Abstract
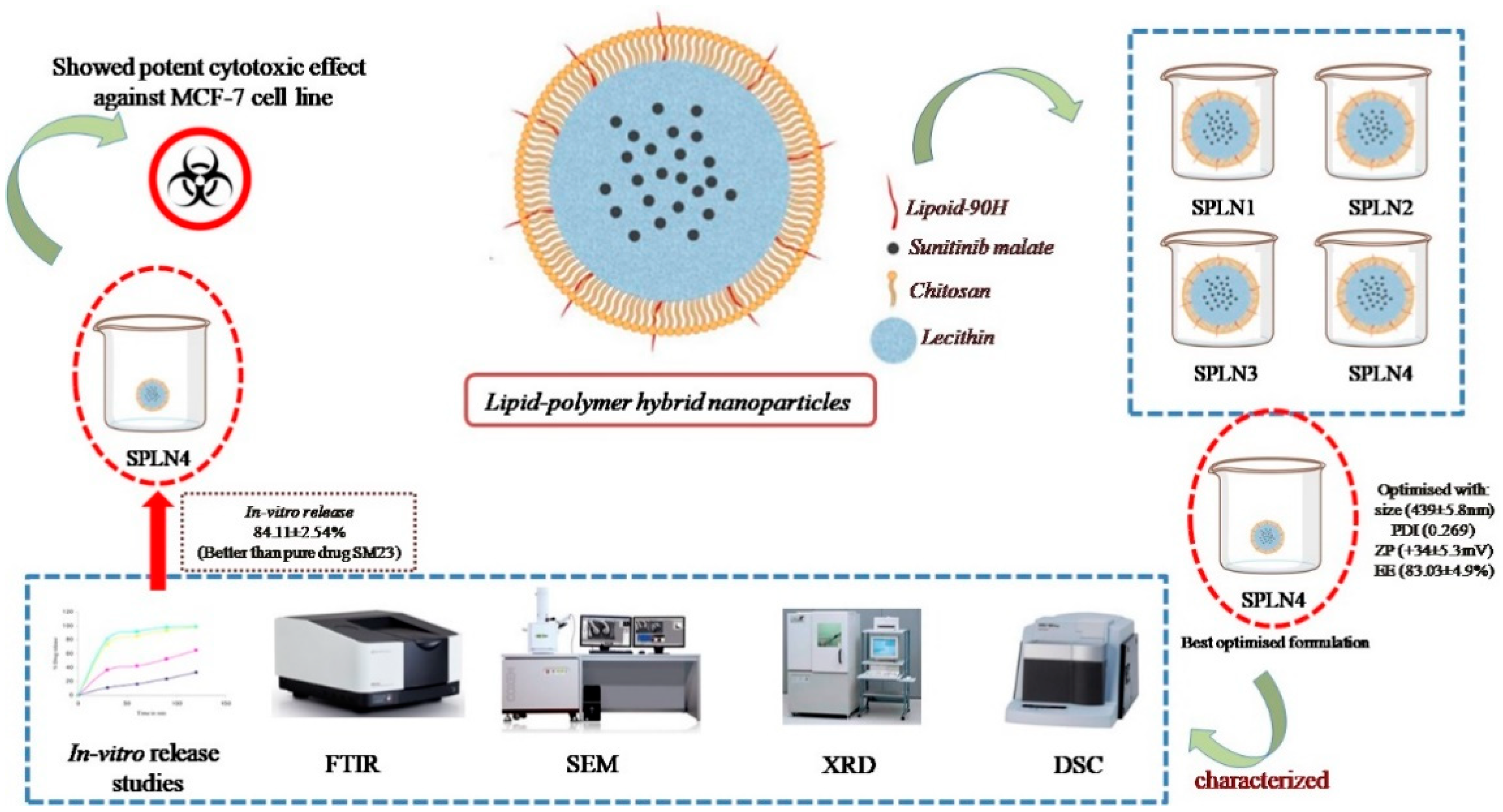
In the current study, lipid-polymer hybrid nanoparticles (LPHNPs) fabricated with lipoid-90H and chitosan, sunitinib malate (SM), an anticancer drug was loaded using lecithin as a stabilizer by employing emulsion solvent evaporation technique. Four formulations (SLPN1–SLPN4) were developed by varying the concentration of chitosan polymer. Based on particle characterization, SLPN4 was optimized with size (439 ± 5.8 nm), PDI (0.269), ZP (+34 ± 5.3 mV), and EE (83.03 ± 4.9%). Further, the optimized formulation was characterized by FTIR, DSC, XRD, SEM, and in vitro release studies. In-vitro release of the drug from SPN4 was found to be 84.11 ± 2.54% as compared with pure drug SM 24.13 ± 2.67%; in 48 h, release kinetics followed the Korsmeyer–Peppas model with Fickian release mechanism. The SLPN4 exhibited a potent cytotoxicity against MCF-7 breast cancer, as evident by caspase 3, 9, and p53 activities. According to the findings, SM-loaded LPHNPs might be a promising therapy option for breast cancer.
1. Introduction
Breast cancer is considered one of the leading types of cancer, surpassing lung cancer, as per worldwide cancer incidence in 2020 [1]. Breast cancer starts with the uncontrolled growth of breast cell in one or both sides. About one in eight women are diagnosed with breast cancer during their lifetime; the good news is that its curable if detected at an early stage [2]. Proliferation, apoptosis, angiogenesis, hypoxia, cancer stem cell activity, epithelial to mesenchymal transition (EMT), and metastasis are all related to the signaling system. Notch receptors and their ligands were shown to be overexpressed in breast cancer [3,4]. Signaling pathways upregulated leading to breast cancer include human epidermal growth factor receptor 2 (HER-2) tyrosine kinase pathway, a member of the ErbB family of transmembrane receptor tyrosine kinases [5]. The Hedgehog signaling pathway is also deregulated in breast cancer which is responsible for proper cell differentiation [6]. p53 mutation leads to aggressive diseases, such as breast cancer, playing a vital role in regulating the cell cycle, and apoptosis mutation of this gene causes cancer and shortens the overall survival. Another pathway actively involved in breast cancer is Phosphatase and tensin homolog (PTEN), reduced expression of which causes deceased formation of an enzyme phosphatase protein that acts as a tumor suppressor [7,8,9].
Sunitinib malate (SM) is a multiple tyrosine kinases inhibitor, used effectively in the cancer of the stomach, bowel, and esophagus, generally called gastrointestinal stromal tumors (GST), an abnormal proliferation of cells in gastrointestinal tract tissues [10,11]. It is a new vascular endothelial growth factor receptor practiced as a first-line therapy for advanced renal cell carcinoma [12] and a first-joint FDA-approved drug for these two indications. SM showed a promising activity against colorectal cancer, advanced non-small cell lung cancer (NSCLC) [13], hepatic cancer [14], and pancreatic neuroendocrine tumors (pNET) [15]. SM is one of the extensively studied antitumor agents in the breast cancer treatment with twenty-eight ongoing clinical trials, specifically sunitinib alone and in combinations. Sunhui et al.’s study showed sunitinib and curcumin have potential anticancer activity against breast cancer [16]. Sunitinib-loaded self-nanoemulsifying formulation has been developed with improved anticancer activity against the MCF-7 breast cancer cell line [17,18,19].
Moreover, breast cancer treatment failure could be surfaced due to poor drug solubility, low-bioavailability, permeability, cell uptake, drug resistance, and systemic toxicity. The cost of therapy and adverse drug effects could be lowered by adopting and aligning with new technologies. Nanotechnologies ensure it reduces adverse systemic toxic effects; thereby, the cost of therapy will also be reduced [20,21,22]. Nanocarriers enhance systemic drug circulation, and improve bioavailability, sustained release kinetics, and drug targeting at the receptor site. Enhanced permeability and retention effect facilitate the targeting of small molecules of nanosize and higher deposition of drug in cancer cells compared with the normal cells. Drug targeting involves the conjugate of chemotherapeutic-loaded nanocarrier with molecules that bind to the target (tumor) cell receptors [23,24,25,26]. Liposomes are thought to be a biocompatible vesicular structure with properties similar to biological membranes. Stability, low drug encapsulation, and burst drug release are the key concerns for vesicular systems. Polymer-based nanoparticles, on the other hand, are more stable than liposomes and also provide longer-lasting drug release. Synthetic (e.g., PLGA) and natural (e.g., chitosan) polymers are used to create polymeric nanoparticles. The preparation of lipid polymer hybrid nanoparticles (LPHNPs), which have both lipid and polymeric carriers, can address the constraints of both liposomes and polymeric nanoparticles. LPHNPs are next-generation core-shell nanostructures that are derived from both liposomes and polymeric nanoparticles (NPs), with a lipid coating encasing the polymer core loaded with drug that helps it to prolong systemic circulation and protect drug mitigation, and does not allow the water to obtain access into the drug-containing core. LPHNPs shows high entrapment, controlled release, cellular targeting, and serum stability. To the best of the author’s knowledge, no study has been conducted or reported on LPHNPs of SM [27,28,29]. Therefore, to facilitate targeting of SM at the tumor cell with higher drug loading, nanocarriers with a conjugate of lipid and polymer were selected in order to achieve higher antineoplastic activity with reduced toxicity to the normal cell at the target region. The objective of the current study was to prepare SM-loaded lipid–polymer hybrid nanoparticles using lipoid-90, chitosan, and evaluated for particle size, drug release, and anticancer activity against MCF7 cell lines.
2. Materials and Methods
2.1. Materials
Sunitininb malate (SM) was purchased from “Mesochem Technology” Beijing, China. Chitosan and polyvinyl alcohol (PVA) were procured from Sigma Aldrich, St. Louis, MO, USA. Lipoid 90H was a generous gift from Lyon, France. Human breast cancer cell line (MCF-7 cells) with estrogen, progesterone, and glucocorticoid receptors was procured from the American Type Culture Collection (ATCC). All other chemicals used were of analytical grade, and Milli-Q water was used wherever needed.
2.2. Preparation of SM-loaded Lipid–Polymer Hybrid Nanoparticles
Lipid–polymer hybrid nanoparticles (LPH-NPs) were prepared by emulsification solvent evaporation technique [30]. Briefly, pure SM (20 mg) and lipoid 90H (40 mg) was dissolved in 10 mL of dichloromethane to obtain the organic phase. Separately, the aqueous phase was prepared by dissolving chitosan (25–100 mg) and soyalecithin (20 mg) in 0.5% w/v acetic acid (Table 1). Further, prepared organic phase was emulsified into aqueous phase (with rate of 0.3 mL/min) using probe sonication (ultrasonic processor, Fisher scientific, Waltham, MA, United States) at power 65%, on–off cycle 5 sec, for 3 min. Formed emulsion was kept on a magnetic stirrer (500 rpm) at room temperature overnight. After complete evaporation of dichloromethane, reduced volume was centrifuged (HermleLabortechnik, Z216MK, Wehingen, Germany) at 6000 rpm for 15 h to obtain the sediment. Sediment pellet was then washed with milli-Q water thrice and lyophilized (Millirock Technology, Kingston, NY, USA) and lyophilized LPHNPs were collected for further analysis (Figure 1).

Table 1.
Composition of SM-loaded LPHNPs.
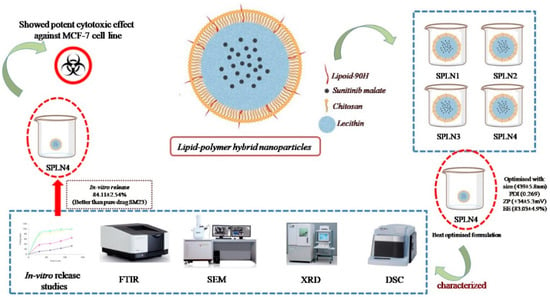
Figure 1.
Schematic diagram of SLPNs synthesis.
2.3. Measurement of Particle Size, Polydispersity Index (PDI) and Zeta Potential (ZP)
Freshly prepared SM-loaded LPHNPs were diluted 200 times with milli-Q water, sonicated for 5 min, and transferred into plastic cuvette, then analyzed for particle size and PDI using Malvern Zetasizer (ZEN-3600, Malvern Instruments Ltd., Worcestershire, UK). Three measurements were carried out for 11 runs with 10 sec durations each run at 25 °C temperature. The same procedure was followed for ZP measurements as particle size except a glass electrode sample holder was used instead of a plastic cuvette [31]. Each sample measurement was performed in triplicate.
2.4. Percent Drug Entrapment Efficiency (%EE)
Percent entrapment efficiency (%EE) was measured indirectly [20]. Freshly prepared SM-loaded LPHNPs dispersion were subjected to the centrifugation at 10,000 rpm for 15 min. Supernatant was then collected, pre-filtered by syringe filter 45 µm, then suitably diluted with methanol and quantified for unentrapped drug. Aliquots were analyzed using UV/Visible spectroscopy at λmax 250 nm [31] (Jasco V630 UV/Visible spectrophotometer, Tokyo, Japan). The %EE was calculated using the following equations:
2.5. Differential Scanning Calorimetry
Drug entrapment in the nanoparticles can be identified by differential scanning calorimetry (DSC). Samples 5 mg (pure SM, Lipoid 90H, chitosan, and optimized LPHNPs) (SLPN4) were packed in a hemispherical aluminium pan, separately. A pan filled with the sample was kept in a heating chamber against the blank. The temperature was raised from 50 °C to 350 °C at a rate of 20 °C/min; additionally, nitrogen gas was supplied at flow rate of 20 mL/min (Sinco 400, Seoul, Korea) [32]. Endothermic peaks were seen at the melting point of the sample; the temperature was then noted and studied.
2.6. FTIR Spectroscopy
FTIR spectra of SM, lipoid 90H, chitosan, and optimized LPHNPs (SLNP4) were taken using the KBr technique. The samples were mixed with crystalline KBr and the mixture was then compressed into transparent pellets using a hand-held compression machine. Thin transparent sample film enclosed in the die was fixed into the sample holder and scanned in the range of 400–4000 cm−1 (Jasco, V750, FTIR spectrophotometer, Tokyo, Japan). Peaks at the fingerprint region were interpreted, and additional or absent peaks were then studied for possible functional group interactions between drug and excipients. Spectrums were then collaged and presented for compatibility study [33].
2.7. XRD Diffraction Study
Sample (pure SM and SLPN4) were characterized by XRD to identify the nature of solid-state. X-ray diffractometer (Ultima IV, Rigaku Inc., Tokyo, Japan) was used with the following set parameters: Cu-kα radiation at 40 kV/40 mA, scan rate of 0.500°/min in the 0–60 (2θ) range, at a fixed monocromator (U4), attached with scintillation detector [33].
2.8. In-Vitro Release Studies
Comparative in vitro release studies of pure SM and optimized LPHNPs (SLPN4) were carried out using a dialysis bag (cut off mol wt. 12 kD) as per our previously reported method [30]. Briefly, pure SM and SLPN4 (equivalent to 20 mg of SM) were dissolved in pre-soaked dialysis containing 10 mL of phosphate buffer (pH 6.8), then dialysis bags were dipped into a beaker containing 40 mL of dissolution media and shaken on biological shaker (LBS-030S-Lab Tech, Kyonggi, Korea) at 100 rpm. At fixed time intervals (0.5, 1, 2, 3, 6, 12, 24, and 48 h), 1 mL of sample was withdrawn and compensated immediately with respective media to maintain sink condition, filtered, diluted, and analyzed at 250 nm [31]. Each sample was analyzed in triplicate. Further study of release mechanism was executed by fixing the release data into the following mathematical equations.
where, Qt and Q0 represents (SM dissolved in media overtime t), (initial amount of SM dissolved in media, i.e., equal to zero). K marked constants of models. Mt and M∞ are cumulative drug release at time t and infinite time, respectively, t is the release time and n denotes the diffusional exponent indicating release mechanism [30].
Qt = Q0 + k0t Zero order
logQt = logQ0 − kt/2.303 First order
Qt = kHt 1/2 Higuchi model
Mt/M∞ = ktn Korsmeyer Peppas model
2.9. Morphology
The morphology and size of images of optimized LPHNPs (SLPN4) were viewed by Scanning Electron Microscopic (SEM) (Zeiss EVO LS10; Cambridge, UK). In a thin film coater under vacuum, the sample was homogeneously dispersed and coated with gold-metal (Quorum Q150R S, Lewes, East Sussex, UK). The pre-treated sample was then bombarded with an electron beam, resulting in the creation of secondary electrons known as auger electrons. Only the electrons scattered at ≥90 degrees were picked from this interaction between the electron beam and the specimen’s atoms, and surface topography was obtained at 15 kV acceleration voltage and 7.58 K × magnification.
2.10. Cell Culture and Treatments
The Human breast cancer cell line (MCF7) was procured from the American Type Cutler Collection (Manassas, VA, USA). The cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) with phenol red supplement with 10% Fetal Bovine Serum (FBS), with Penicillin (100 units/mL), Streptomycin (100 μg/mL), and Amphotericin B (250 ng/mL), Gibco® (New York, NY, USA). The cells were grown at 37 °C in 50 cm2 tissue culture flasks in a 5% CO2 humidified incubator. The cells were seeded into 96-well cell culture plates in DMEM.
2.11. MTT Assay on MCF7 Cells
To determine the dose dependent cell viability of MCF-7 cells, they were incubated with SM and SLPN4 ranging from 0.78 to 100 µg/mL (containing equivalent amount of SM drug) for 48h. The data presented demonstrate relative cell viability after the treatment, since MTT assay determines the viable cells through activity of mitochondria. This is primarily targeted through mitochondria-mediated apoptosis. Thus, this approach was adopted to determine the activity of SM and SLPN4. The IC50 values were calculated using Log (inhibitor) versus normalized response on variable slope by GraphPad Prism V-5.1 (San Diego, CA, USA).
2.12. Morphological Changes on MCF-7 Cells
The cytotoxic effect of pure STB and SLPN4 was also determined by visualizing the morphological changes in MCF7 cells [34]. The IC50 value of SM (10.79 µg/mL) equivalent to SLPN4 (8.24 µg/mL) was taken as the dose of treatment and morphological features were manifested by phase-contrast microscopy. Morphological features such as membrane blebbing, cell shrinkage, and necrosis were determined.
2.13. Caspase-3, Caspase-9 and p53 Assay by ELISA
Caspase-3, caspase-9, and p53 assay ELISA kits were used to measure caspase activity [35]. The MCF-7 cells (50,000 cell/well) were seeded in 96-well plates. The cells were cultured for 24 h at 37 °C in a humidified incubator with 5% CO2. The SM, SLPN4-treated, and untreated control cells were then allowed to equilibrate at room temperature in 96-well plates. Each well of plate (SM, SLPN4-treated, and control) containing 100 µL of culture media received 100 µL of caspase-3 and 9 reagents. The plate was covered and the contents were stirred at 500 rpm for 30 s. After 30 min of incubation at room temperature, the optical density was measured at 405 nm using an an ELISA reader.
2.14. Stability Study
To analyze the change in formulation over storage or shelf life, a stability study was conducted for optimized LPHNPs (SLPN4). The formulation was sealed in a glass vial and stored for three months at 25 ± 0.5 °C/65 ± 5% RH and 40 ± 2 °C/75 ± 5% RH in a stability chamber, and physical appearance, particle size, PDI, ZP and entrapment were examined in samples obtained at 0, 1, 2, and 3 months [36,37].
2.15. Statistical Analysis
The experimental data were analyzed statistically using one way ANOVA followed by Tukey’s multiple comparison test using SPSS 16 software (version 25.0; SPSS Inc., Chicago, IL, USA) (p < 0.01) was considered significant.
3. Results and Discussion
3.1. Measurement of Particle Size, Polydispersity Index (PDI) and Zeta Potential (ZP)
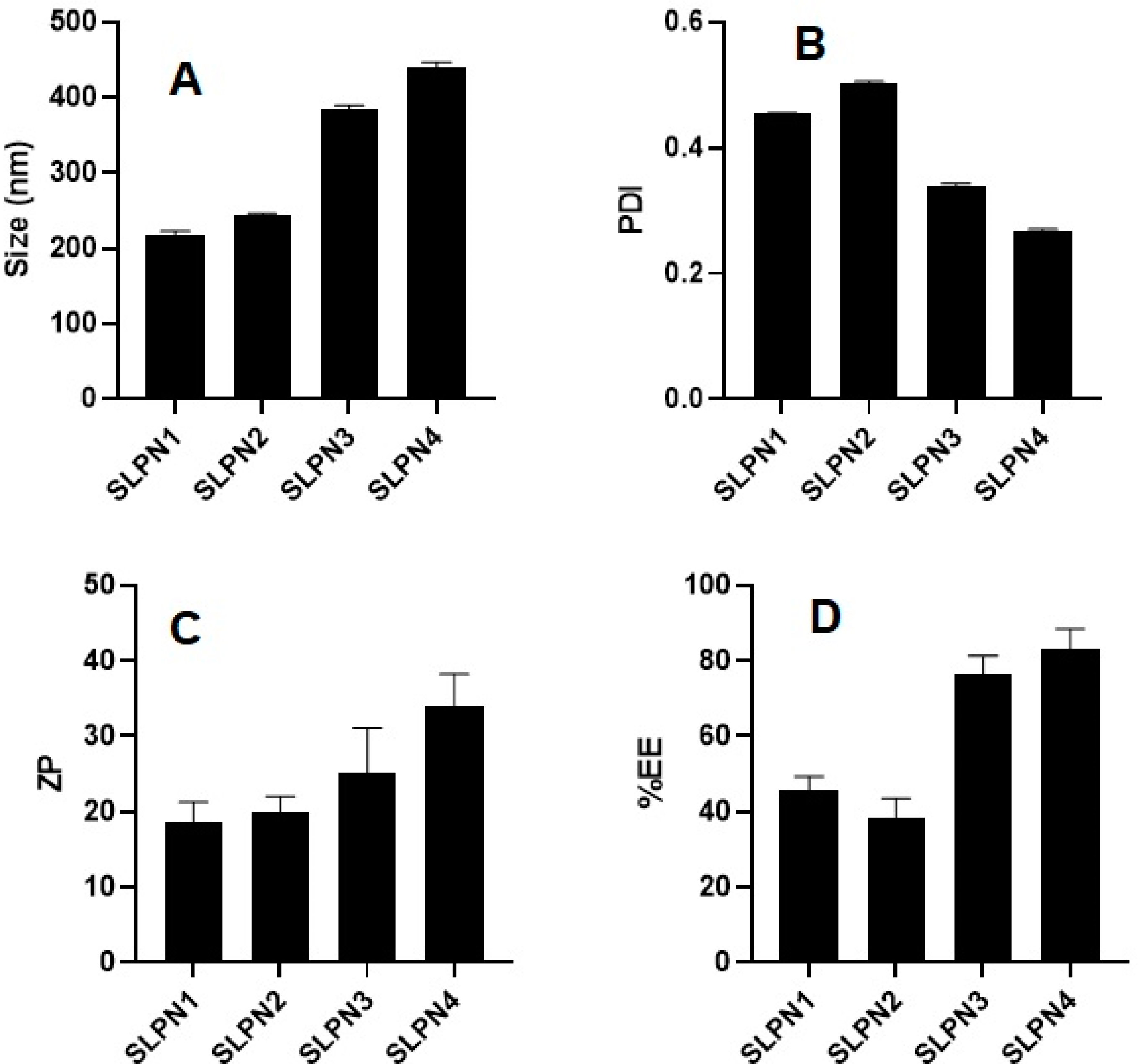
SM-loaded LPHNPs (SLPN1-SLPN4) were prepared by the single emulsification method. The developed LPHNPs were characterized for their particle size, PDI, and ZP and measured in the range of 218–439 nm, 0.269–0.504, and +18 to +34 mV, respectively (Figure 2). According to studies, nanoparticles with a size range of 40 to 400 nm are appropriate for ensuring long circulation duration and increased accumulation of drug in tumors with limited renal clearance [38,39]. The positive values of ZP was measured due to the amino group present on the surface of chitosan polymer [40,41]. From the results, it was observed that increase in concentration of chitosan in formulations increased the size of particles.
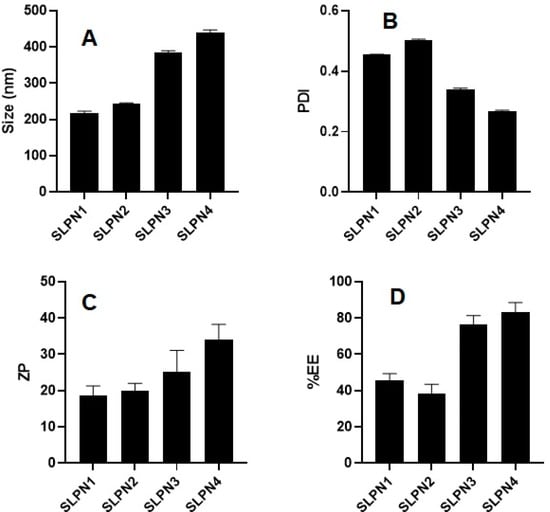
Figure 2.
Particle size, PDI, ZP, and %EE of developed SM-loaded lipid polymer nanoparticles (SLPN1–SLPN4) tested with one way ANOVA followed by Tukey’s multiple comparison between formulations. (A) Particle size—significant difference (** p < 0.01) among formulations. (B) PDI—significant difference (** p < 0.001) among formulations. (C) Zeta potential—results are not significant among (SLPN1 vs. SLPN2, SLPN1 vs. SLPN3, SLPN2 vs. SLPN3, and SLPN3 vs. SLPN4 formulations) and significant (** p < 0.01) between SLPN1 vs. SLPN4 and SLPN2 vs. SLPN4. (D) %EE—results are not significant among (SLPN1 vs. SLPN2 and SLPN3 vs. SLPN4 formulations) and the rest are significant (** p < 0.01).
3.2. Percent Drug Entrapment Efficiency (%EE)
The %EE of SM-loaded LPHNPs (SLPN1–SLPN4) were measured in the range of 45.71 ± 3.3–83.03 ± 4.9% (Figure 2). The highest drug entrapment (83.03 ± 4.9%) was measured in SLPN4, the large amount of chitosan (100 mg) used in this formulae expected to prevent leakage of drug from polymeric core, thereby improving the entrapment efficiency of drug [42]. Among the developed LPHNPs, SLPN4 was optimized with size (439 ± 5.8 nm), PDI (0.269), ZP (+34 ± 5.3 mV), and EE (83.03 ± 4.9%) and further evaluated.
3.3. Differential Scanning Calorimetry
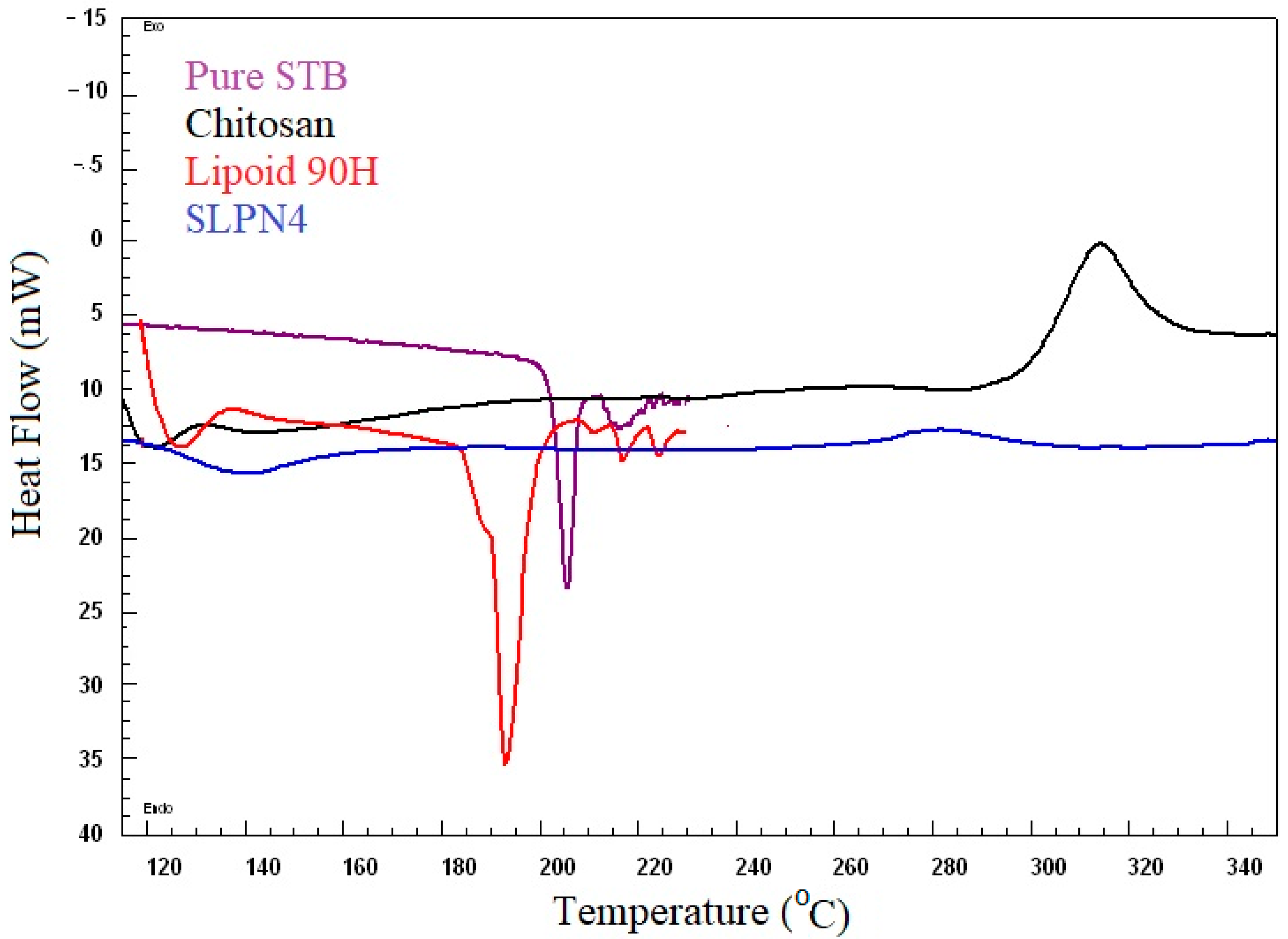
DSC spectra of pure SM, Lipoid 90H, chitosan, and optimized LPHNPs (SLPN4) are presented in Figure 3. DSC studies confirmed the crystallinity and amorphocity nature of the sample, which indicates the encapsulation of drugs in nanoparticles [20,43]. Pure drug SM exhibited a sharp endothermic peak at 205 °C, which indicates its melting temperature [44,45]. The SM peak completely disappeared in DSC spectra of SLPN4, confirming SM entrapment in LPHNPs. Sharp endothermic and broad exothermic peaks of lipoid 90H and chitosan could be seen near 193 °C and 320 °C, respectively.
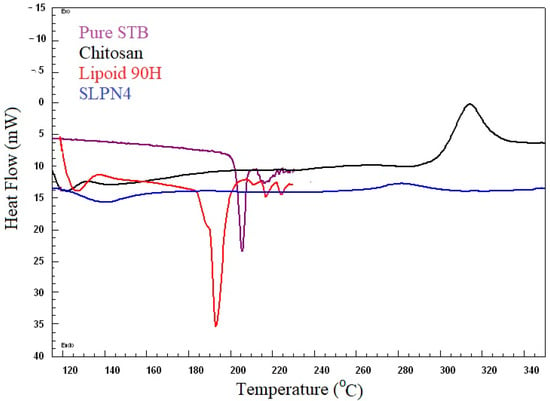
Figure 3.
Comparative DSC spectra of pure SM, SLPN4, lipoid 90H, and chitosan.
3.4. FTIR Spectroscopy
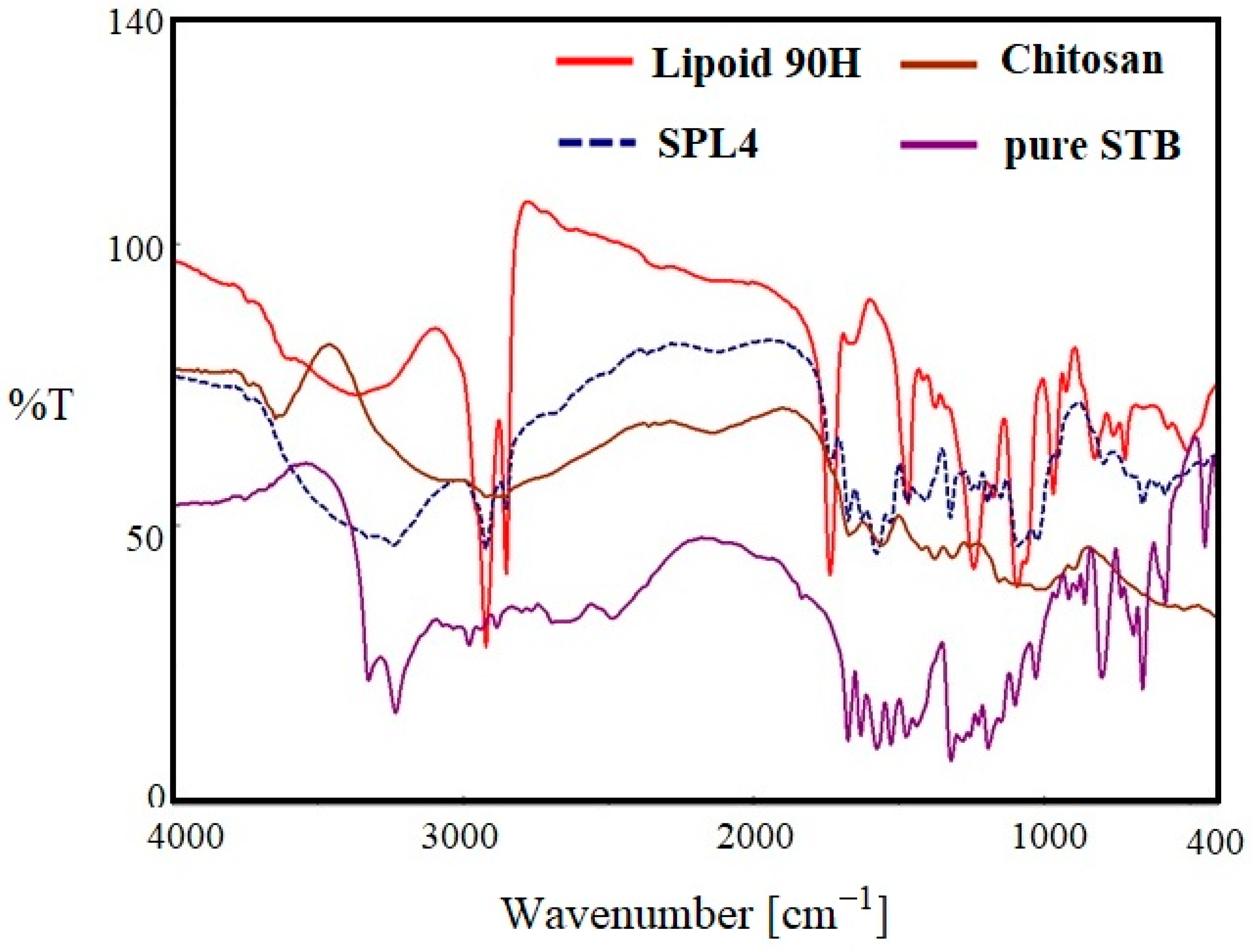
Figure 4 indicates the FTIR spectra of pure SM, lipoid 90H, chitosan, and optimized LPHNPs (SLPN4) in the wavelength range of 400–4000 cm−1. The FTIR spectra of pure SM showed many intense peaks at 3324 cm−1 for the acidic O-H, 2981 cm−1 for the acidic –CH=CH- (aryl) str, 2884 cm−1 for C-H (alkyl) str, 1635 cm−1 for the –NHCO str, and 1073 cm−1 bands correspond to the (C–F stretching) [41]. FTIR spectra of chitosan showed a broad peak at 3579 cm−1 (-OH str), 2873 cm−1 (CH2 str) [46]. The FTIR spectra of phospholipon 90H exhibited characteristic peaks at 2934 cm−1 and 2861 cm−1 for –C-H- str present in long fatty acid chain, 1722 cm−1 for –C=O str, and 972 cm−1 for P=O str [47]. In the optimized LPHNPs (SLP4), peaks at 3324 cm−1, 2981 cm−1, and 2884 cm−1 were found to have disappeared. The FTIR peaks of SLPN4 represents no significant modifications in the functional peaks of the SM in the lipid–polymeric NPs matrix, thereby retaining drug’s physicochemical properties and efficient chemical stability for the encapsulated SM in the fabricated nanocarriers. As there is no evidence of drug–polymers interaction, the selected lipid, polymer, and drug have the compatibility and suitability for the SM-loaded HNPs.
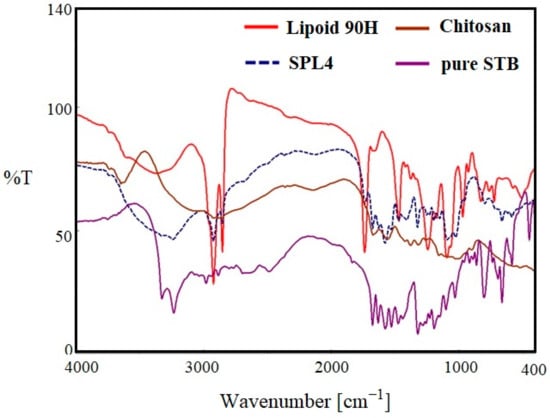
Figure 4.
Comparative FTIR spectra of pure SM, SLPN4, lipoid 90H, and chitosan.
3.5. XRD Diffraction Study
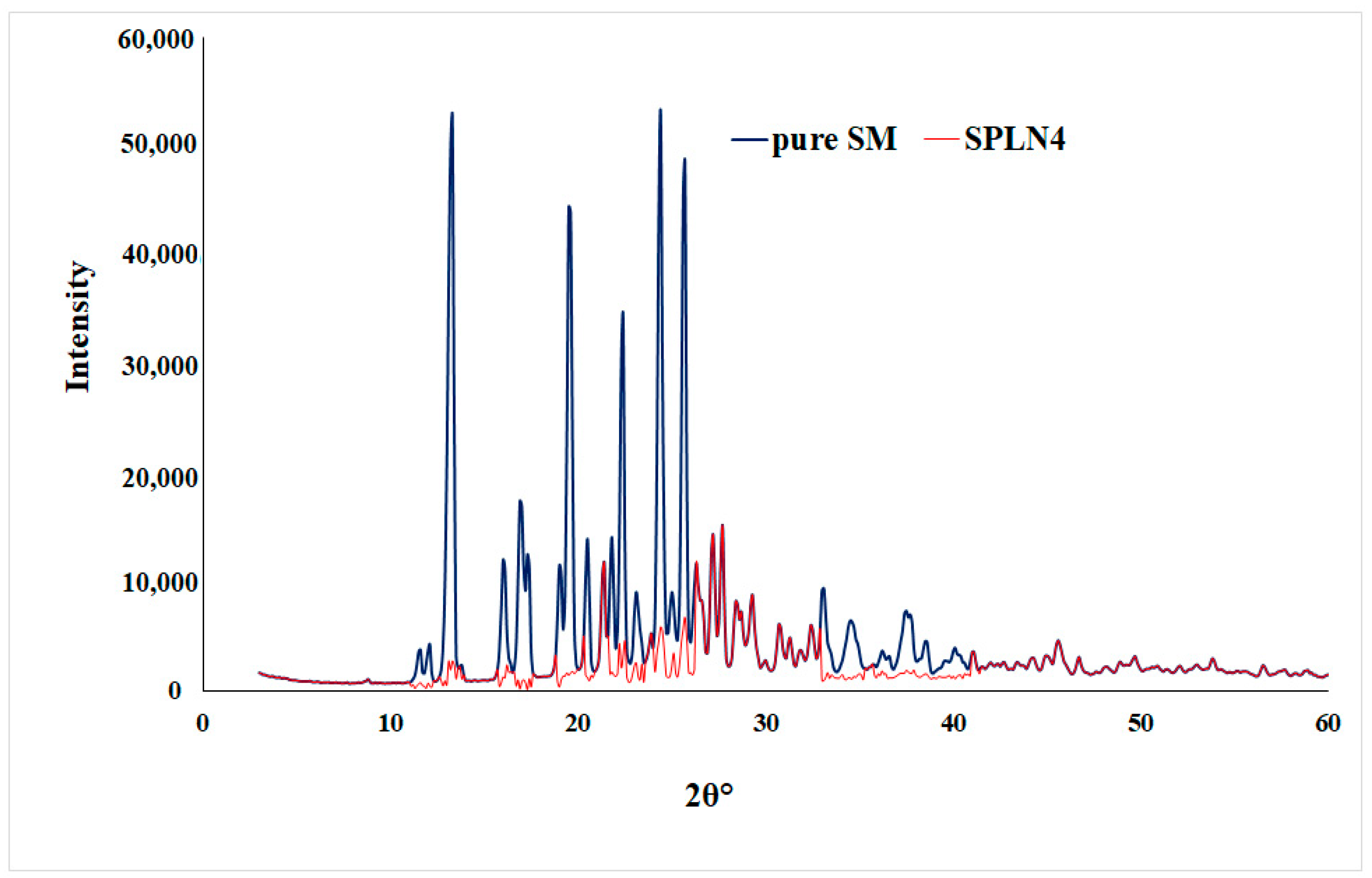
Pure drug sunitinib (SM) showed intense X-ray diffractions at 13.3°, 19.5°, 22.4°, and 25.6° at 2θ, which represents the crystalline state of the drug [48]. However, the optimized LPHNPs (SLPN4) diffractogram also showed few peaks with reduced intensities, which are attributed to the amorphous state of drug in the nanoparticles due to the destruction of crystalline nature of SM and molecular dispersion of polymers, lipid, and drug (Figure 5).
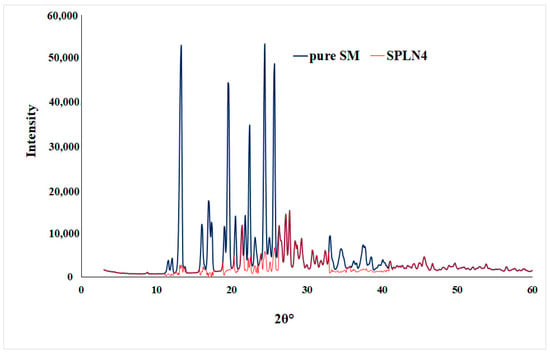
Figure 5.
XRD spectra of pure SM and optimized LPHNPs (SLPN4).
3.6. In-Vitro Release Studies
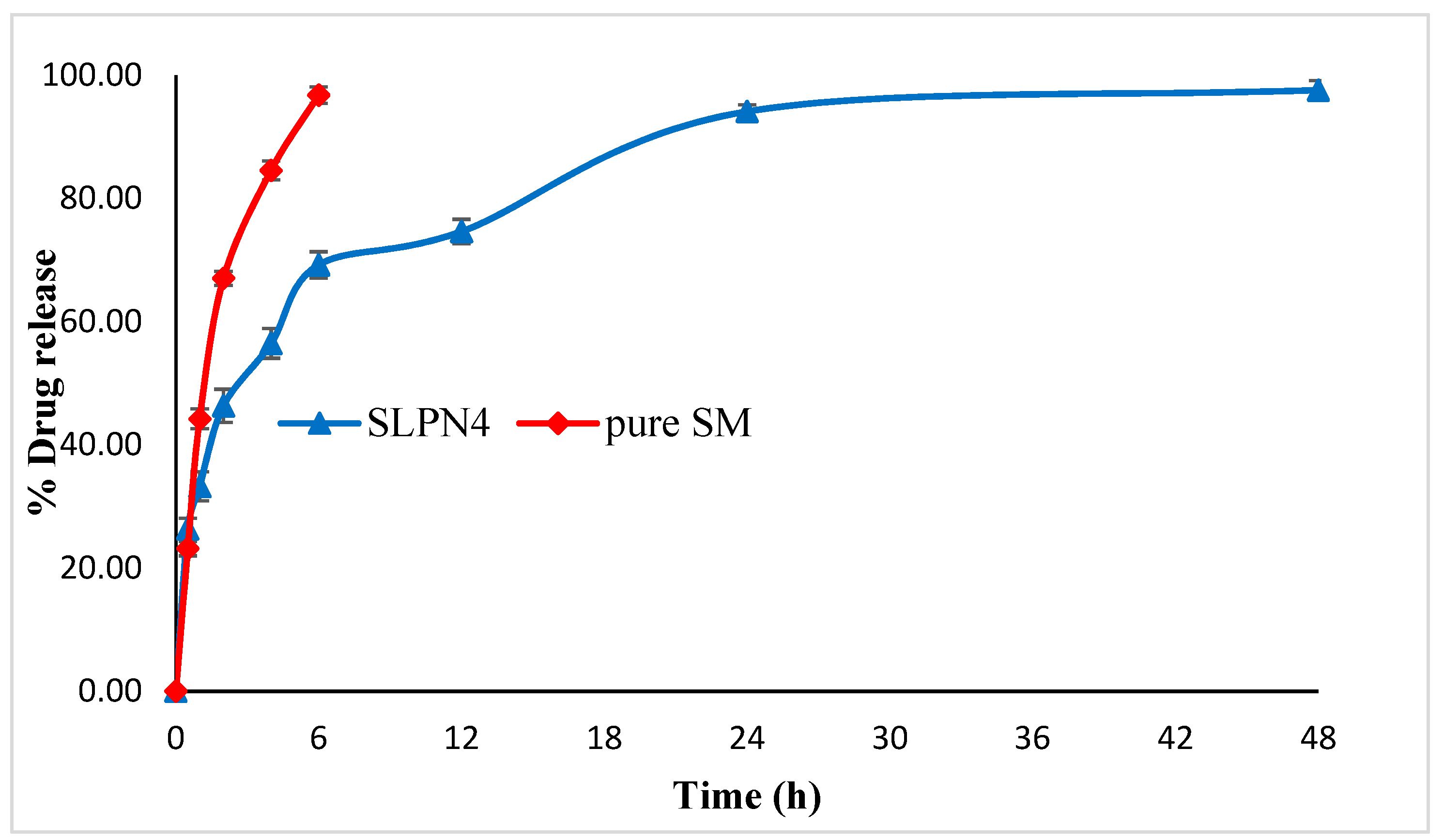
In vitro drug release studies were conducted at pH 6.8, i.e., the pH of cancer cells [49]. A rapid drug release (98.45%) was exhibited by pure SM at the end of 6 h as compared with SLPN4. The optimized formulation (SLPN4) showed an initial rapid release of the drug for the first 6 h of study, followed by a sustained drug release. Initial rapid release could be due to the adsorbed drug over the polymer, which dissociated easily in the diffusion medium and released [20] (Figure 6).
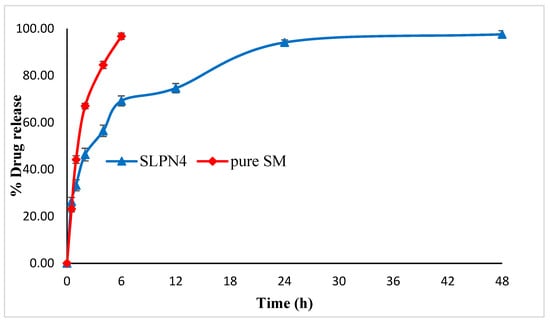
Figure 6.
Comparative in vitro release profile of pure SM and optimized LPHNPs (SLPN4).
Release kinetics were assessed for optimized LPHNPs (SLPN4). The goodness of fit models was selected by evaluating R2 value in the prediction of the release mechanism. The kinetics analysis of regression coefficient of all the four models used indicated R2 values for zero order (0.612), first order (0.9359), Higuchi model (0.8494), and Korsmeyer–Peppas (0.9406) with diffusion coefficient n (0.271). The optimized LPHNPs (SLPN4) followed the Fickian diffusion (n < 0.5) and mechanism of release from the Korsmeyer–Peppas kinetic model [30].
3.7. Morphology
SEM images of optimized LPHNPs (SLPN4) are shown in the Figure 7. It was confirmed that optimized LPHNPs (SLNP4) were small and spherical in shape with aggregation, probably due to the presence of lipids. The size observed by SEM was approximately same as that measured by the DLS method.

Figure 7.
SEM images of optimized LPHNPs (SLPN4).
3.8. MTT Assay on MCF7 Cells
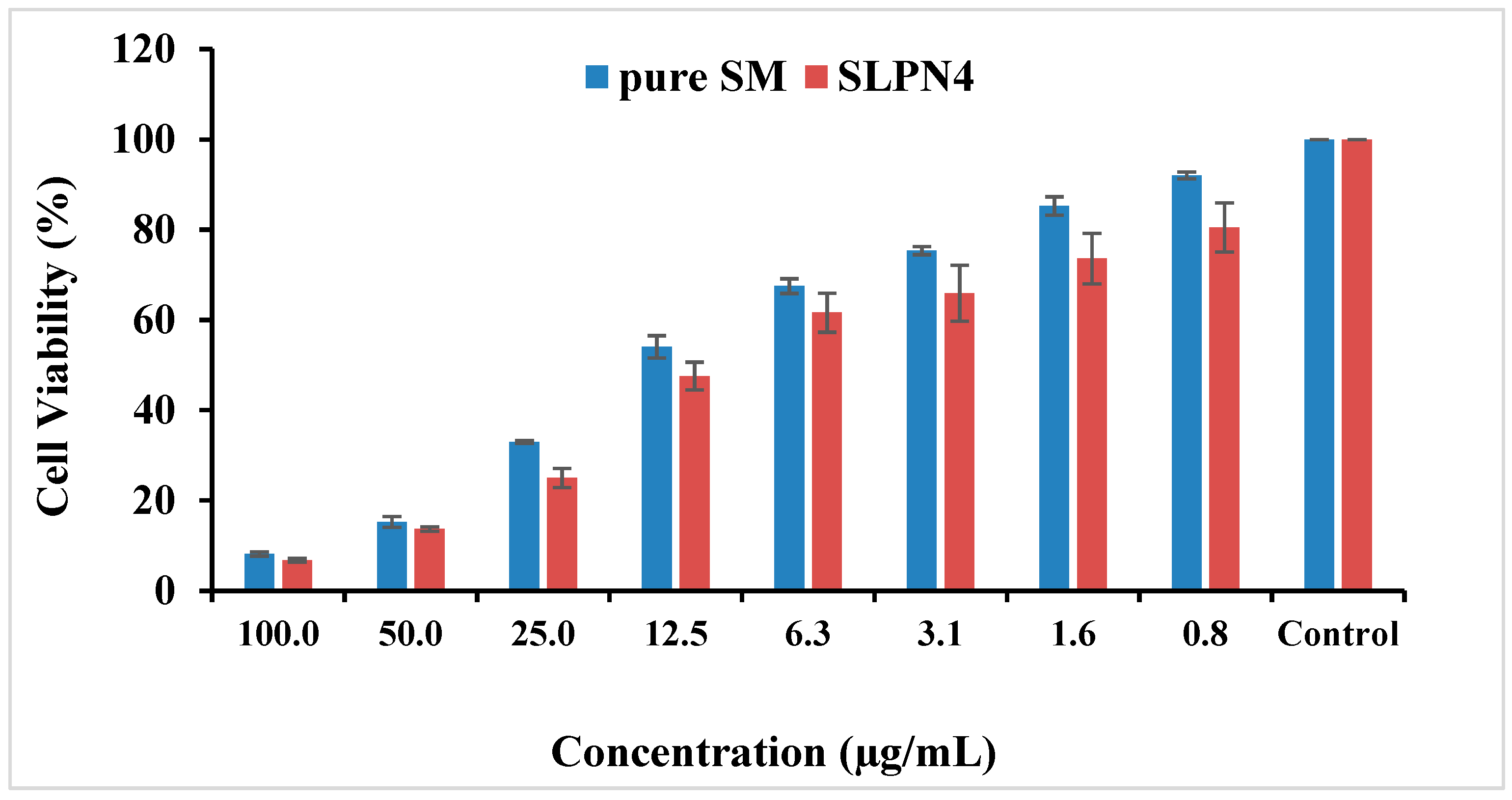
The MTT assay showed concentration-dependent reduction in cell viability for SM and optimized SM-loaded LPHNPs (SLPN4) against MCF-7 cell lines (Table 2 and Figure 8). The IC50 values for pure drug SM and SLP4 were found as 10.79 and 8.24 µg/mL for MCF-7 cells, respectively. The formulation SLP4 showed a significant reduction in cell viability (80.52%, 73.58%, 65.89%, 61.60%, 47.53%, 24.97%, 13.63%, and 6.79% at 0.8, 1.6, 3.1, 6.3, 12.5, 25, 50, and 100 µg/mL) in comparison with pure drug SM (92.05%, 85.26%, 75.35%, 67.49%, 54.04%, 32.94%, 15.24%, and 8.17% at 0.8, 1.6, 3.1, 6.3, 12.5, 25, 50, and 100 µg/mL), respectively, against MCF-7 cells. Based on the results of MTT assay, it was observed that SLP4 exhibited potential anticancer activity against breast cancer cell lines, probably due to the enhancement of the release of SM from the SLP4 formulation. SM-loaded LPHNPs (SLPN4) could be used as a potent carrier for the treatment of breast cancer.

Table 2.
Percent cell viability against concentration.
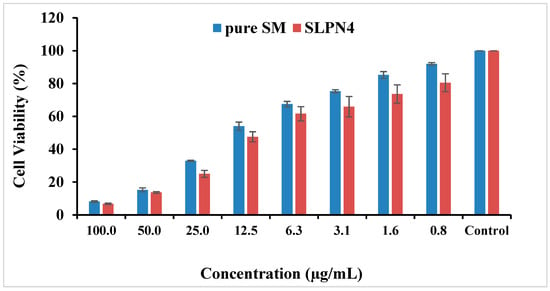
Figure 8.
Cytotoxicity of pure SM and SLPN4 after 48 h incubation with MCF-7 cells at concentrations 0.8–100 µg/mL.
3.9. Morphological Changes on MCF-7 Cells
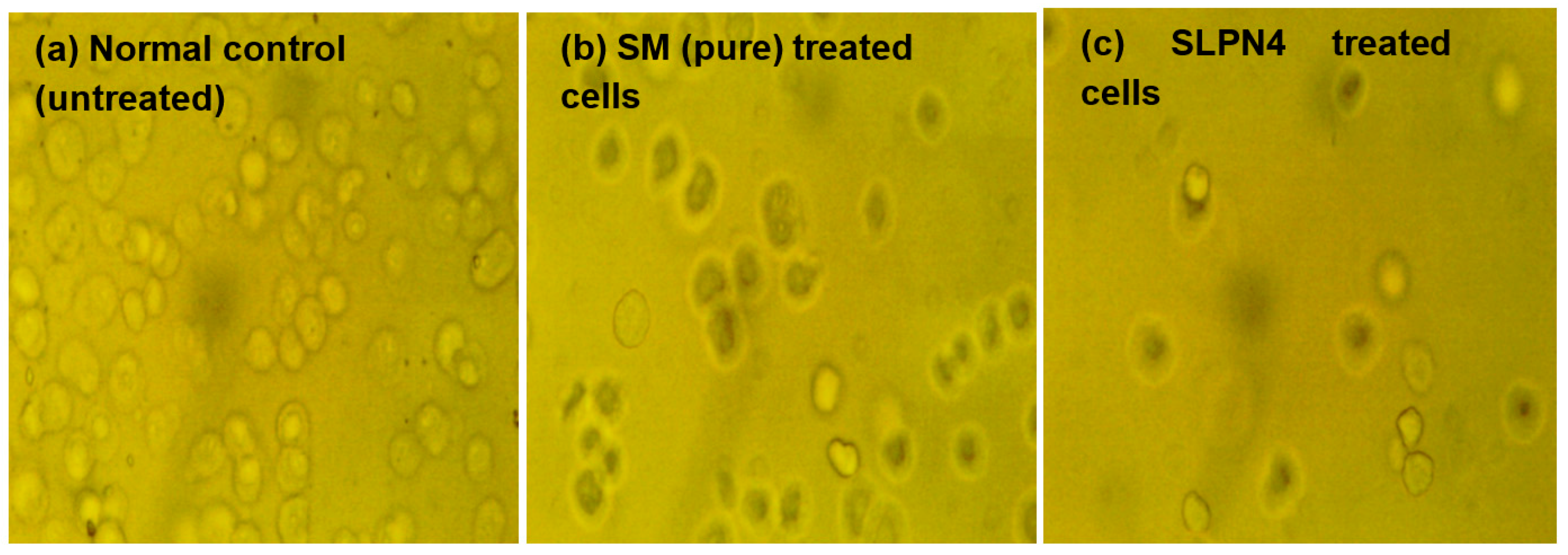
The morphological changes on MCF-7 cell lines after treatment of control, pure SM, and SLPN4 are presented in Figure 9, and after 24 h of incubation, SLPN4-treated cells evidenced maximum cell death in comparison with pure SM and control. The morphological changes observed in MCF-7 by SLPN4 were due to damage in cell organelles. The morphological changes were observed in MCF7 cells by SM as it is a tyrosine kinase inhibitor and has an anti-proliferative effect on breast cancer cell lines (MCF-7). The SM and SLPN4 treatments cause concentration-dependent cell growth suppression due to apoptosis, as made evident by Caspase-3, p53, and Caspase-9 levels in MCF7. These results are consistent and similar in previous reports [50,51].
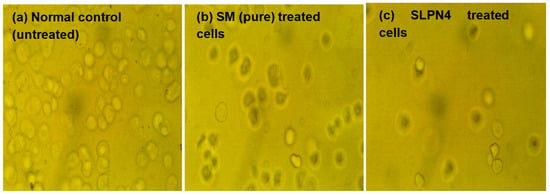
Figure 9.
Morphological changes in MCF7 cells after exposure to Sunitinib-containing formulations. Untreated cells/control, cells incubated with pure SM, and SLPN4.
3.10. Caspase-3, Caspase-9 and p53 Assay by ELISA
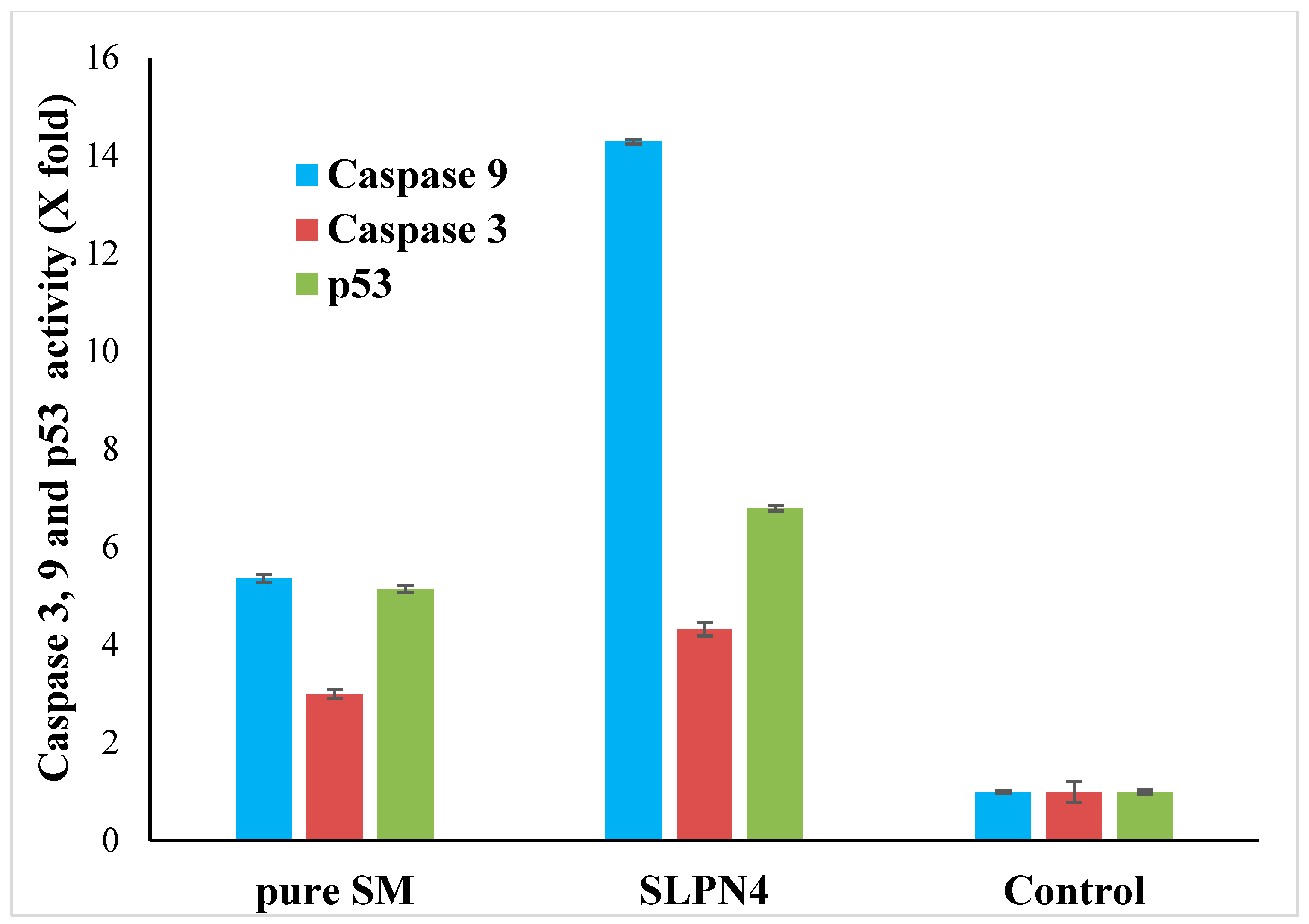
Apoptosis is a type of programmed cell death that involves the disassembly of intracellular components while avoiding injury and inflammation to nearby cells [52,53]. The activation of caspase-3, -9, and p53 are mainly responsible for cancer cell apoptosis [54,55]. In this study, an increase in caspase 3, 9, and p53 production in pure SM- and SLPN4-treated MCF-7 cells was compared to the untreated control group to confirm apoptosis. When MCF-7 cells were exposed to pure SM and SLPN4, their caspase 3, 9, and p53 activity was found four, fourteen, and seven times higher compared with control (untreated) cells, respectively (Figure 10). Enhanced effectiveness of SM in LPHNPs suggests a possible reason for induction of apoptosis in cancer cells.
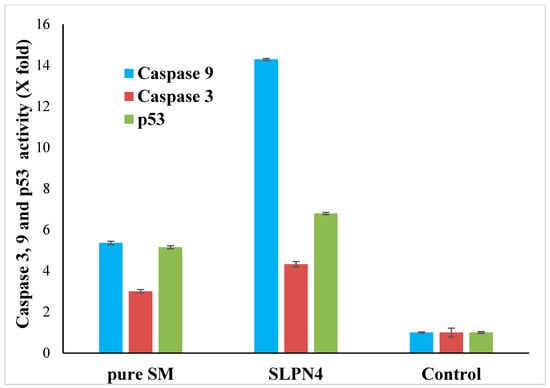
Figure 10.
Activation of caspase 3, 9, and p53 in pure SM- and SLPN4-treated MCF7 cells was compared to the untreated control. SLPN4 (p < 0.05) vs. free SM and control group.
3.11. Stability Study
Following ICH guidelines, the stability of optimized LPHNPs (SLPN4) was assessed for three months in terms of particle size, PDI, ZP, and entrapment efficiency after storage. Table 3 shows the stability study parameters for LPHNPs (SLPN4), which were all within acceptable ranges, indicating that the developed formulation was stable for three months. A significant change (** p < 0.01) in particle size and PDI were noted when stored at 25 ± 0.5 °C/65 ± 5% RH and 40 ± 2 °C/75 ± 5% RH, which indicated that high temperature can lead to aggregation of the nanoparticles.

Table 3.
Stability data of optimized LPHNPs (SLPN4).
4. Conclusions
No research has been reported on SM as a breast cancer treatment using lipid–polymer nanoparticles. In vitro release and cell line investigations showed that the developed optimized SM-loaded LPHNPs (SLPN4) significantly enhanced the release and accessibility of SM at the breast cancer cells. Importantly, the potential cytotoxicity of SLPN4 on the MCF-7 breast cancer cell line was determined using the MTT test for cytotoxicity, ELISA activity of caspase-3, -9, and p53 in comparison to free SM or control. Thus, the developed formulation could provide an attractive nanoplatform for the treatment of breast cancer and may be the focus for the future chemotherapeutic investigations.
Author Contributions
Conceptualization, M.M.A. and M.K.A.; methodology, F.F.; software, A.S.A.; validation, M.A.K. and M.F.A.; investigation, F.F.; resources, A.A.; data curation, M.M.A.; writing—original draft preparation, M.K.A.; writing—review and editing, M.K.A. and A.I.A.; supervision, M.K.A.; project administration, M.M.A.; funding acquisition, M.M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Deanship for Research & Innovation, Ministry of Education in Saudi Arabia, grant number IF/PSAU-2021/03/18862.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The authors extend their appreciation to the Deanship for Research & Innovation, Ministry of Education in Saudi Arabia, for funding the research work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Yekedüz, E.; Dizdar, Ö.; Kertmen, N.; Aksoy, S. Comparison of Clinical and Pathological Factors Affecting Early and Late Recurrences in Patients with Operable Breast Cancer. J. Clin. Med. 2022, 11, 2332. [Google Scholar] [CrossRef]
- Acar, A.; Simoes, B.M.; Clarke, R.B.; Brennan, K. A role for Notch signaling in breast cancer and endocrine resistance. Stem Cells Int. 2016, 2016, 2498764. [Google Scholar] [CrossRef]
- Ling, H.; Sylvestre, J.R.; Jolicoeur, P. Notch1-induced mammary tumor development is cyclin D1-dependent and correlates with expansion of pre-malignant multipotent duct-limited progenitors. Oncogene 2010, 29, 4543–4554. [Google Scholar] [CrossRef][Green Version]
- Alladin, A.; Chaible, L.; Garcia Del Valle, L.; Sabine, R.; Loeschinger, M.; Wachsmuth, M.; Hériché, J.K.; Tischer, C.; Jechlinger, M. Tracking cells in epithelial acini by light sheet microscopy reveals proximity effects in breast cancer initiation. eLife 2020, 9, e54066. [Google Scholar] [CrossRef]
- Sari, I.N.; Phi, L.; Jun, N.; Wijaya, Y.T.; Lee, S.; Kwon, H.Y. Hedgehog Signaling in Cancer: A Prospective Therapeutic Target for Eradicating Cancer Stem Cells. Cells 2018, 7, 208. [Google Scholar] [CrossRef]
- Muñoz-Fontela, C.; Mandinova, A.; Aaronson, S.A.; Lee, S.W. Emerging roles of p53 and other tumour-suppressor genes in immune regulation. Nature reviews. Immunology 2016, 16, 741–750. [Google Scholar] [CrossRef]
- Alimonti, A.; Carracedo, A.; Clohessy, J.G.; Trotman, L.C.; Nardella, C.; Egia, A.; Salmena, L.; Sampieri, K.; Haveman, W.J.; Brogi, E.; et al. Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 2010, 42, 454–458. [Google Scholar] [CrossRef]
- Antico Arciuch, V.G.; Russo, M.A.; Kang, K.S.; Di Cristofano, A. Inhibition of AMPK and Krebs cycle gene expression drives metabolic remodeling of Pten-deficient preneoplastic thyroid cells. Cancer Res. 2013, 73, 5459–5472. [Google Scholar] [CrossRef]
- Wen, J.; Li, H.Z.; Ji, Z.G.; Jin, J. Human urothelial carcinoma cell response to Sunitinib malate therapy in vitro. Cancer Cell Int. 2015, 15, 26. [Google Scholar] [CrossRef]
- Grimaldi, A.M.; Guida, T.; D’Attino, R.; Perrotta, E.; Otero, M.; Masala, A.; Cartenì, G. Sunitinib: Bridging present and future cancer treatment. Ann. Oncol. 2007, 18, vi31–vi34. [Google Scholar] [CrossRef] [PubMed]
- Sangwan, S.; Panda, T.; Thiamattam, R.; Dewan, S.K.; Thaper, R.K. Novel Salts of Sunitinib an Anticancer Drug with Improved Solubility. Int. Res. J. Pure Appl. Chem. 2014, 5, 352–365. [Google Scholar] [CrossRef]
- Papaetis, G.S.; Syrigos, K.N. Sunitinib: A multitargeted receptor tyrosine kinase inhibitor in the era of molecular cancer therapies. BioDrugs. 2009, 23, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Ramazani, F.; Hiemstra, C.; Steendam, R.; Kazazi-Hyseni, F.; Van Nostrum, C.F.; Storm, G.; Kiessling, F.; Lammers, T.; Hennink, W.E.; Kok, R.J. Sunitinib microspheres based on [PDLLA-PEG-PDLLA]-b-PLLA multi-block copolymers for ocular drug delivery. Eur. J. Pharm. Biopharm. 2015, 95 (Pt B), 368–377. [Google Scholar] [CrossRef]
- Raymond, E.; Dahan, L.; Raoul, J.L.; Bang, Y.J.; Borbath, I.; Lombard-Bohas, C.; Valle, J.; Metrakos, P.; Smith, D.; Vinik, A.; et al. Sunitinib malate for the treatment of pancreatic neuroendocrine tumors. N. Engl. J. Med. 2011, 364, 501–513. [Google Scholar] [CrossRef]
- Chen, S.; Liang, Q.; Liu, E.; Yu, Z.; Sun, L.; Ye, J.; Shin, M.; Wang, J.; He, H. Curcumin/sunitinib co-loaded BSA-stabilized SPIOs for synergistic combination therapy for breast cancer. J. Mater. Chem. B 2017, 5, 4060–4072. [Google Scholar] [CrossRef]
- Nazari-Vanani, R.; Azarpira, N.; Heli, H.; Karimian, K.; Sattarahmady, N. A novel self-nanoemulsifying formulation for sunitinib: Evaluation of anticancer efficacy. Colloids Surfaces B Biointerfaces 2017, 160, 65–72. [Google Scholar] [CrossRef]
- Adams, V.R.; Leggas, M. Sunitinib malate for the treatment of metastatic renal cell carcinoma and gastrointestinal stromal tumors. Clin. Ther. 2007, 29, 1338–1353. [Google Scholar] [CrossRef]
- Alshahrani, S.M.; Alshetaili, A.S.; Alalaiwe, A.; Alsulays, B.B.; Anwer, M.K.; Al-Shdefat, R.; Imam, F.; Shakeel, F. Anticancer Efficacy of Self-Nanoemulsifying Drug Delivery System of Sunitinib Malate. AAPS PharmSciTech 2018, 19, 123–133. [Google Scholar] [CrossRef]
- Anwer, M.K.; Ahmed, M.M.; Ezzeldin, E.; Fatima, F.; Alalaiwe, A.; Iqbal, M. Preparation of sustained release apremilast-loaded PLGA nanoparticles: In vitro characterization and in vivo pharmacokinetic study in rats. Int. J. Nanomed. 2019, 14, 1587–1595. [Google Scholar] [CrossRef]
- Jamil, A.; Aamir Mirza, M.; Anwer, M.K.; Thakur, P.S.; Alshahrani, S.M.; Alshetaili, A.S.; Telegaonkar, S.; Panda, A.K.; Iqbal, Z. Co-delivery of gemcitabine and simvastatin through PLGA polymeric nanoparticles for the treatment of pancreatic cancer: In-vitro characterization, cellular uptake, and pharmacokinetic studies. Drug Dev. Ind. Pharm. 2019, 45, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Anwer, M.K.; Al-Shdefat, R.; Ezzeldin, E.; Alshahrani, S.M.; Alshetaili, A.S.; Iqbal, M. Preparation, Evaluation and Bioavailability Studies of Eudragit Coated PLGA Nanoparticles for Sustained Release of Eluxadoline for the Treatment of Irritable Bowel Syndrome. Front. Pharm. 2017, 8, 844. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, N.; Borkar, S.B.; Nandanwar, S.K.; Panda, P.K.; Choi, E.H.; Kaushik, N.K. Nanocarrier cancer therapeutics with functional stimuli-responsive mechanisms. J. Nanobiotechnol. 2022, 20, 152. [Google Scholar] [CrossRef]
- Anwer, M.K.; Mohammad, M.; Iqbal, M.; Ansari, M.N.; Ezzeldin, E.; Fatima, F.; Alshahrani, S.M.; Aldawsari, M.F.; Alalaiwe, A.; Alzahrani, A.A.; et al. Sustained release and enhanced oral bioavailability of rivaroxaban by PLGA nanoparticles with no food effect. J. Thromb. Thrombolysis 2020, 49, 404–412. [Google Scholar] [CrossRef]
- Raza, F.; Zhu, Y.; Chen, L.; You, X.; Zhang, J.; Khan, A.; Khan, M.W.; Hasnat, M.; Zafar, H.; Wu, J.; et al. Paclitaxel-loaded pH responsive hydrogel based on self-assembled peptides for tumor targeting. Biomater. Sci. 2019, 7, 2023–2036. [Google Scholar] [CrossRef]
- Raza, F.; Zafar, H.; You, X.; Khan, A.; Wu, J.; Ge, L. Cancer nanomedicine: Focus on recent developments and self-assembled peptide nanocarriers. J. Mater. Chem. B 2019, 7, 7639–7655. [Google Scholar] [CrossRef]
- Anwer, M.K.; Iqbal, M.; Muharram, M.M.; Mohammad, M.; Ezzeldin, E.; Aldawsari, M.F.; Alalaiwe, A.; Imam, F. Development of Lipomer Nanoparticles for the Enhancement of Drug Release, Anti-microbial Activity and Bioavailability of Delafloxacin. Pharmaceutics 2020, 12, 252. [Google Scholar] [CrossRef]
- Khan, M.M.; Madni, A.; Torchilin, V.; Filipczak, N.; Pan, J.; Tahir, N.; Shah, H. Lipid-chitosan hybrid nanoparticles for controlled delivery of cisplatin. Drug Deliv. 2019, 26, 765–772. [Google Scholar] [CrossRef]
- Dong, W.; Wang, X.; Liu, C.; Zhang, X.; Zhang, X.; Chen, X.; Kou, Y.; Mao, S. Chitosan based polymer-lipid hybrid nanoparticles for oral delivery of enoxaparin. Int. J. Pharm. 2018, 547, 499–505. [Google Scholar] [CrossRef]
- Anwer, M.K.; Ali, E.A.; Iqbal, M.; Ahmed, M.M.; Aldawsari, M.F.; Saqr, A.A.; Ansari, M.N.; Aboudzadeh, M.A. Development of Sustained Release Baricitinib Loaded Lipid-Polymer Hybrid Nanoparticles with Improved Oral Bioavailability. Molecules 2022, 27, 168. [Google Scholar] [CrossRef]
- Joseph, J.J.; Sangeetha, D.; Gomathi, T. Sunitinib loaded chitosan nanoparticles formulation and its evaluation. Int. J. Biol. Macromol. 2016, 82, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Khuroo, T.; Verma, D.; Khuroo, A.; Ali, A.; Iqbal, Z. Simultaneous delivery of paclitaxel and erlotinib from dual drug loaded PLGA nanoparticles: Formulation development, thorough optimization and in vitro release. J. Mol. Liq. 2018, 257, 52–68. [Google Scholar] [CrossRef]
- Alshetaili, A.S.; Anwer, M.K.; Alshahrani, S.M.; Alalaiwe, A.; Alsulays, B.B.; Ansari, M.J.; Imam, F.; Alshehri, S. Characteristics and anticancer properties of Sunitinib malate-loaded poly-lactic-co-glycolic acid nanoparticles against human colon cancer HT-29 cells lines. Trop. J. Pharm. Sci. 2018, 17, 1263. [Google Scholar] [CrossRef]
- Alhakamy, N.A.; Fahmy, U.A.; Badr-Eldin, S.M.; Ahmed, O.A.A.; Asfour, H.Z.; Aldawsari, H.M.; Algandaby, M.M.; Eid, B.G.; Abdel-Naim, A.B.; Awan, Z.A.; et al. Optimized Icariin Phytosomes Exhibit Enhanced Cytotoxicity and Apoptosis-Inducing Activities in Ovarian Cancer Cells. Pharmaceutics. 2020, 12, 346. [Google Scholar] [CrossRef]
- Md, S.; Alhakamy, N.A.; Alharbi, W.S.; Ahmad, J.; Shaik, R.A.; Ibrahim, I.M.; Ali, J. Development and Evaluation of Repurposed Etoricoxib Loaded Nanoemulsion for Improving Anticancer Activities against Lung Cancer Cells. Int. J. Mol. Sci. 2021, 22, 13284. [Google Scholar] [CrossRef] [PubMed]
- Anwer, M.K.; Ahmed, M.M.; Aldawsari, M.F.; Alshahrani, S.; Fatima, F.; Ansari, M.N.; Rehman, N.U.; Al-Shdefat, R.I. Eluxadoline Loaded Solid Lipid Nanoparticles for Improved Colon Targeting in Rat Model of Ulcerative Colitis. Pharmaceuticals 2020, 13, 255. [Google Scholar] [CrossRef] [PubMed]
- Sahu, A.R.; Bothara, S.B. Formulation and evaluation of phytosome drug delivery system of boswellia Serrata extract. Int. J. Res. Med. 2015, 4, 94–99. [Google Scholar]
- Liechty, W.B.; Peppas, N.A. Expert opinion: Responsive polymer nanoparticles in cancer therapy. Eur. J. Pharm. Biopharm. 2012, 80, 241–246. [Google Scholar] [CrossRef]
- Subhan, M.A.; Yalamarty, S.S.K.; Filipczak, N.; Parveen, F.; Torchilin, V.P. Recent Advances in Tumor Targeting via EPR Effect for Cancer Treatment. J. Pers. Med. 2021, 11, 571. [Google Scholar] [CrossRef]
- Silva, M.M.; Calado, R.; Marto, J.; Bettencourt, A.; Almeida, A.J.; Gonçalves, L. Chitosan Nanoparticles as a Mucoadhesive Drug Delivery System for Ocular Administration. Mar. Drugs 2017, 15, 370. [Google Scholar] [CrossRef]
- Alshetaili, A.S. Gefitinib loaded PLGA and chitosan coated PLGA nanoparticles with magnified cytotoxicity against A549 lung cancer cell lines. Saudi J. Biol. Sci. 2021, 28, 5065–5073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.I.; Zhang, L. Lipid–polymer hybrid nanoparticles: Synthesis, Characterization and Applications. Nano Life 2010, 1, 163–173. [Google Scholar] [CrossRef]
- Anzar, N.; Mirza, M.A.; Anwer, M.K.; Khuroo, T.; Alshetaili, A.S.; Alshahrani, S.M.; Meena, J.; Hasan, N.; Talegaonkar, S.; Panda, A.K.; et al. Preparation, evaluation and pharmacokinetic studies of spray dried PLGA polymeric submicron particles of simvastatin for the effective treatment of breast cancer. J. Mol. Liq. 2018, 249, 609–616. [Google Scholar] [CrossRef]
- Bhatt, P.; Narvekar, P.; Lalani, R.; Chougule, M.B.; Pathak, Y.; Sutariya, V. An in vitro Assessment of Thermo-Reversible Gel Formulation Containing Sunitinib Nanoparticles for Neovascular Age-Related Macular Degeneration. AAPS PharmSciTech 2019, 20, 281. [Google Scholar] [CrossRef] [PubMed]
- Razmimanesh, F.; Sodeifian, G.; Sajadian, S.A. An investigation into Sunitinib malate nanoparticle production by US- RESOLV method: Effect of type of polymer on dissolution rate and particle size distribution. J. Supercrit Fluids 2021, 170, 105163. [Google Scholar] [CrossRef]
- Varma, R.; Vasudevan, S. Extraction, Characterization, and Antimicrobial Activity of Chitosan from Horse Mussel Modiolus modiolus. ACS Omega 2020, 5, 20224–20230. [Google Scholar] [CrossRef]
- Saoji, S.D.; Raut, N.A.; Dhore, P.W.; Borkar, C.D.; Popielarczyk, M.; Dave, V.S. Preparation and Evaluation of Phospholipid-Based Complex of Standardized Centella Extract (SCE) for the Enhanced Delivery of Phytoconstituents. AAPS J. 2016, 18, 102–114. [Google Scholar] [CrossRef]
- Selic, L. New Crystal Form of Sunitinib Malate. European Patent Number EP2362873B1, 3 June 2015. [Google Scholar]
- Arora, S.; Saharan, R.; Kaur, H.; Kaur, I.; Bubber, P.; Bharadwaj, L.M. Attachment of Docetaxel to Multiwalled Carbon Nanotubes for Drug Delivery Applications. Adv. Sci. Lett. 2012, 17, 70–75. [Google Scholar] [CrossRef]
- Korashy, H.M.; Maayah, Z.H.; Al Anazi, F.E.; Alsaad, A.M.; Alanazi, I.O.; Belali, O.M.; Al-Atawi, F.O.; Alshamsan, A. Sunitinib Inhibits Breast Cancer Cell Proliferation by Inducing Apoptosis, Cell-cycle Arrest and DNA Repair While Inhibiting NF-κB Signaling Pathways. Anticancer Res. 2017, 37, 4899–4909. [Google Scholar] [CrossRef]
- Maayah, Z.H.; El Gendy, M.A.; El-Kadi, A.O.; Korashy, H.M. Sunitinib, a tyrosine kinase inhibitor, induces cytochrome P450 1A1 gene in human breast cancer MCF7 cells through ligand-independent aryl hydrocarbon receptor activation. Arch. Toxicol. 2013, 87, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Ullah, I.; Khalil, A.T.; Ali, M.; Iqbal, J.; Ali, W.; Alarifi, S.; Shinwari, Z.K. Green-Synthesized Silver Nanoparticles Induced Apoptotic Cell Death in MCF-7 Breast Cancer Cells by Generating Reactive Oxygen Species and Activating Caspase 3 and 9 Enzyme Activities. Oxid. Med. Cell. Longev. 2020, 2020, 1215395. [Google Scholar] [CrossRef] [PubMed]
- McIlwain, D.R.; Berger, T.; Mak, T.W. Caspase functions in cell death and disease. Cold Spring Harb. Perspect. Biol. 2013, 5, a008656. [Google Scholar] [CrossRef]
- Boatright, K.M.; Renatus, M.; Scott, F.L.; Sperandio, S.; Shin, H.; Pedersen, I.M.; Ricci, J.E.; Edris, W.A.; Sutherlin, D.P.; Green, D.R.; et al. A unified model for apical caspase activation. Mol. Cell. 2003, 11, 529–541. [Google Scholar] [CrossRef]
- Riedl, S.J.; Shi, Y. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 2004, 5, 897–907. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).