In Silico Study: Combination of α-Mangostin and Chitosan Conjugated with Trastuzumab against Human Epidermal Growth Factor Receptor 2
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
- Program ChemOffice 2012 (PerkinElmer Informatics, Waltham, MA, USA) contains the program ChemDraw 12.0 and Chem3D 12.0 was used to draw a 2D structure and 3D structure of the molecule [25];
- The PACKMOL-Memgen (IQ-UNICAMP, University of Campinas, Campinas, SP, Brazil) program was used to package α-mangostin, sodium tripolyphosphate, and chitosan compounds into combined particle form [28];
- The SWISS-MODEL program (Swiss Institute of Bioinformatics Biozentrum, Klingelbergstrase, Basel, Switzerland), which was accessed online from 2nd–20th February 2021 via https://swissmodel.expasy.org/ (accessed on 20 April 2022), was used in the modeling and validation of trastuzumab structure [29,30];
- The PatchDock program (Sackler Institute of Molecular Medicine, Tel Aviv University, Tel Aviv, Israel) was accessed online on 2nd–20th February 2021 at https://bioinfo3d.cs.tau.ac.il/PatchDock/ (accessed on 12 March 2022) was used for the trastuzumab conjugation process and molecular docking simulation [31,32];
2.2. Methods
2.2.1. Structure Preparation of α-Mangostin, Sodium Tripolyphosphate, and Chitosan
2.2.2. Packaging of α-Mangostin, Sodium Tripolyphosphate, and Chitosan into Combined Particles
NA = 6.02 × 1023
2.2.3. Structure Modeling and Validation of Trastuzumab
2.2.4. HER2 Preparation as Receptor
2.2.5. Particle Conjugation of α-Mangostin and Chitosan Combination against Trastuzumab
2.2.6. Molecular Docking Simulation of Particles of a Combination of α-Mangostin and Chitosan Conjugated with Trastuzumab against HER2
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Breast Cancer. Available online: https://www.who.int/cancer/prevention/diagnosis-screening/breast-cancer/en/ (accessed on 19 December 2020).
- Moo, T.A.; Sanford, R.; Dang, C.; Morrow, M. Overview of Breast Cancer Therapy. PET Clin. 2018, 13, 339–354. [Google Scholar] [CrossRef] [PubMed]
- WHO. Guideline for Management of Breast Cancer; WHO: Geneva, Switzerland, 2006; pp. 16–32. [Google Scholar]
- Schmidt, M. Chemotherapy in early breast cancer: When, how and which one? Breast Care 2014, 9, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, D.; Yuan, S. Tyrosine Kinase Inhibitors in the Combination Therapy of HER2 Positive Breast Cancer. Technol. Cancer Res. Treat. 2020, 19, 1533033820962140. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S. Late effects of cancer treatment in breast cancer survivors. South Asian J. Cancer 2014, 3, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Moiseenko, F.; Volkov, N.; Bogdanov, A.; Dubina, M.; Moiseyenko, F. Resistance mechanisms to drug therapy in breast cancer and other solid tumors: An opinion. F1000 Res. 2017, 6, 288. [Google Scholar] [CrossRef]
- Lee, H.N.; Jang, H.Y.; Kim, H.J.; Shin, S.A.; Choo, G.S.; Park, Y.S.; Kim, S.K.; Jung, J.Y. Antitumor and apoptosis-inducing effects of α-mangostin extracted from the pericarp of the mangosteen fruit (Garcinia mangostana L.) in YD-15 tongue mucoepidermoid carcinoma cells. Int. J. Mol. Med. 2016, 37, 939–948. [Google Scholar] [CrossRef]
- Akao, Y.; Nakagawa, Y.; Iinuma, M.; Nozawa, Y. Anti-cancer effects of xanthones from pericarps of mangosteen. Int. J. Mol. Sci. 2008, 9, 355–370. [Google Scholar] [CrossRef]
- Zhang, K.J.; Gu, Q.L.; Yang, K.; Ming, X.J.; Wang, J.X. Anticarcinogenic Effects of α-Mangostin: A Review. Planta Med. 2017, 83, 188–202. [Google Scholar] [CrossRef]
- Li, P.; Tian, W.; Ma, X. Alpha-mangostin inhibits intracellular fatty acid synthase and induces apoptosis in breast cancer cells. Mol. Cancer 2014, 13, 138. [Google Scholar] [CrossRef]
- Kritsanawong, S.; Innajak, S.; Imoto, M.; Watanapokasin, R. Antiproliferative and apoptosis induction of α-mangostin in T47D breast cancer cells. Int. J. Oncol. 2016, 48, 2155–2165. [Google Scholar] [CrossRef]
- Verma, R.K.; Yu, W.; Shrivastava, A.; Shankar, S.; Srivasta, R.K. α-Mangostin-encapsulated PLGA nanoparticles inhibit pancreatic carcinogenesis by targeting cancer stem cells in human, and transgenic (Kras(G12D), and Kras(G12D)/tp53R270H) mice. Sci. Rep. 2016, 6, 32743. [Google Scholar] [CrossRef] [PubMed]
- Bumrung, J.; Chanchao, C.; Intasanta, V.; Palaga, T.; Wanichwecharungruang, S. Water-dispersible unadulterated α-mangostin particles for biomedical applications. R. Soc. Open Sci. 2020, 7, 200543. [Google Scholar] [CrossRef] [PubMed]
- Gunasekaran, T.; Haile, T.; Nigusse, T.; Dhanaraju, M.D. Nanotechnology: An effective tool for enhancing bioavailability and bioactivity of phytomedicine. Asian Pac. J. Trop. Biomed. 2014, 4, S1–S7. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.A.; Syeda, J.T.M.; Wasan, K.M.; Wasan, E.K. An Overview of Chitosan Nanoparticles and Its Application in Non-Parenteral Drug Delivery. Pharmaceuticals 2017, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Ballesta, M.; Gil-Izquierdo, Á.; García-Viguera, C.; Domínguez-Perles, R. Nanoparticles and Controlled Delivery for Bioactive Compounds: Outlining Challenges for New “Smart-Foods” for Health. Foods 2018, 7, 72. [Google Scholar] [CrossRef] [PubMed]
- Goktas, Z.; Zu, Y.; Abbasi, M.; Galyean, S.; Wu, D.; Fan, Z.; Wang, S. Recent Advances in Nanoencapsulation of Phytochemicals to Combat Obesity and Its Comorbidities. J. Agric. Food Chem. 2020, 68, 8119–8131. [Google Scholar] [CrossRef]
- Wathoni, N.; Rusdin, A.; Febriani, E.; Purnama, D.; Daulay, W.; Azhary, S.Y.; Panatarani, C.; Joni, I.M.; Lesmana, R.; Motoyama, K.; et al. Formulation and characterization of α-mangostin in chitosan nanoparticles coated by sodium alginate, sodium silicate, and polyethylene glycol. J. Pharm. Bioallied Sci. 2019, 11, S619–S627. [Google Scholar] [CrossRef]
- Kim, E.G.; Kim, K.M. Strategies and Advancement in Antibody-Drug Conjugate Optimization for Targeted Cancer Therapeutics. Biomol. Ther. 2015, 23, 493–509. [Google Scholar] [CrossRef]
- Spitzer, D.; Simon, P.O., Jr.; Kashiwagi, H.; Xu, J.; Zeng, C.; Vangveravong, S.; Zhou, D.; Chang, K.; McDunn, J.E.; Hornick, J.R.; et al. Use of Multifunctional Sigma-2 Receptor Ligand Conjugates to Trigger Cancer-Selective Cell Death Signaling. Cancer Res. 2012, 72, 201–209. [Google Scholar] [CrossRef]
- Thomas, A.; Teicher, B.A.; Hassan, R. Antibody-drug conjugates for cancer therapy. Lancet Oncol. 2016, 17, e254–e262. [Google Scholar] [CrossRef]
- You, Y.; Xu, Z.; Chen, Y. Doxorubicin conjugated with a trastuzumab epitope and an MMP-2 sensitive peptide linker for the treatment of HER2-positive breast cancer. Drug Deliv. 2018, 25, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Badkas, A.; Stevenson, M.; Lee, J.; Leung, Y. Herceptin conjugated PLGA-PHis-PEG pH sensitive nanoparticles for targeted and controlled drug delivery. Int. J. Pharm. 2015, 487, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Helgren, T.R.; Hagen, T.J. Demonstration of AutoDock as an Educational Tool for Drug Discovery. J. Chem. Educ. 2017, 94, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Torres, P.H.M.; Sodero, A.C.R.; Jofily, P.; Silva-Jr, F.P. Key Topics in Molecular Docking for Drug Design. Int. J. Mol. Sci. 2019, 20, 4574. [Google Scholar] [CrossRef]
- Kemmish, H.; Fasnacht, M.; Yan, L. Fully automated antibody structure prediction using BIOVIA tools: Validation study. PLoS ONE 2017, 12, e0177923. [Google Scholar] [CrossRef] [PubMed]
- Martínez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. PACKMOL: A package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucl. Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Stude, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, 363–367. [Google Scholar] [CrossRef]
- Pagadala, N.S.; Syed, K.; Tuszynski, J. Software for molecular docking: A review. Biophys. Rev. 2017, 9, 91–102. [Google Scholar] [CrossRef]
- Atkins, P.W. Physical Chemistry, 5th ed.; Oxford University Press: Oxford, UK, 1994. [Google Scholar]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo—Distance constraints applied on model quality estimation. Struct. Bioinform. 2020, 36, 2647. [Google Scholar] [CrossRef]
- Pantsar, T.; Poso, A. Binding Affinity via Docking: Fact and Fiction. Molecules 2018, 23, 1899. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, J.; Ríos-Vera, C.; Melo, F.; Schüller, A. Calculation of accurate interatomic contact surface areas for the quantitative analysis of non-bonded molecular interactions. Bioinformatics 2019, 35, 3499–3501. [Google Scholar] [CrossRef] [PubMed]
- Vanommeslaeghe, K.; Guvench, O.; MacKerell, A.D., Jr. Robustness in the fitting of molecular mechanics parameters. Curr. Pharm. Des. 2015, 20, 3281–3292. [Google Scholar] [CrossRef] [PubMed]
- Lim, V.T.; Hahn, D.F.; Tresadern, G.; Bayly, C.I.; Mobley, D.L. Benchmark assessment of molecular geometries and energies from small molecule force fields. F1000Research 2020, 9, 1390. [Google Scholar] [CrossRef]
- Chaudhari, M.I.; Holleran, S.A.; Ashbaugh, H.S.; Pratt, L.R. Molecular-scale hydrophobic interactions between hard-sphere reference solutes are attractive and endothermic. Proc. Natl. Acad. Sci. USA 2013, 110, 20557–20562. [Google Scholar] [CrossRef]
- Yousefpour, P.; Atyabi, F.; Vasheghani-Farahani, E.; Mousavi, A.; Dinarvand, R. Targeted delivery of doxorubicin-utilizing chitosan nanoparticles surface-functionalized with anti-Her2 trastuzumab. Int. J. Nanomed. 2011, 6, 1977–1990. [Google Scholar] [CrossRef][Green Version]
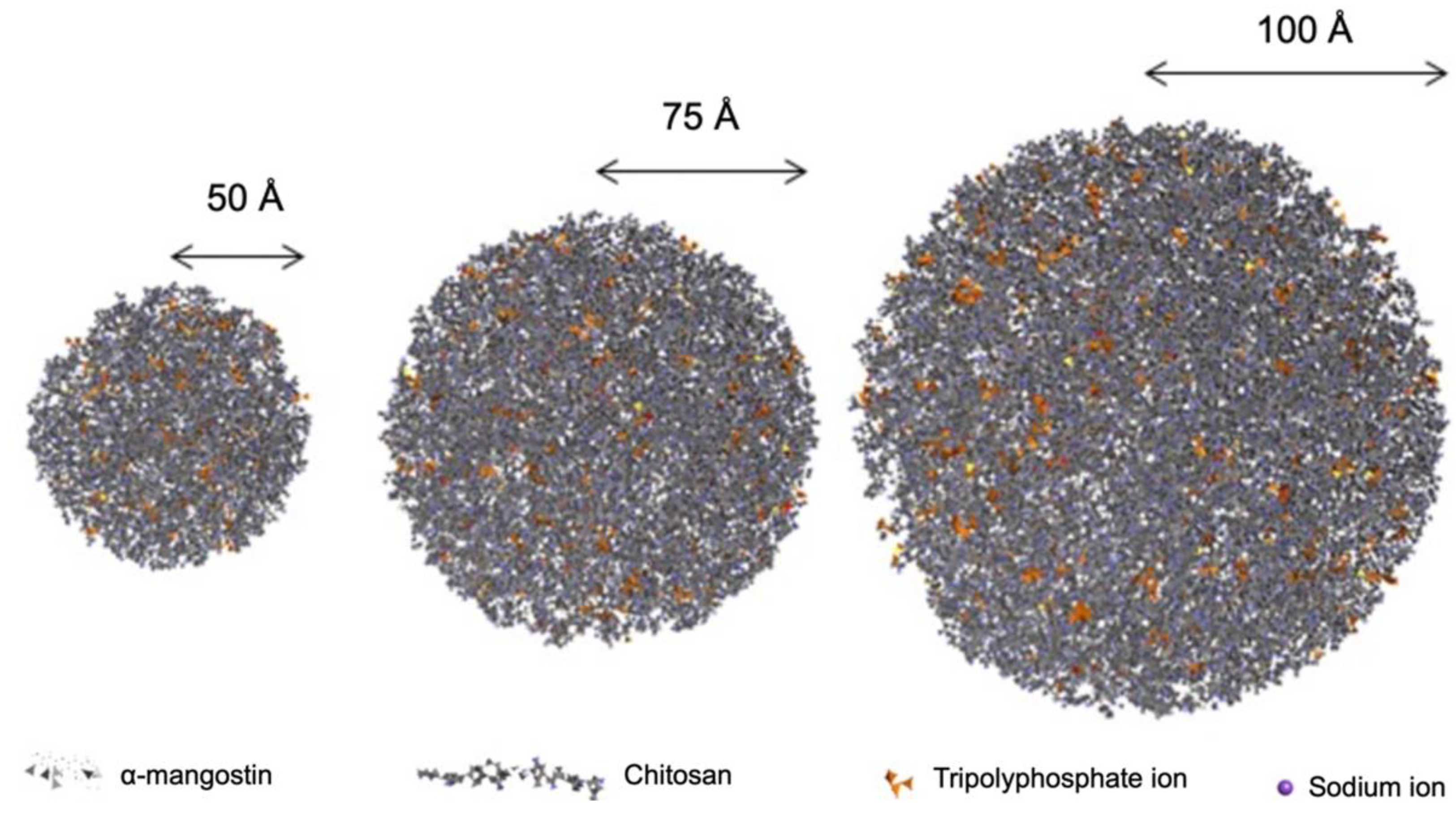


| Structure Radius | Number of Molecules | |||
|---|---|---|---|---|
| α-Mangostin | Chitosan | Sodium Ions | Tripolyphosphate Ions | |
| 50 Å | 60 | 161 | 469 | 94 |
| 75 Å | 203 | 545 | 1.582 | 316 |
| 100 Å | 480 | 1.291 | 3.749 | 750 |
| Compound | Chemical Formula | 3D Structure |
|---|---|---|
| α-mangostin | C24H26O6 |  |
| Chitosan-ionated | C56H112N9O399+ |  |
| Sodium ion | Na+ |  |
| Tripolyphosphate ion | P3O105− |  |
| Model | %Similarity | GMQE | QMEAN |Z-Score| |
|---|---|---|---|
| Trastuzumab | 82.60% | 0.85 | 0.05 |
| Compound | Docking Score | Hydrogen Bond Interactions |
|---|---|---|
| Combined particle (50 Å) | 26,606 | B:Glu275, B:Trp280, B:Lys291, B:Lys293, B:Tyr303, B:Val306, C:Gln147, C:Asn152, C:Thr197, C:Ser202, C:Ser203, D:Arg295, D:Tyr303 |
| Combined particle (75 Å) | 30,532 | B:Leu254, B:Ser257, B:His288, B:Ala290, B:Thr292, B:Pro294, B:Glu297, B:Gln298, B:Tyr303, B:Arg304, C:Asp151, C:His198, C:Leu201, C:Ser203, C:Pro204, D:Glu261, D:Thr259, D:Glu296, D:Thr310, D:Glu383, D:Ser386, D:Asn387, D:Gln389, D:Arg419 |
| Combined particle (100 Å) | 30,994 | C:Ser12, C:Thr109, C:Val110, C:Tyr140, C:Lys149, C:Glu195, C:Gln199, C:Ser202, C:Ser203, C:Thr206, D:Thr259, D:His288, D:Glu296, D:Thr310, D:Gln389, D:Pro390, D:Tyr439, D:Gln441 |
| Compound | Docking Score | Area | ACE (kcal/mol) |
|---|---|---|---|
| α-mangostin | 5416 | 683.30 | −420.39 |
| Chitosan | 10,378 | 1504.70 | −643.54 |
| Tripolyphosphate ion | 2688 | 309.20 | 71.12 |
| Trastuzumab | 21,440 | 3853.40 | 196.60 |
| Combined particle (50 Å) | 23,044 | 3663.40 | −875.02 |
| Combined particle (50 Å) conjugated with trastuzumab | 26,872 | 4146.30 | −582.33 |
| Combined particle (75 Å) | 27,858 | 4008.60 | −766.09 |
| Combined particle (75 Å) conjugated with trastuzumab | 26,520 | 4044.80 | −580.04 |
| Combined particle (100 Å) | 29,884 | 4389.40 | −279.12 |
| Combined particle (100 Å) conjugated with trastuzumab | 29,996 | 4643.70 | −424.13 |
| Compound | Hydrogen Bond Interactions |
|---|---|
| α-mangostin | - |
| Chitosan | Val3, Thr5, Tyr281, Tyr387, Leu414, Ser441 |
| Tripolyphosphate ion | Thr5 |
| Trastuzumab | Asn89, Glu216, Tyr252, Cys289, Glu330 |
| Combined particle (50 Å) | Thr83, Asp88, Asn89, Asn155, Leu157, Gln217, Val250, Tyr252, Thr256, Asp285, His296, Asn297, Lys311, Cys312, Ser313, Lys314, Arg318, Glu326, His327, Glu330 |
| Combined particle (50 Å) conjugated with trastuzumab | Gln2, Leu249, Tyr267, Thr275, Ala276, Thr306, Asp461, Arg465, Arg495, Val507, Cys509, Gln511, Val533, Asn534, Arg536, Gly550, Phe555, Asp560, Glu597 |
| Combined particle (75 Å) | Lys153, Gln156, Leu157, Ala158, Cys202, Gln217, Leu244, Tyr252, Asp255, Thr256, Val292, Cys293, Cys316, Ala317, Val319, Tyr321, His327 |
| Combined particle (75 Å) conjugated with trastuzumab | Thr223, Ser239, Leu249, Met260, Pro263, Arg266, Ser272, Thr275, Tyr279, Asn508, Cys509, Ser510, Gln511, Asn534, Ala535 |
| Combined particle (100 Å) | Thr1, Asp22, Gln29, Gln32, Asn46, Ser50, Gln53, Arg76, Cys190, Thr223, Asp229, Thr275, Asp461, Gln462, Phe464, Leu471, His473, Gln491 |
| Combined particle (100 Å) conjugated with trastuzumab | Thr1, Gln29, Gln32, Asn46, Ser50, Gln53, Arg76, Glu188, Cys190, Ser192, Thr223, Asp229, Cys230, Asn237, Thr275, Asp461, Phe464, Leu471, His473, Glu479, Gln491, Arg495, Ser510 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Megantara, S.; Wathoni, N.; Mohammed, A.F.A.; Suhandi, C.; Ishmatullah, M.H.; Putri, M.F.F.D. In Silico Study: Combination of α-Mangostin and Chitosan Conjugated with Trastuzumab against Human Epidermal Growth Factor Receptor 2. Polymers 2022, 14, 2747. https://doi.org/10.3390/polym14132747
Megantara S, Wathoni N, Mohammed AFA, Suhandi C, Ishmatullah MH, Putri MFFD. In Silico Study: Combination of α-Mangostin and Chitosan Conjugated with Trastuzumab against Human Epidermal Growth Factor Receptor 2. Polymers. 2022; 14(13):2747. https://doi.org/10.3390/polym14132747
Chicago/Turabian StyleMegantara, Sandra, Nasrul Wathoni, Ahmed Fouad Abdelwahab Mohammed, Cecep Suhandi, Maryam H. Ishmatullah, and Melisa F. F. D. Putri. 2022. "In Silico Study: Combination of α-Mangostin and Chitosan Conjugated with Trastuzumab against Human Epidermal Growth Factor Receptor 2" Polymers 14, no. 13: 2747. https://doi.org/10.3390/polym14132747
APA StyleMegantara, S., Wathoni, N., Mohammed, A. F. A., Suhandi, C., Ishmatullah, M. H., & Putri, M. F. F. D. (2022). In Silico Study: Combination of α-Mangostin and Chitosan Conjugated with Trastuzumab against Human Epidermal Growth Factor Receptor 2. Polymers, 14(13), 2747. https://doi.org/10.3390/polym14132747







