Characterization of the Interaction of Polymeric Micelles with siRNA: A Combined Experimental and Molecular Dynamics Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of siRNA-Loaded Micelleplexes
2.3. Characterization of Size and Zeta Potential Measurements
2.4. Gel Retardation Assay
2.5. Capillary Zone Electrophoresis (CZE)
2.5.1. Instrumentation
2.5.2. Standard Solutions and Samples
2.6. RiboGreen® Fluorescence-Based Assay
2.7. Molecular Dynamics Simulations
2.8. Estimation of Binding Affinity
3. Results
3.1. Particle Size, Zeta Potential and Morphology
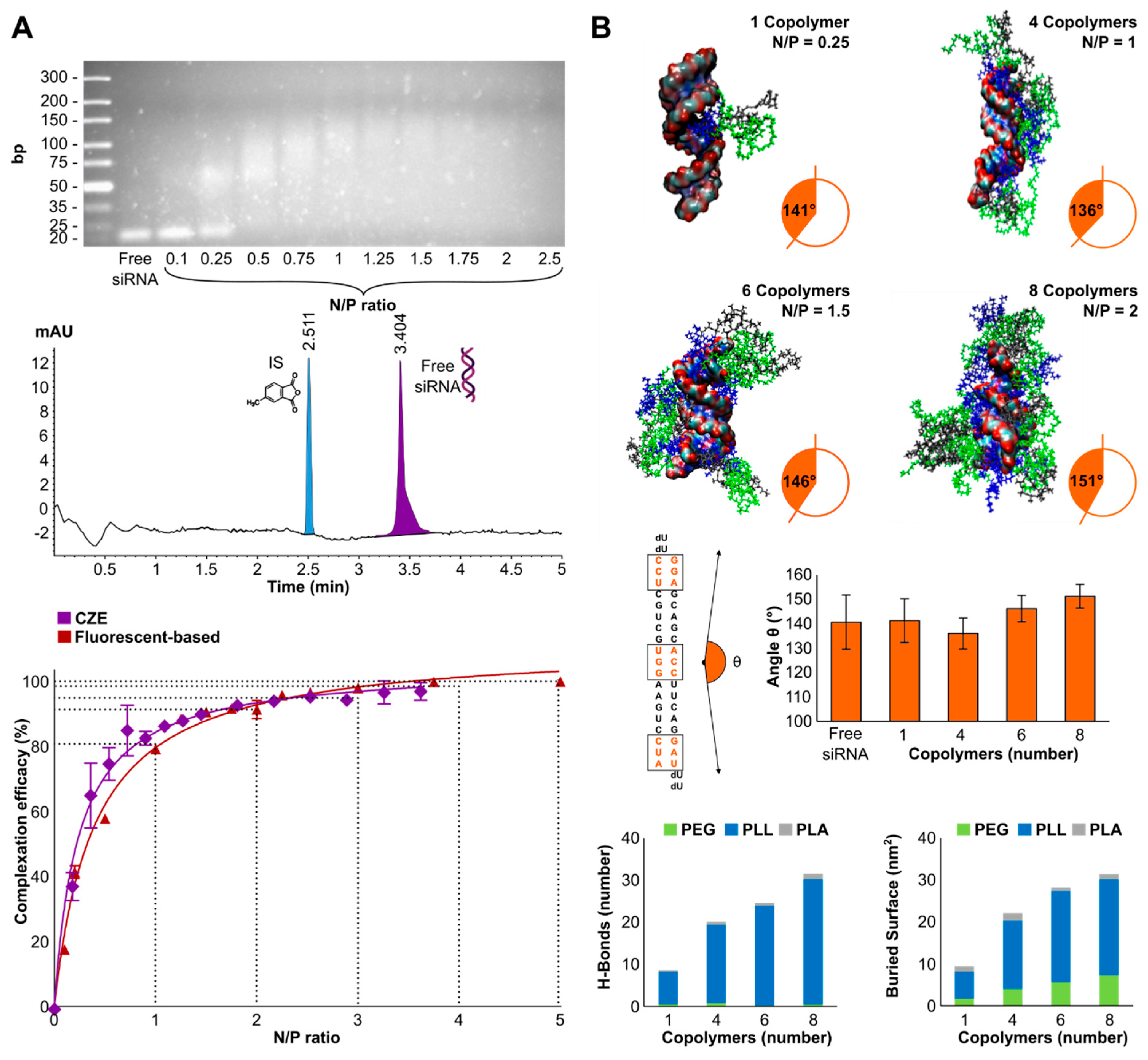
3.2. Complexation Efficacy and Molecular Mechanism of Interaction
3.3. Complexation Efficacy and Molecular Mechanism of Interaction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.J. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Hou, X.; Vick, O.G.; Dong, Y. RNA delivery biomaterials for the treatment of genetic and rare diseases. Biomaterials 2019, 217, 119291. [Google Scholar] [CrossRef] [PubMed]
- Mahmoodi Chalbatani, G.; Dana, H.; Gharagouzloo, E.; Grijalvo, S.; Eritja, R.; Logsdon, C.D.; Memari, F.; Miri, S.R.; Rad, M.R.; Marmari, V. Small interfering RNAs (siRNAs) in cancer therapy: A nano-based approach. Int. J. Nanomed. 2019, 14, 3111–3128. [Google Scholar] [CrossRef] [PubMed]
- Lachelt, U.; Wagner, E. Nucleic acid therapeutics using polyplexes: A journey of 50 years (and beyond). Chem. Rev. 2015, 115, 11043–11078. [Google Scholar] [CrossRef] [PubMed]
- Surgucheva, I.; Shestopalov, V.I.; Surguchov, A. Effect of gamma-synuclein silencing on apoptotic pathways in retinal ganglion cells. J. Biol. Chem. 2008, 283, 36377–36385. [Google Scholar] [CrossRef]
- Bholakant, R.; Qian, H.; Zhang, J.; Huang, X.; Huang, D.; Feijen, J.; Zhong, Y.; Chen, W. Recent advances of polycationic siRNA vectors for cancer therapy. Biomacromolecules 2020, 21, 2966–2982. [Google Scholar] [CrossRef]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef]
- Gary, D.J.; Min, J.; Kim, Y.; Park, K.; Won, Y.Y. The effect of N/P ratio on the in vitro and in vivo interaction properties of PEGylated poly[2-(dimethylamino)ethyl methacrylate]-based siRNA complexes. Macromol. Biosci. 2013, 13, 1059–1071. [Google Scholar] [CrossRef]
- Marquet, F.; Borchard, G. Polymeric micelles for drug delivery in oncology with an emphasis on siRNA conveyance. In Polymeric Micelles for Drug Delivery; Kesharwani, P., Greish, K., Eds.; Woodhead Publishing (Elsevier): Cambridge, UK, 2022; pp. 199–284. [Google Scholar] [CrossRef]
- Marquet, F.; Patrulea, V.; Borchard, G. Comparison of triblock copolymeric micelles based on α- and ε-poly(L-lysine): A Cornelian choice. Polym. J. 2021, 54, 199–209. [Google Scholar] [CrossRef]
- Grasso, G.; Deriu, M.A.; Patrulea, V.; Borchard, G.; Moller, M.; Danani, A. Free energy landscape of siRNA-polycation complexation: Elucidating the effect of molecular geometry, polymer flexibility, and charge neutralization. PLoS ONE 2017, 12, e0186816. [Google Scholar] [CrossRef]
- Furst, T.; Bettonville, V.; Farcas, E.; Frere, A.; Lechanteur, A.; Evrard, B.; Fillet, M.; Piel, G.; Servais, A.C. Capillary electrophoresis method to determine siRNA complexation with cationic liposomes. Electrophoresis 2016, 37, 2685–2691. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tuffery, P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction. J. Chem. Theory Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef]
- Deriu, M.A.; Tsapis, N.; Noiray, M.; Grasso, G.; El Brahmi, N.; Mignani, S.; Majoral, J.P.; Fattal, E.; Danani, A. Elucidating the role of surface chemistry on cationic phosphorus dendrimer-siRNA complexation. Nanoscale 2018, 10, 10952–10962. [Google Scholar] [CrossRef] [PubMed]
- Stojceski, F.; Grasso, G.; Pallante, L.; Danani, A. Molecular and coarse-grained modeling to characterize and optimize dendrimer-based nanocarriers for short interfering RNA delivery. ACS Omega 2020, 5, 2978–2986. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Kutzner, C.; Pall, S.; Fechner, M.; Esztermann, A.; de Groot, B.L.; Grubmuller, H. More bang for your buck: Improved use of GPU nodes for GROMACS 2018. J. Comput. Chem. 2019, 40, 2418–2431. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Postma, J.P.M.; Gunsteren, W.F.; Di Nola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Johnson, J.K.; Zollweg, J.A.; Gubbins, K.E. The Lennard-Jones equation of state revisited. Mol. Phys. 1993, 78, 591–618. [Google Scholar] [CrossRef]
- Miller, B.R., 3rd; McGee, T.D., Jr.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Zheng, M.; Pavan, G.M.; Neeb, M.; Schaper, A.K.; Danani, A.; Klebe, G.; Merkel, O.M.; Kissel, T. Targeting the blind spot of polycationic nanocarrier-based siRNA delivery. ACS Nano 2012, 6, 9447–9454. [Google Scholar] [CrossRef]
- Veziroglu, E.M.; Mias, G.I. Characterizing extracellular vesicles and their diverse RNA contents. Front. Genet. 2020, 11, 700. [Google Scholar] [CrossRef]
- Kwok, A.; McCarthy, D.; Hart, S.L.; Tagalakis, A.D. Systematic Comparisons of Formulations of Linear Oligolysine Peptides with siRNA and Plasmid DNA. Chem. Biol. Drug Des. 2016, 87, 747–763. [Google Scholar] [CrossRef]
- Prozeller, D.; Morsbach, S.; Landfester, K. Isothermal titration calorimetry as a complementary method for investigating nanoparticle-protein interactions. Nanoscale 2019, 11, 19265–19273. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.T.; Hom, K.; Zhang, D.; Leng, Q.; Tricoli, L.J.; Hustedt, J.M.; Lee, A.; Shapiro, M.J.; Seog, J.; Kahn, J.D.; et al. Enhanced silencing and stabilization of siRNA polyplexes by histidine-mediated hydrogen bonds. Biomaterials 2014, 35, 846–855. [Google Scholar] [CrossRef] [PubMed]
- Lundbäck, T.; Härd, T. Salt dependence of the free energy, enthalpy, and entropy of nonsequence Specific DNA Binding. J. Phys. Chem. 1996, 100, 17690–17695. [Google Scholar] [CrossRef]
- Mascotti, D.P.; Lohman, T.M. Thermodynamic extent of counterion release upon binding oligolysines to single-stranded nucleic acids. Proc. Natl. Acad. Sci. USA 1990, 87, 3142–3146. [Google Scholar] [CrossRef] [PubMed]
- Marini, R.D.; Rozet, E.; Montes, M.L.; Rohrbasser, C.; Roht, S.; Rheme, D.; Bonnabry, P.; Schappler, J.; Veuthey, J.L.; Hubert, P.; et al. Reliable low-cost capillary electrophoresis device for drug quality control and counterfeit medicines. J. Pharm. Biomed. Anal. 2010, 53, 1278–1287. [Google Scholar] [CrossRef] [PubMed]
- Tobolkina, E.; Rudaz, S. Capillary electrophoresis instruments for medical applications and falsified drug analysis/quality control in developing countries. Anal. Chem. 2021, 93, 8107–8115. [Google Scholar] [CrossRef]
- Caputo, F.; Clogston, J.; Calzolai, L.; Rosslein, M.; Prina-Mello, A. Measuring particle size distribution of nanoparticle enabled medicinal products, the joint view of EUNCL and NCI-NCL. A step by step approach combining orthogonal measurements with increasing complexity. J. Control. Release 2019, 299, 31–43. [Google Scholar] [CrossRef]
- Halamoda-Kenzaoui, B.; Holzwarth, U.; Roebben, G.; Bogni, A.; Bremer-Hoffmann, S. Mapping of the available standards against the regulatory needs for nanomedicines. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2019, 11, e1531. [Google Scholar] [CrossRef]
- Caputo, F.; Arnould, A.; Bacia, M.; Ling, W.L.; Rustique, E.; Texier, I.; Mello, A.P.; Couffin, A.C. Measuring particle size distribution by asymmetric flow field flow fractionation: A powerful method for the preclinical characterization of lipid-based nanoparticles. Mol. Pharm. 2019, 16, 756–767. [Google Scholar] [CrossRef]
- Mildner, R.; Hak, S.; Parot, J.; Hyldbakk, A.; Borgos, S.E.; Some, D.; Johann, C.; Caputo, F. Improved multidetector asymmetrical-flow field-flow fractionation method for particle sizing and concentration measurements of lipid-based nanocarriers for RNA delivery. Eur. J. Pharm. Biopharm. 2021, 163, 252–265. [Google Scholar] [CrossRef]
- Santos, I.C.; Brodbelt, J.S. Recent developments in the characterization of nucleic acids by liquid chromatography, capillary electrophoresis, ion mobility, and mass spectrometry (2010–2020). J. Sep. Sci. 2021, 44, 340–372. [Google Scholar] [CrossRef]
- Talmadge, K.W.; Zhu, M.; Olech, L.; Siebert, C. Oligonucleotide analysis by capillary polymer sieving electrophoresis using acryloylaminoethoxyethanol-coated capillaries. J. Chromatogr. A 1996, 744, 347–354. [Google Scholar] [CrossRef]
- Przybylski, C.; Benito, J.M.; Bonnet, V.; Mellet, C.O.; Garcia Fernandez, J.M. Revealing cooperative binding of polycationic cyclodextrins with DNA oligomers by capillary electrophoresis coupled to mass spectrometry. Anal. Chim. Acta 2018, 1002, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Mironov, G.; Berezovski, M.V. Direct detection of endogenous MicroRNAs and their post-transcriptional modifications in cancer serum by capillary electrophoresis-mass spectrometry. Anal. Bioanal. Chem. 2016, 408, 2891–2899. [Google Scholar] [CrossRef] [PubMed]
- Pero-Gascon, R.; Sanz-Nebot, V.; Berezovski, M.V.; Benavente, F. Analysis of circulating microRNAs and their post-transcriptional modifications in cancer serum by on-line solid-phase extraction-capillary electrophoresis-mass spectrometry. Anal. Chem. 2018, 90, 6618–6625. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]

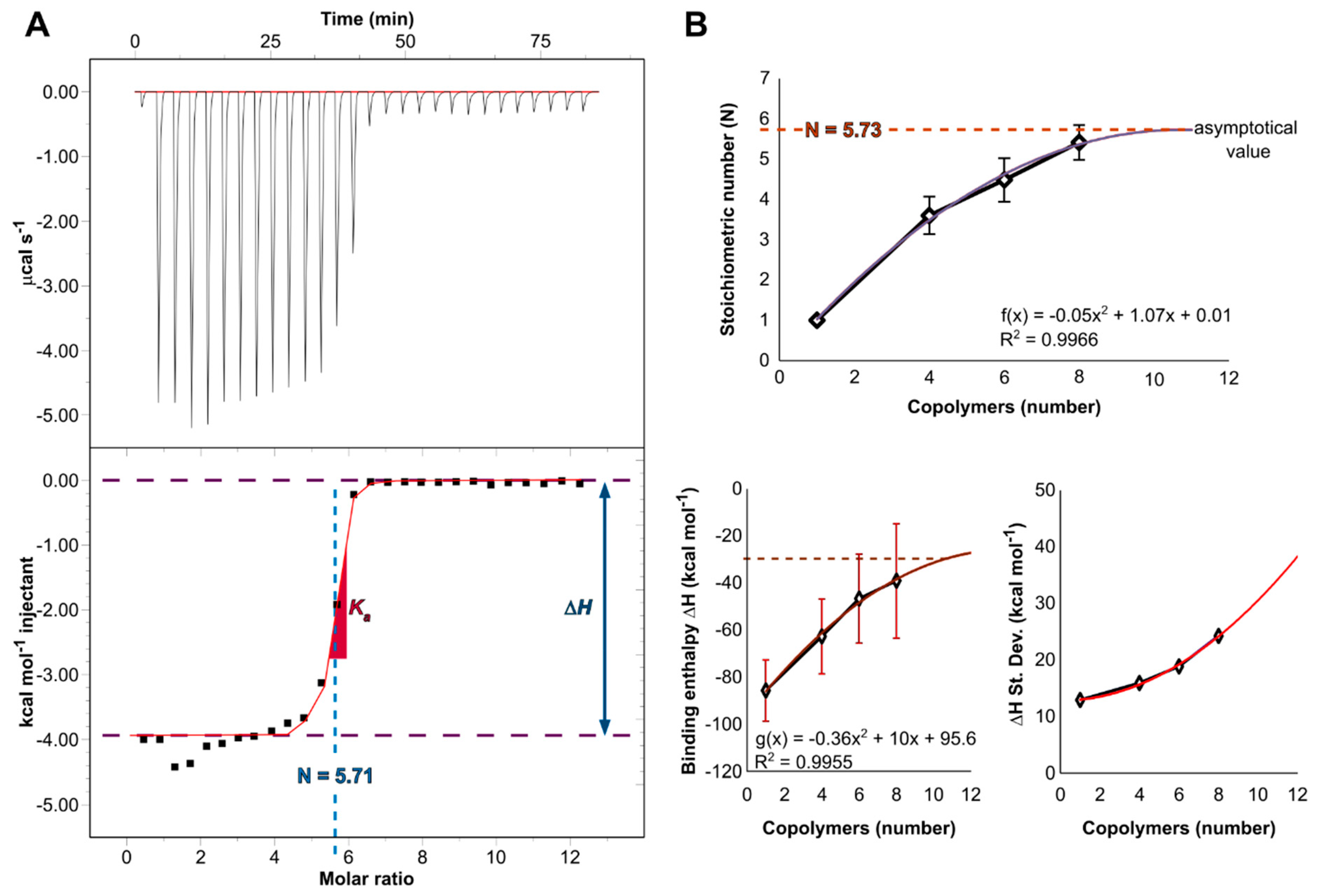
| N (sites) | Ka (M−1) | Kd (nM) | ΔH (kJ mol−1) | ΔS (kJ K−1 mol−1) | ΔG (kJ mol−1) | |
|---|---|---|---|---|---|---|
| Experimental | 5.71 ± 0.15 | 2.6 · 107 | 38.5 | −16.5 | 0.087 | −42.3 |
| Computational | 5.73 | N/A | N/A | −132.2 | N/A | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marquet, F.; Stojceski, F.; Grasso, G.; Patrulea, V.; Danani, A.; Borchard, G. Characterization of the Interaction of Polymeric Micelles with siRNA: A Combined Experimental and Molecular Dynamics Study. Polymers 2022, 14, 4409. https://doi.org/10.3390/polym14204409
Marquet F, Stojceski F, Grasso G, Patrulea V, Danani A, Borchard G. Characterization of the Interaction of Polymeric Micelles with siRNA: A Combined Experimental and Molecular Dynamics Study. Polymers. 2022; 14(20):4409. https://doi.org/10.3390/polym14204409
Chicago/Turabian StyleMarquet, Franck, Filip Stojceski, Gianvito Grasso, Viorica Patrulea, Andrea Danani, and Gerrit Borchard. 2022. "Characterization of the Interaction of Polymeric Micelles with siRNA: A Combined Experimental and Molecular Dynamics Study" Polymers 14, no. 20: 4409. https://doi.org/10.3390/polym14204409
APA StyleMarquet, F., Stojceski, F., Grasso, G., Patrulea, V., Danani, A., & Borchard, G. (2022). Characterization of the Interaction of Polymeric Micelles with siRNA: A Combined Experimental and Molecular Dynamics Study. Polymers, 14(20), 4409. https://doi.org/10.3390/polym14204409








