. epidermidis 22 ** | 3.15 (16) | 6.3 (32) | 4830 (1024) | >4830 (>1024) |
| S. epidermidis 180 *** | 3.15 (16) | 6.3 (32) | 2415 (512) | 4830 (1024) |
| S. epidermidis 181 *** | 3.15 (16) | 6.3 (32) | 4830 (1024) | >4830 (>1024) |
| S. haemoliticus 193 ** | 3.15 (16) | 3.15 (16) | 1209 (256) | 1209 (256) |
| S. hominis 125 ** | 3.15 (16) | 3.15 (16) | 604 (128) | 604 (128) |
| S. lugdunensis 129 | 3.15 (16) | 3.15 (16) | 604 (128) | 1209 (256) |
| S. warneri 74 | 3.15 (16) | 6.3 (32) | 1209 (256) | 1209 (256) |
| S. saprophyticus 41 | 3.15 (16) | 3.15 (16) | 604(128) | 604 (128) |
| S. simulans 163 ** | 3.15 (16) | 3.15 (16) | 2415 (512) | 2415 (512) |
| Gram-positive sporogenic isolate | ||||
| B. subtilis | 6.3 (32) | 12.5 (64) | 2415 (512) | (4832) 1024 |
| Gram-negative species of Enterobacteriaceae family | ||||
| E. coli 224 S | 6.3 (32) | 6.3 (32) | 2415 (512) | >2415 (>512) |
| E. coli 238 # | 6.3 (32) | 6.3 (32) | >2415 (>512) | nt3 |
| E. coli 246§ | 6.3 (32) | 12.6 (64) | 2415 (512) | 2415 (512) |
| Y. enterocolitica 342 | 25.1 (128) | 50.2 (256) | 1209 (256) | 2415 (512) |
| S. marcescens 228 | 100.4 (512) | nt 3 | >2415 (>512) | >2415 (>512) |
| P. mirabilis 254 | 25.1 (128) | 25.1 (128) | >2415 (>512) | nt 3 |
| M. morganii 372 | 50.2 (256) | 100.4 (512) | >2415 (>512) | nt 3 |
| K. oxytoca 252 | 12.6 (64) | 12.6 (64) | 2415 (512) | 2415 (512) |
| K. pneumoniae 231 # | 12.6 (64) | 12.6 (64) | 2415 (512) | >2415 (>512) |
| K. pneumoniae 232 # | 12.6 (64) | 25.1 (128) | 2415 (512) | >2415 (>512) |
| K. pneumoniae 233 # | 6.3 (32) | 12.6 (64) | 2415 (512) | 2415 (512) |
| K. pneumoniae 260 # | 12.6 (64) | 25.1 (128) | 2415 (512) | >2415 (>512) |
| K. pneumoniae 366 # | 12.6 (64) | 12.6 (64) | 2415 (512) | 2415 (512) |
| K. pneumoniae 367 # | 12.6 (64) | 12.6 (64) | 2415 (512) | >2415 (>512) |
| K. pneumoniae 369 | 12.6 (64) | 12.6 (64) | 2415 (512) | >2415 (>512) |
| K. pneumoniae 377 S | 6.3 (32) | 12.6 (64) | 2415 (512) | 2415 (512) |
| P. stuartii 374 | 25.1 (128) | 25.1 (128) | >2415 (>512) | nt 3 |
| Salmonella gr.B 227 | 12.6 (64) | 12.6 (64) | 2415 (512) | 2415 (512) |
| Non-fermenting Gram-negative species | ||||
| P. aeruginosa 229 | 3.15 (16) | 6.3 (32) | 2415 (512) | >2415 (>512) |
| P. aeruginosa 247 | 6.3 (32) | 6.3 (32) | 2415 (512) | >2415 (>512) |
| P. aeruginosa 256 | 6.3 (32) | 6.3 (32) | >2415 (>512) | nt 3 |
| P. aeruginosa 259 | 6.3 (32) | 12.6 (64) | >2415 (>512) | nt 3 |
| P. aeruginosa 268 | 6.3 (32) | 12.6 (64) | >2415 (>512) | nt 3 |
| P. aeruginosa 269 | 6.3 (32) | 6.3 (32) | 2415 (512) | 2415 (512) |
| P. fluorescens 278 | 3.15 (16) | 6.3 (32) | 2415 (512) | 2415 (512) |
| P. putida 262 | 6.3 (32) | 6.3 (32) | 2415 (512) | 2415 (512) |
| A. baumannii 257 | 6.3 (32) | 12.6 (64) | 1209 (256) | 2415 (512) |
| A. baumannii 279 | 6.3 (32) | 6.3 (32) | 1209 (256) | 2415 (512) |
| A. baumannii 236 | 6.3 (32) | 12.6 (64) | 2415 (512) | 2415 (512) |
| A. baumannii 245 | 6.3 (32) | 12.6 (64) | 1209 (256) | 2415 (512) |
| A. baumannii 279 | 12.6 (64) | 25.1 (128) | 2415 (512) | 4830 (1024) |
| A. pitti 272 | 6.3 (32) | 6.3 (32) | 2415 (512) | 4830 (1024) |
| S. maltophylia 255 | 25.1 (128) | 50.2 (256) | 1209 (256) | 2415 (512) |
| S. maltophylia 280 | 6.3 (32) | 6.3 (32) | 1209 (256) | 2415 (512) |
| S. maltophylia 2 | 6.3 (32) | 12.6 (64) | 1209 (256) | 2415 (512) |
| S. maltophylia 3 | 3.15 (16) | 6.3 (32) | 604 (128) | 604 (128) |
| S. maltophylia 4 | 6.3 (32) | 12.6 (64) | 1209 (256) | 1209 (256) |
| S. maltophylia 5 | 12.6 (64) | 25.1 (128) | 1209 (256) | 1209 (256) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alfei, S.; Piatti, G.; Caviglia, D.; Schito, A.M. Synthesis, Characterization, and Bactericidal Activity of a 4-Ammoniumbuthylstyrene-Based Random Copolymer. Polymers 2021, 13, 1140. https://doi.org/10.3390/polym13071140
Alfei S, Piatti G, Caviglia D, Schito AM. Synthesis, Characterization, and Bactericidal Activity of a 4-Ammoniumbuthylstyrene-Based Random Copolymer. Polymers. 2021; 13(7):1140. https://doi.org/10.3390/polym13071140
Chicago/Turabian StyleAlfei, Silvana, Gabriella Piatti, Debora Caviglia, and Anna Maria Schito. 2021. "Synthesis, Characterization, and Bactericidal Activity of a 4-Ammoniumbuthylstyrene-Based Random Copolymer" Polymers 13, no. 7: 1140. https://doi.org/10.3390/polym13071140
APA StyleAlfei, S., Piatti, G., Caviglia, D., & Schito, A. M. (2021). Synthesis, Characterization, and Bactericidal Activity of a 4-Ammoniumbuthylstyrene-Based Random Copolymer. Polymers, 13(7), 1140. https://doi.org/10.3390/polym13071140