Time Estimation of Polymer Translocation through Nano-Membrane
Abstract
:1. Introduction
2. Theoretical Background
Average Time of Polymer Translocation
3. Materials and Methods
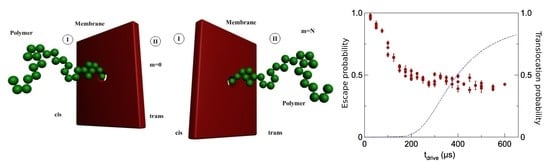
The Polymer Escape Probability
4. Results Discussions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mackenzie, R.J. DNA vs. RNA—5 Key Differences and Comparison; Technology Networks: Sudbury, UK, 2020; pp. 1–6. [Google Scholar]
- Sung, W.; Park, P.J. Polymer Translocation through a Pore in a Membrane. Phys. Rev. Lett. 1996, 77, 783–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paun, V.P. Theoretical study of the polymer transport through nanopores. Rev. Chim. 2006, 57, 221–223. [Google Scholar]
- Muthukumar, M. Polymer translocation through a hole. J. Chem. Phys. 1999, 111, 10371–10374. [Google Scholar] [CrossRef]
- Kong, C.Y.; Muthukumar, M. Monte Carlo study of adsorption of a polyelectrolyte onto charged surfaces. J. Chem. Phys. 1998, 109, 1522–1527. [Google Scholar] [CrossRef]
- Nichita, M.V.; Paun, M.A.; Paun, V.A.; Paun, V.P. Fractal Analysis of Brain Glial Cells. Fractal Dimension and Lacunarity. Univ. Politeh. Buchar. Sci. Bull. Ser. A Appl. Math. Phys. 2019, 81, 273–284. [Google Scholar]
- Bordescu, D.; Paun, M.A.; Paun, V.A.; Paun, V.P. Fractal analysis of Neuroimagistic. Lacunarity degree, a precious indicator in the detection of Alzheimer’s disease. Univ. Politeh. Buchar. Sci. Bull. Ser. A Appl. Math. Phys. 2018, 80, 309–320. [Google Scholar]
- Paun, V.P. An estimation of the polymer translocation time through membrane. Mater. Plast. 2006, 43, 57–58. [Google Scholar]
- Paun, V.P. Relaxation model for polymeric materials in the hereditary theory of elasticity. Mater. Plast. 2003, 40, 81–82. [Google Scholar]
- Ambjörnsson, T.; Apell, S.P.; Konkoli, Z.; Di Marzio, E.A.; Kasianowicz, J.J. Charged polymer membrane translocation. J. Chem. Phys. 2002, 117, 4063–4073. [Google Scholar] [CrossRef]
- Chen, S.; Olson, E.; Jiang, S.; Yong, X. Nanoparticle assembly modulated by polymer chain conformation in composite mate-rials. Nanoscale 2020, 12, 14560–14572. [Google Scholar] [CrossRef]
- Luo, K.; Ala-Nissila, T.; Ying, S.-C.; Bhattacharya, A. Sequence Dependence of DNA Translocation through a Nanopore. Phys. Rev. Lett. 2008, 100, 058101. [Google Scholar] [CrossRef] [Green Version]
- Paun, V.P.; Paun, M.A.; Toma, A.; Ciucu, C.; Popentiu, F. Transport Phenomenon Simulation for Linear Polymers through Nanometer Pores. Mater. Plast. 2008, 45, 57–60. [Google Scholar]
- Pusca, S.; Paun, M.-A.; Toma, C. Viscoelastic behaviour analysis of the technical polymers by bidimensional pulses generation. Mater. Plast. 2007, 44, 39–42. [Google Scholar]
- Moghimikheirabadi, A.; Kröger, M.; Karatrantos, A.V. Insights from modeling into structure, entanglements, and dynamics in attractive polymer nanocomposites. Soft. Matter. 2021, 17, 6362–6373. [Google Scholar]
- Paun, V.P. Polymer dynamics simulation at nanometer scale in a 2D diffusion model. Mater. Plast. 2007, 44, 393–395. [Google Scholar]
- Kasianowicz, J.J.; Henrickson, S.E.; Weetall, H.H.; Robertson, B. Simultaneous multianalyte detection with a nanometer-scale pore. Anal Chem. 2001, 73, 2268–2272. [Google Scholar]
- Muthukumar, M. Translocation of a Confined Polymer through a Hole. Phys. Rev. Lett. 2001, 86, 3188–3191. [Google Scholar] [CrossRef]
- Doi, M.; Edwards, S.F. Theory of Polymer Dynamics; Clarendon Press: Oxford, UK, 1986. [Google Scholar]
- Lubensky, D.K.; Nelson, D.R. Driven Polymer Translocation Through a Narrow Pore. Biophys. J. 1999, 77, 1824–1838. [Google Scholar] [CrossRef] [Green Version]
- Bates, M.; Burns, M.; Meller, A. Dynamics of DNA Molecules in a Membrane Channel Probed by Active Control Techniques. Biophys. J. 2003, 84, 2366–2372. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.K. Statistical Mechanic; World Scientific: Singapore, 1985. [Google Scholar]
- Hamidabad, M.N.; Asgari, S.; Abdolvahab, R.H. Nanoparticle-assisted polymer translocation through a nanopore. Polymer 2020, 204, 122847. [Google Scholar] [CrossRef]
- Kasianowicz, J.J.; Brandin, E.; Branton, D.; Deamer, D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA 1996, 93, 13770–13773. [Google Scholar] [CrossRef] [Green Version]
- Bezrukov, S.M.; Vodyanoy, I.; Brutyan, R.A.; Kasianowicz, J.J. Dynamics and Free Energy of Polymers Partitioning into a Nanoscale Pore. Macromolecules 1996, 29, 8517–8522. [Google Scholar] [CrossRef]
- Meller, A.; Nivon, L.; Branton, D. Voltage-driven DNA trans- locations through a nanopore. Phys. Rev. Lett. 2001, 86, 3435–3438. [Google Scholar] [CrossRef] [Green Version]
- Meller, A.; Branton, D. Single Molecule Measurements of DNA Transport through a Nanopore. Electrophoresis 2002, 23, 2583–2591. [Google Scholar] [CrossRef]
- Slonkina, E.; Kolomeisky, A. Polymer translocation through a long nanopore. J. Chem. Phys. 2003, 118, 7112–7118. [Google Scholar] [CrossRef] [Green Version]
- Hamidabad, M.N.; Abdolvahab, R.H. Translocation through a narrow pore under a pulling force. Sci. Rep. 2019, 9, 17885. [Google Scholar] [CrossRef] [Green Version]
- Menais, T. Polymer translocation under a pulling force: Scaling arguments and threshold forces. Phys. Rev. E 2018, 97, 022501. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wang, R.; Lu, Y.; An, L.; Shi, A.-C.; Wang, Z.-G. Mechanisms of Flow-Induced Polymer Translocation. Macromolecules 2022, 55, 3602–3612. [Google Scholar] [CrossRef]
- Sarabadani, J.; Metzler, R.; Ala-Nissila, T. Driven polymer translocation into a channel: Iso-flux tension propagation theory and Langevin dynamics simulations. arXiv 2022, arXiv:2202.08128v2. [Google Scholar]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paun, M.-A.; Paun, V.-A.; Paun, V.-P. Time Estimation of Polymer Translocation through Nano-Membrane. Polymers 2022, 14, 2090. https://doi.org/10.3390/polym14102090
Paun M-A, Paun V-A, Paun V-P. Time Estimation of Polymer Translocation through Nano-Membrane. Polymers. 2022; 14(10):2090. https://doi.org/10.3390/polym14102090
Chicago/Turabian StylePaun, Maria-Alexandra, Vladimir-Alexandru Paun, and Viorel-Puiu Paun. 2022. "Time Estimation of Polymer Translocation through Nano-Membrane" Polymers 14, no. 10: 2090. https://doi.org/10.3390/polym14102090
APA StylePaun, M.-A., Paun, V.-A., & Paun, V.-P. (2022). Time Estimation of Polymer Translocation through Nano-Membrane. Polymers, 14(10), 2090. https://doi.org/10.3390/polym14102090