Polyelectrolytes for Enzyme Immobilization and the Regulation of Their Properties
Abstract
1. Introduction
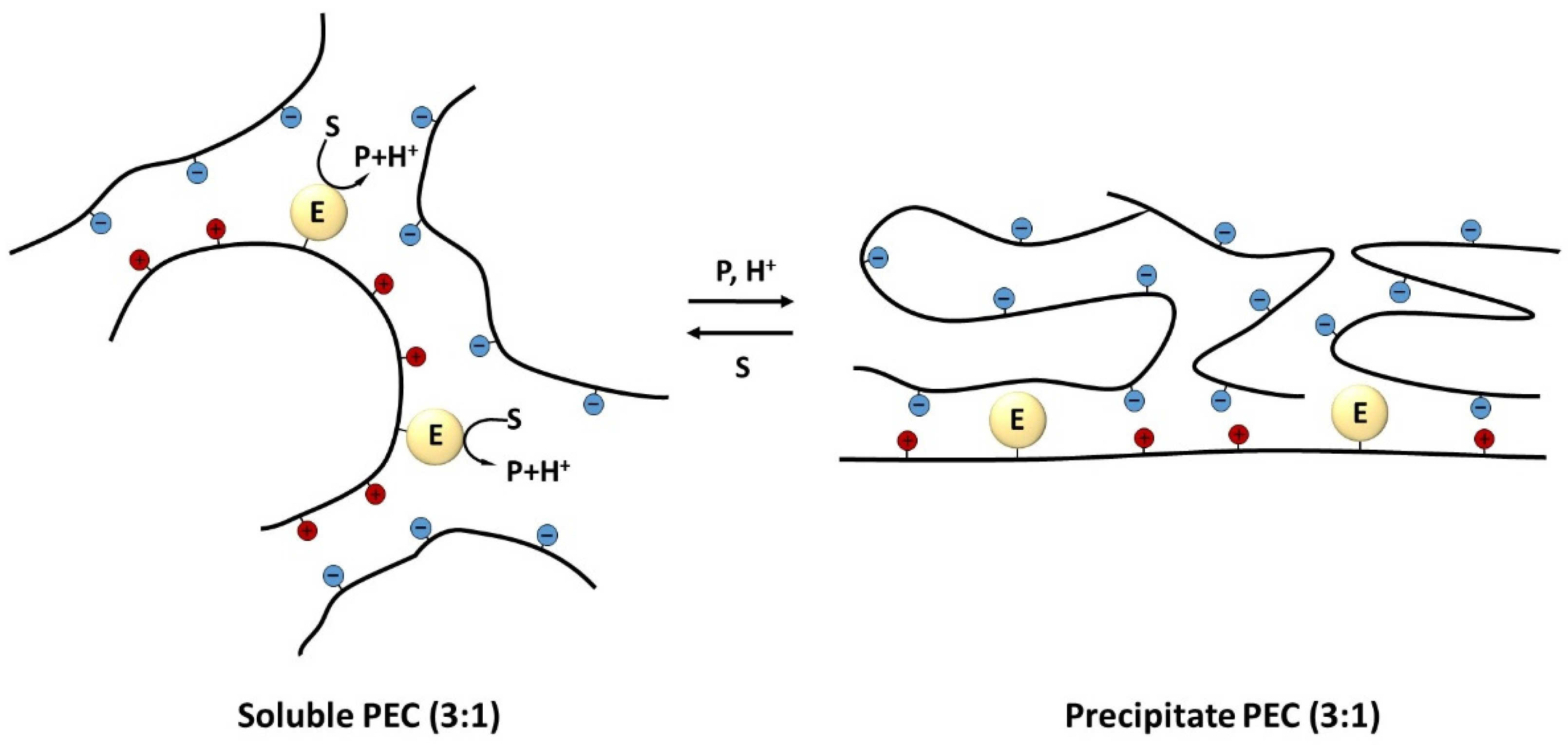
2. Polyelectrolyte Complexes with Immobilized Enzymes for Obtaining Self-Regulating Biocatalysts
3. Immobilization of Enzyme via Polymers
4. Effect of Immobilization on Enzyme Activity
5. Application of Polyelectrolytes and Polyelectrolyte Complexes as Artificial Chaperones
6. Modeling of the Complexation
7. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cooper, C.L.; Dubin, P.L.; Kayitmazer, A.B.; Turksen, S. Polyelectrolyte–Protein Complexes. Curr. Opin. Colloid Interface Sci. 2005, 10, 52–78. [Google Scholar] [CrossRef]
- Laschewsky, A. Recent Trends in the Synthesis of Polyelectrolytes. Curr. Opin. Colloid Interface Sci. 2012, 17, 56–63. [Google Scholar] [CrossRef]
- Kayitmazer, A.B.; Seeman, D.; Minsky, B.B.; Dubin, P.L.; Xu, Y. Protein-Polyelectrolyte Interactions. Soft Matter 2013, 9, 2553–2583. [Google Scholar] [CrossRef]
- Becker, A.L.; Henzler, K.; Welsch, N.; Ballauff, M.; Borisov, O. Proteins and Polyelectrolytes: A Charged Relationship. Curr. Opin. Colloid Interface Sci. 2012, 17, 90–96. [Google Scholar] [CrossRef]
- Shchukin, D.G.; Sukhorukov, G.B. Nanoparticle Synthesis in Engineered Organic Nanoscale Reactors. Adv. Mater. 2004, 16, 671–682. [Google Scholar] [CrossRef]
- Stuart, M.A.C.; Huck, W.T.S.; Genzer, J.; Müller, M.; Ober, C.; Stamm, M.; Sukhorukov, G.B.; Szleifer, I.; Tsukruk, V.V.; Urban, M.; et al. Emerging Applications of Stimuli-Responsive Polymer Materials. Nat. Mater. 2010, 9, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Förster, S.; Abetz, V.; Müller, A.H.E. Polyelectrolyte Block Copolymer Micelles. In Polyelectrolytes with Defined Molecular Architecture II; Schmidt, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2004; Volume 166, pp. 173–210. ISBN 978-3-540-00556-8. [Google Scholar]
- Kabanov, V.A. Polyelectrolyte Complexes in Solution and in Bulk. Russ. Chem. Rev. 2005, 74, 3–20. [Google Scholar] [CrossRef]
- Chen, F.; Stenzel, M.H. Polyion Complex Micelles for Protein Delivery. Aust. J. Chem. 2018, 71, 768–780. [Google Scholar] [CrossRef]
- Izumrudov, V.A.; Kasaikin, V.A.; Yermakova, L.N.; Zezin, A.B. Study of Water Soluble Polyelectrolyte Complexes of Nonequimolar Composition. Polym. Sci. USSR 1978, 20, 452–460. [Google Scholar] [CrossRef]
- Kharenko, O.A.; Kharenko, A.V.; Kalyuzhnaya, R.I.; Izumrudov, V.A.; Kasaikin, V.A.; Zezin, A.B.; Kabanov, V.A. Non-Stoichiometric Polyelectrolyte Complexes: New, Water Soluble Macromolecular Compounds. Polym. Sci. USSR 1979, 21, 3002–3008. [Google Scholar] [CrossRef]
- Kharenko, O.A.; Kharenko, A.V.; Kasaikin, V.A.; Zeizin, A.B.; Kabanov, V.A. The Structure of Non-Stoichiometric, Water Soluble Polyelectrolyte Complexes. Polym. Sci. USSR 1979, 21, 3009–3017. [Google Scholar] [CrossRef]
- Margolin, A.L.; Izumrudov, V.A.; Švedas, V.K.; Zezin, A.B.; Kabanov, V.A.; Berezin, I.V. Preparation and Properties of Penicillin Amidase Immobilized in Polyelectrolyte Complexes. Biochim. Biophys. Acta BBA Enzymol. 1981, 660, 359–365. [Google Scholar] [CrossRef]
- Margolin, A.L.; Izumrudov, V.A.; Švedas, V.K.; Zezin, A.B. Soluble-Insoluble Immobilized Enzymes. Biotechnol. Bioeng. 1982, 24, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Margolin, A.L.; Izumrudov, V.A.; Sherstiuk, S.F.; Zezin, A.B.; Shviadas, V.K. Enzymes incorporated in polyelectrolyte complexes. Effect of matrix conformational changes and phase transitions in solutions on catalytic properties. Mol. Biol. (Mosk.) 1983, 17, 1001–1008. [Google Scholar]
- Margolin, A.L.; Sherstyuk, S.F.; Izumrudov, V.A.; Zezin, A.B.; Kabanov, V.A. Enzymes in Polyelectrolyte Complexes. The Effect of Phase Transition on Thermal Stability. Eur. J. Biochem. 1985, 146, 625–632. [Google Scholar] [CrossRef]
- Margolin, A.L.; Sherstiuk, S.F.; Izumrudov, V.A.; Shviadas, V.K.; Zezin, A.B. Protein-protein interactions in systems containing synthetic polyelectrolytes. Dokl. Akad. Nauk SSSR 1985, 284, 997–1001. [Google Scholar]
- Galaev, I.Y.; Mattiasson, B. “Smart” Polymers and What They Could Do in Biotechnology and Medicine. Trends Biotechnol. 1999, 17, 335–340. [Google Scholar] [CrossRef]
- Roy, I.; Sharma, S.; Gupta, M.N. Smart Biocatalysts: Design and Applications. Adv. Biochem. Eng. Biotechnol. 2004, 86, 159–189. [Google Scholar] [CrossRef]
- Shakya, A.K.; Nandakumar, K.S. An Update on Smart Biocatalysts for Industrial and Biomedical Applications. J. R. Soc. Interface 2018, 15, 20180062. [Google Scholar] [CrossRef]
- Decher, G. Fuzzy Nanoassemblies: Toward Layered Polymeric Multicomposites. Science 1997, 277, 1232–1237. [Google Scholar] [CrossRef]
- Ariga, K.; Hill, J.P.; Ji, Q. Layer-by-Layer Assembly as a Versatile Bottom-up Nanofabrication Technique for Exploratory Research and Realistic Application. Phys. Chem. Chem. Phys. 2007, 9, 2319–2340. [Google Scholar] [CrossRef] [PubMed]
- Onda, M.; Lvov, Y.; Ariga, K.; Kunitake, T. Sequential Reaction and Product Separation on Molecular Films of Glucoamylase and Glucose Oxidase Assembled on an Ultrafilter. J. Ferment. Bioeng. 1996, 82, 502–506. [Google Scholar] [CrossRef]
- Sigolaeva, L.V.; Dubacheva, G.V.; Porus, M.V.; Eremenko, A.V.; Rudakova, E.V.; Makhaeva, G.F.; Richardson, R.J.; Kurochkin, I.N. A Layer-by-Layer Tyrosinase Biosensor for Assay of Carboxylesterase and Neuropathy Target Esterase Activities in Blood. Anal. Methods 2013, 5, 3872–3879. [Google Scholar] [CrossRef]
- Zhang, Y.; Arugula, M.A.; Williams, S.T.; Minteer, S.D.; Simonian, A.L. Layer-by-Layer Assembly of Carbon Nanotubes Modified with Invertase/Glucose Dehydrogenase Cascade for Sucrose/O2 Biofuel Cell. J. Electrochem. Soc. 2016, 163, F449. [Google Scholar] [CrossRef]
- Shutava, T.G.; Kommireddy, D.S.; Lvov, Y.M. Layer-by-Layer Enzyme/Polyelectrolyte Films as a Functional Protective Barrier in Oxidizing Media. J. Am. Chem. Soc. 2006, 128, 9926–9934. [Google Scholar] [CrossRef]
- Haynie, D.T.; Zhang, L.; Zhao, W.; Rudra, J.S. Protein-Inspired Multilayer Nanofilms: Science, Technology and Medicine. Nanomed. Nanotechnol. Biol. Med. 2006, 2, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zheng, B.; Haynie, D.T. A Molecular Dynamics Study of the Physical Basis of Stability of Polypeptide Multilayer Nanofilms. Langmuir 2006, 22, 6668–6675. [Google Scholar] [CrossRef]
- Jewell, C.M.; Zhang, J.; Fredin, N.J.; Lynn, D.M. Multilayered Polyelectrolyte Films Promote the Direct and Localized Delivery of DNA to Cells. J. Control. Release 2005, 106, 214–223. [Google Scholar] [CrossRef]
- Becker, A.L.; Welsch, N.; Schneider, C.; Ballauff, M. Adsorption of RNase A on Cationic Polyelectrolyte Brushes: A Study by Isothermal Titration Calorimetry. Biomacromolecules 2011, 12, 3936–3944. [Google Scholar] [CrossRef]
- Henzler, K.; Haupt, B.; Rosenfeldt, S.; Harnau, L.; Narayanan, T.; Ballauff, M. Interaction Strength between Proteins and Polyelectrolyte Brushes: A Small Angle X-Ray Scattering Study. Phys. Chem. Chem. Phys. 2011, 13, 17599–17605. [Google Scholar] [CrossRef]
- Ferrand-Drake del Castillo, G.; Koenig, M.; Müller, M.; Eichhorn, K.-J.; Stamm, M.; Uhlmann, P.; Dahlin, A. Enzyme Immobilization in Polyelectrolyte Brushes: High Loading and Enhanced Activity Compared to Monolayers. Langmuir 2019, 35, 3479–3489. [Google Scholar] [CrossRef] [PubMed]
- Rühe, J.; Ballauff, M.; Biesalski, M.; Dziezok, P.; Gröhn, F.; Johannsmann, D.; Houbenov, N.; Hugenberg, N.; Konradi, R.; Minko, S.; et al. Polyelectrolyte Brushes. In Polyelectrolytes with Defined Molecular Architecture I; Schmidt, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 79–150. ISBN 978-3-540-36433-7. [Google Scholar]
- Zhou, Z.; Yu, P.; Geller, H.M.; Ober, C.K. Biomimetic Polymer Brushes Containing Tethered Acetylcholine Analogs for Protein and Hippocampal Neuronal Cell Patterning. Biomacromolecules 2013, 14, 529–537. [Google Scholar] [CrossRef] [PubMed]
- de Kruif, C.G.; Weinbreck, F.; de Vries, R. Complex Coacervation of Proteins and Anionic Polysaccharides. Curr. Opin. Colloid Interface Sci. 2004, 9, 340–349. [Google Scholar] [CrossRef]
- Svenson, S.; Tomalia, D.A. Dendrimers in Biomedical Applications—Reflections on the Field. Adv. Drug Deliv. Rev. 2005, 57, 2106–2129. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Fukushima, S.; Bae, Y.; Hiki, S.; Ishii, T.; Kataoka, K. A Protein Nanocarrier from Charge-Conversion Polymer in Response to Endosomal PH. J. Am. Chem. Soc. 2007, 129, 5362–5363. [Google Scholar] [CrossRef]
- Batrakova, E.V.; Li, S.; Reynolds, A.D.; Mosley, R.L.; Bronich, T.K.; Kabanov, A.V.; Gendelman, H.E. A Macrophage−Nanozyme Delivery System for Parkinson’s Disease. Bioconjug. Chem. 2007, 18, 1498–1506. [Google Scholar] [CrossRef]
- Lindhoud, S.; Voorhaar, L.; de Vries, R.; Schweins, R.; Cohen Stuart, M.A.; Norde, W. Salt-Induced Disintegration of Lysozyme-Containing Polyelectrolyte Complex Micelles. Langmuir 2009, 25, 11425–11430. [Google Scholar] [CrossRef]
- Weber, C.; Drogoz, A.; David, L.; Domard, A.; Charles, M.-H.; Verrier, B.; Delair, T. Polysaccharide-Based Vaccine Delivery Systems: Macromolecular Assembly, Interactions with Antigen Presenting Cells, and in Vivo Immunomonitoring. J. Biomed. Mater. Res. A 2010, 93A, 1322–1334. [Google Scholar] [CrossRef]
- Vermonden, T.; Censi, R.; Hennink, W.E. Hydrogels for Protein Delivery. Chem. Rev. 2012, 112, 2853–2888. [Google Scholar] [CrossRef]
- Lu, Y.; Ballauff, M. Thermosensitive Core–Shell Microgels: From Colloidal Model Systems to Nanoreactors. Prog. Polym. Sci. 2011, 36, 767–792. [Google Scholar] [CrossRef]
- Wu, Q.; Su, T.; Mao, Y.; Wang, Q. Thermal Responsive Microgels as Recyclable Carriers to Immobilize Active Proteins with Enhanced Nonaqueous Biocatalytic Performance. Chem. Commun. 2013, 49, 11299–11301. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Liu, J.; Xin, Y.; Zhan, M.; Xiao, J.; Lu, L.; Peng, S. Temperature and Salt Responsive Zwitterionic Polysulfamide-Based Nanogels with Surface Regeneration Ability and Controlled Drug Release. Polym. Chem. 2019, 10, 6423–6431. [Google Scholar] [CrossRef]
- Sigolaeva, L.V.; Pergushov, D.V.; Gladyr, S.Y.; Kurochkin, I.N.; Richtering, W. Microgels in Tandem with Enzymes: Tuning Adsorption of a PH- and Thermoresponsive Microgel for Improved Design of Enzymatic Biosensors. Adv. Mater. Interfaces 2022, 9, 2200310. [Google Scholar] [CrossRef]
- Yaroslavov, A.A.; Kaplan, I.B.; Erokhina, T.N.; Morozov, S.Y.; Solovyev, A.G.; Leshchiner, A.D.; Rakhnyanskaya, A.A.; Malinin, A.S.; Stepanova, L.A.; Kiselev, O.I.; et al. A New Method for Producing Biologically Active Nanocomplexes by a Noncovalent Conjugation of Proteins with Viral Particles. Russ. J. Bioorganic Chem. 2011, 37, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Sedlák, E.; Fedunová, D.; Veselá, V.; Sedláková, D.; Antalík, M. Polyanion Hydrophobicity and Protein Basicity Affect Protein Stability in Protein-Polyanion Complexes. Biomacromolecules 2009, 10, 2533–2538. [Google Scholar] [CrossRef]
- Stogov, S.V.; Izumrudov, V.A.; Muronetz, V.I. Structural Changes of a Protein Bound to a Polyelectrolyte Depend on the Hydrophobicity and Polymerization Degree of the Polyelectrolyte. Biochem. (Mosc.) 2010, 75, 437–442. [Google Scholar] [CrossRef]
- Semenyuk, P.I.; Muronetz, V.I.; Haertlé, T.; Izumrudov, V.A. Effect of Poly(Phosphate) Anions on Glyceraldehyde-3-Phosphate Dehydrogenase Structure and Thermal Aggregation: Comparison with Influence of Poly(Sulfoanions). Biochim. Biophys. Acta BBA Gen. Subj. 2013, 1830, 4800–4805. [Google Scholar] [CrossRef]
- Shiraki, K.; Kurinomaru, T.; Tomita, S. Wrap-and-Strip Technology of Protein-Polyelectrolyte Complex for Biomedical Application. Curr. Med. Chem. 2016, 23, 276–289. [Google Scholar] [CrossRef]
- Semenyuk, P.I.; Kurochkina, L.P.; Mäkinen, L.; Muronetz, V.I.; Hietala, S. Thermocontrolled Reversible Enzyme Complexation-Inactivation-Protection by Poly(N-Acryloyl Glycinamide). Polymers 2021, 13, 3601. [Google Scholar] [CrossRef]
- Kurinomaru, T.; Tomita, S.; Hagihara, Y.; Shiraki, K. Enzyme Hyperactivation System Based on a Complementary Charged Pair of Polyelectrolytes and Substrates. Langmuir 2014, 30, 3826–3831. [Google Scholar] [CrossRef]
- Thiele, M.J.; Davari, M.D.; König, M.; Hofmann, I.; Junker, N.O.; Mirzaei Garakani, T.; Vojcic, L.; Fitter, J.; Schwaneberg, U. Enzyme–Polyelectrolyte Complexes Boost the Catalytic Performance of Enzymes. ACS Catal. 2018, 8, 10876–10887. [Google Scholar] [CrossRef]
- Manukhov, I.; Kotova, V.; Zavil’genskiĭ, G. Role of GroEL/GroES chaperonin system and Lon protease in regulation of expression Vibrio fischeri lux genes in Escherichia coli cells. Mol. Biol. (Mosk.) 2006, 40, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Shalova, I.N.; Naletova, I.N.; Saso, L.; Muronetz, V.I.; Izumrudov, V.A. Interaction of Polyelectrolytes with Proteins, 3. Influence of Complexing Polycations on the Thermoaggregation of Oligomeric Enzymes. Macromol. Biosci. 2007, 7, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.; Ma, D.; Herbet, A.; Boquet, D.; Winnik, F.M.; Tribet, C. Prevention of Thermally Induced Aggregation of IgG Antibodies by Noncovalent Interaction with Poly(Acrylate) Derivatives. Biomacromolecules 2014, 15, 2952–2962. [Google Scholar] [CrossRef] [PubMed]
- Ivinova, O.N.; Izumrudov, V.A.; Muronetz, V.I.; Galaev, I.Y.; Mattiasson, B. Influence of Complexing Polyanions on the Thermostability of Basic Proteins. Macromol. Biosci. 2003, 3, 210–215. [Google Scholar] [CrossRef]
- Delcroix, M.F.; Huet, G.L.; Conard, T.; Demoustier-Champagne, S.; Du Prez, F.E.; Landoulsi, J.; Dupont-Gillain, C.C. Design of Mixed PEO/PAA Brushes with Switchable Properties Toward Protein Adsorption. Biomacromolecules 2013, 14, 215–225. [Google Scholar] [CrossRef]
- Wang, S.; Chen, K.; Li, L.; Guo, X. Binding between Proteins and Cationic Spherical Polyelectrolyte Brushes: Effect of PH, Ionic Strength, and Stoichiometry. Biomacromolecules 2013, 14, 818–827. [Google Scholar] [CrossRef]
- Kamiya, N.; Klibanov, A.M. Controling the Rate of Protein Release from Polyelectrolyte Complexes. Biotechnol. Bioeng. 2003, 82, 590–594. [Google Scholar] [CrossRef]
- Shalova, I.N.; Asryants, R.A.; Sholukh, M.V.; Saso, L.; Kurganov, B.I.; Muronetz, V.I.; Izumrudov, V.A. Interaction of Polyanions with Basic Proteins, 2(a): Influence of Complexing Polyanions on the Thermo-Aggregation of Oligomeric Enzymes. Macromolar Biosci. 2005, 5, 1184–1192. [Google Scholar] [CrossRef]
- Ganguli, S.; Yoshimoto, K.; Tomita, S.; Sakuma, H.; Matsuoka, T.; Shiraki, K.; Nagasaki, Y. Regulation of Lysozyme Activity Based on Thermotolerant Protein/Smart Polymer Complex Formation. J. Am. Chem. Soc. 2009, 131, 6549–6553. [Google Scholar] [CrossRef]
- Evstafyeva, D.B.; Izumrudov, V.A.; Muronetz, V.I.; Semenyuk, P.I. Tightly Bound Polyelectrolytes Enhance Enzyme Proteolysis and Destroy Amyloid Aggregates. Soft Matter 2018, 14, 3768–3773. [Google Scholar] [CrossRef] [PubMed]
- De, M.; Rotello, V.M. Synthetic “Chaperones”: Nanoparticle-Mediated Refolding of Thermally Denatured Proteins. Chem. Commun. 2008, 30, 3504–3506. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Mazzawi, M.; Chen, K.; Sun, L.; Dubin, P.L. Protein Purification by Polyelectrolyte Coacervation: Influence of Protein Charge Anisotropy on Selectivity. Biomacromolecules 2011, 12, 1512–1522. [Google Scholar] [CrossRef] [PubMed]
- Shimada, N.; Nakayama, M.; Kano, A.; Maruyama, A. Design of UCST Polymers for Chilling Capture of Proteins. Biomacromolecules 2013, 14, 1452–1457. [Google Scholar] [CrossRef]
- Hagemann, A.; Giussi, J.M.; Longo, G.S. Use of PH Gradients in Responsive Polymer Hydrogels for the Separation and Localization of Proteins from Binary Mixtures. Macromolecules 2018, 51, 8205–8216. [Google Scholar] [CrossRef]
- Lu, D.; Liu, Z.; Zhang, M.; Wang, X.; Liu, Z. Dextran-Grafted-PNIPAAm as an Artificial Chaperone for Protein Refolding. Biochem. Eng. J. 2006, 27, 336–343. [Google Scholar] [CrossRef]
- Semenyuk, P.; Tiainen, T.; Hietala, S.; Tenhu, H.; Aseyev, V.; Muronetz, V. Artificial Chaperones Based on Thermoresponsive Polymers Recognize the Unfolded State of the Protein. Int. J. Biol. Macromol. 2019, 121, 536–545. [Google Scholar] [CrossRef]
- Muronetz, V.I.; Korpela, T. Isolation of Antigens and Antibodies by Affinity Chromatography. J. Chromatogr. B 2003, 790, 53–66. [Google Scholar] [CrossRef]
- Grigorieva, J.A.; Dainiak, M.B.; Katrukha, A.G.; Muronetz, V.I. Antibodies to the Nonnative Forms of D-Glyceraldehyde-3-Phosphate Dehydrogenase: Identification, Purification, and Influence on the Renaturation of the Enzyme. Arch. Biochem. Biophys. 1999, 369, 252–260. [Google Scholar] [CrossRef]
- Kuravsky, M.L.; Schmalhausen, E.V.; Pozdnyakova, N.V.; Muronetz, V.I. Isolation of Antibodies against Different Protein Conformations Using Immunoaffinity Chromatography. Anal. Biochem. 2012, 426, 47–53. [Google Scholar] [CrossRef]
- Dainiak, M.B.; Izumrudov, V.A.; Muronetz, V.I.; Galaev, I.Y.; Mattiasson, B. Conjugates of Monoclonal Antibodies with Polyelectrolyte Complexes—An Attempt to Make an Artificial Chaperone. Biochim. Biophys. Acta BBA Gen. Subj. 1998, 1381, 279–285. [Google Scholar] [CrossRef]
- Dainiak, M.B.; Izumrudov, V.A.; Muronetz, V.I.; Galaev, I.Y.; Mattiasson, B. Reactivation of Glyceraldehyde-3-Phosphate Dehydrogenase Using Conjugates of Monoclonal Antibodies with Polyelectrolyte Complexes. An Attempt to Make an Artificial Chaperone. J. Mol. Recognit. 1998, 11, 25–27. [Google Scholar] [CrossRef]
- Muronetz, V.I.; Kazakov, S.V.; Dainiak, M.B.; Izumrudov, V.A.; Galaev, I.Y.; Mattiasson, B. Interaction of Antibodies and Antigens Conjugated with Synthetic Polyanions: On the Way of Creating an Artificial Chaperone. Biochim. Biophys. Acta 2000, 1475, 141–150. [Google Scholar] [CrossRef]
- Semenyuk, P.I.; Moiseeva, E.V.; Stroylova, Y.Y.; Lotti, M.; Izumrudov, V.A.; Muronetz, V.I. Sulfated and Sulfonated Polymers Are Able to Solubilize Efficiently the Protein Aggregates of Different Nature. Arch. Biochem. Biophys. 2015, 567, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.V.; Gendrault, J.-L.; Wolff, C.-M. Poly-l-Lysine Dissolves Fibrillar Aggregation of the Alzheimer β-Amyloid Peptide in Vitro. Biochem. Biophys. Res. Commun. 2002, 291, 764–768. [Google Scholar] [CrossRef]
- Klajnert, B.; Cortijo-Arellano, M.; Cladera, J.; Bryszewska, M. Influence of Dendrimer’s Structure on Its Activity against Amyloid Fibril Formation. Biochem. Biophys. Res. Commun. 2006, 345, 21–28. [Google Scholar] [CrossRef]
- Sorokina, S.A.; Stroylova, Y.Y.; Shifrina, Z.B.; Muronetz, V.I. Disruption of Amyloid Prion Protein Aggregates by Cationic Pyridylphenylene Dendrimers. Macromol. Biosci. 2016, 16, 266–275. [Google Scholar] [CrossRef]
- Yu, S.; Xu, X.; Yigit, C.; van der Giet, M.; Zidek, W.; Jankowski, J.; Dzubiella, J.; Ballauff, M. Interaction of Human Serum Albumin with Short Polyelectrolytes: A Study by Calorimetry and Computer Simulations. Soft Matter 2015, 11, 4630–4639. [Google Scholar] [CrossRef]
- Xu, X.; Ran, Q.; Dey, P.; Nikam, R.; Haag, R.; Ballauff, M.; Dzubiella, J. Counterion-Release Entropy Governs the Inhibition of Serum Proteins by Polyelectrolyte Drugs. Biomacromolecules 2018, 19, 409–416. [Google Scholar] [CrossRef]
- Xu, X.; Angioletti-Uberti, S.; Lu, Y.; Dzubiella, J.; Ballauff, M. Interaction of Proteins with Polyelectrolytes: Comparison of Theory to Experiment. Langmuir 2018, 35, 5373–5391. [Google Scholar] [CrossRef]
- Achazi, K.; Haag, R.; Ballauff, M.; Dernedde, J.; Kizhakkedathu, J.N.; Maysinger, D.; Multhaup, G. Understanding the Interaction of Polyelectrolyte Architectures with Proteins and Biosystems. Angew. Chem. Int. Ed. 2021, 60, 3882–3904. [Google Scholar] [CrossRef] [PubMed]
- Sapay, N.; Cabannes, E.; Petitou, M.; Imberty, A. Molecular Modeling of the Interaction between Heparan Sulfate and Cellular Growth Factors: Bringing Pieces Together. Glycobiology 2011, 21, 1181–1193. [Google Scholar] [CrossRef] [PubMed]
- Samsonov, S.A.; Gehrcke, J.-P.; Pisabarro, M.T. Flexibility and Explicit Solvent in Molecular-Dynamics-Based Docking of Protein–Glycosaminoglycan Systems. J. Chem. Inf. Model. 2014, 54, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kett, W.C.; Severin, I.C.; Agyekum, I.; Duan, J.; Amster, I.J.; Proudfoot, A.E.I.; Coombe, D.R.; Woods, R.J. The Interaction of Heparin Tetrasaccharides with Chemokine CCL5 Is Modulated by Sulfation Pattern and PH. J. Biol. Chem. 2015, 290, 15421–15436. [Google Scholar] [CrossRef] [PubMed]
- Salbach-Hirsch, J.; Samsonov, S.A.; Hintze, V.; Hofbauer, C.; Picke, A.-K.; Rauner, M.; Gehrcke, J.-P.; Moeller, S.; Schnabelrauch, M.; Scharnweber, D.; et al. Structural and Functional Insights into Sclerostin-Glycosaminoglycan Interactions in Bone. Biomaterials 2015, 67, 335–345. [Google Scholar] [CrossRef]
- Kurinomaru, T.; Kuwada, K.; Tomita, S.; Kameda, T.; Shiraki, K. Noncovalent PEGylation through Protein–Polyelectrolyte Interaction: Kinetic Experiment and Molecular Dynamics Simulation. J. Phys. Chem. B 2017, 121, 6785–6791. [Google Scholar] [CrossRef]
- Nandy, B.; Saurabh, S.; Sahoo, A.K.; Dixit, N.M.; Maiti, P.K. The SPL7013 Dendrimer Destabilizes the HIV-1 Gp120–CD4 Complex. Nanoscale 2015, 7, 18628–18641. [Google Scholar] [CrossRef]
- Sofronova, A.A.; Izumrudov, V.A.; Muronetz, V.I.; Semenyuk, P.I. Similarly Charged Polyelectrolyte Can Be the Most Efficient Suppressor of the Protein Aggregation. Polymer 2017, 108, 281–287. [Google Scholar] [CrossRef]
- Sofronova, A.A.; Evstafyeva, D.B.; Izumrudov, V.A.; Muronetz, V.I.; Semenyuk, P.I. Protein-Polyelectrolyte Complexes: Molecular Dynamics Simulations and Experimental Study. Polymer 2017, 113, 39–45. [Google Scholar] [CrossRef]
- Valle-Delgado, J.J.; Alfonso-Prieto, M.; de Groot, N.S.; Ventura, S.; Samitier, J.; Rovira, C.; Fernàndez-Busquets, X. Modulation of Aβ42 Fibrillogenesis by Glycosaminoglycan Structure. FASEB J. 2010, 24, 4250–4261. [Google Scholar] [CrossRef]
- Semenyuk, P.; Evstafyeva, D.; Izumrudov, V.; Muronetz, V. Synthetic Sulfated Polymers Control Amyloid Aggregation of Ovine Prion Protein and Decrease Its Toxicity. Polymers 2022, 14, 1478. [Google Scholar] [CrossRef] [PubMed]
- Khurshid, B.; Rehman, A.U.; Luo, R.; Khan, A.; Wadood, A.; Anwar, J. Heparin-Assisted Amyloidogenesis Uncovered through Molecular Dynamics Simulations. ACS Omega 2022, 7, 15132–15144. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Moreno, I.; Bordes, I.; Castillo, R.; Ruiz-Pernía, J.J.; Moliner, V.; García-Junceda, E. Tuning the Phosphoryl Donor Specificity of Dihydroxyacetone Kinase from ATP to Inorganic Polyphosphate. An Insight from Computational Studies. Int. J. Mol. Sci. 2015, 16, 27835–27849. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.E.S.; Morikis, D. Molecular Mechanisms of Macular Degeneration Associated with the Complement Factor H Y402H Mutation. Biophys. J. 2018, 116, 215–226. [Google Scholar] [CrossRef]
- Pol-Fachin, L.; Verli, H. Structural Glycobiology of Heparin Dynamics on the Exosite 2 of Coagulation Cascade Proteases: Implications for Glycosaminoglycans Antithrombotic Activity. Glycobiology 2014, 24, 97–105. [Google Scholar] [CrossRef]
- Sofronova, A.A.; Pozdyshev, D.V.; Barinova, K.V.; Muronetz, V.I.; Semenyuk, P.I. Glycation of Glyceraldehyde-3-Phosphate Dehydrogenase Inhibits the Binding with α-Synuclein and RNA. Arch. Biochem. Biophys. 2021, 698, 108744. [Google Scholar] [CrossRef]
- Semenyuk, P.; Muronetz, V. Protein Interaction with Charged Macromolecules: From Model Polymers to Unfolded Proteins and Post-Translational Modifications. Int. J. Mol. Sci. 2019, 20, 1252. [Google Scholar] [CrossRef]


| Method of Immobilization | Polyelectrolyte | References |
|---|---|---|
| Covalent binding of enzyme or antibody | poly(4-vinyl-N-ethylpyridinium bromide), poly(methacrylic acid), poly(acrylic acid) | [13,14,15,16,17] |
| Layer-by-layer | poly(styrenesulfonate), poly(ethyleneimine), poly(dimethyldiallylammonium) | [23,24,25,26] |
| Electrostatic binding to polyanion or polycation | poly(acrylic acid), poly(methacrylic acid) | [32,52,53,58,60] |
| poly[2-(methacryloyloxy)ethyl]trimethylammonium | [30] | |
| poly(diethylamino)methyl methacrylate | [32] | |
| poly(allylamine) | [52] | |
| poly(l-γ-glutamic acid) | [53] | |
| poly(2-aminoethylmethacrylate hydrochloride) | [59] | |
| Carboxymethylcellulose, poly(L-aspartate), poly(vinylsulfonate), heparin, dextran sulfate, poly(styrene sulfonate) | [60,63] | |
| poly(N,N-diethylaminoethyl methacrylate)-graft-poly(ethylene glycol) | [62] | |
| poly(allylurea-co-allylamine) and succinylated and acetylated derivatives | [66] | |
| Incorporation into microgel | poly(N-isopropylacrylamide) | [42,43] |
| poly(N-isopropylacrylamide-co-N-(3-dimethylaminopropyl)methacrylamide) | [45] | |
| poly(2-((2-(methacryloyloxy)ethyl)dimethylammonio)acetyl)(phenylsulfonyl)amide | [44] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muronetz, V.I.; Pozdyshev, D.V.; Semenyuk, P.I. Polyelectrolytes for Enzyme Immobilization and the Regulation of Their Properties. Polymers 2022, 14, 4204. https://doi.org/10.3390/polym14194204
Muronetz VI, Pozdyshev DV, Semenyuk PI. Polyelectrolytes for Enzyme Immobilization and the Regulation of Their Properties. Polymers. 2022; 14(19):4204. https://doi.org/10.3390/polym14194204
Chicago/Turabian StyleMuronetz, Vladimir I., Denis V. Pozdyshev, and Pavel I. Semenyuk. 2022. "Polyelectrolytes for Enzyme Immobilization and the Regulation of Their Properties" Polymers 14, no. 19: 4204. https://doi.org/10.3390/polym14194204
APA StyleMuronetz, V. I., Pozdyshev, D. V., & Semenyuk, P. I. (2022). Polyelectrolytes for Enzyme Immobilization and the Regulation of Their Properties. Polymers, 14(19), 4204. https://doi.org/10.3390/polym14194204






