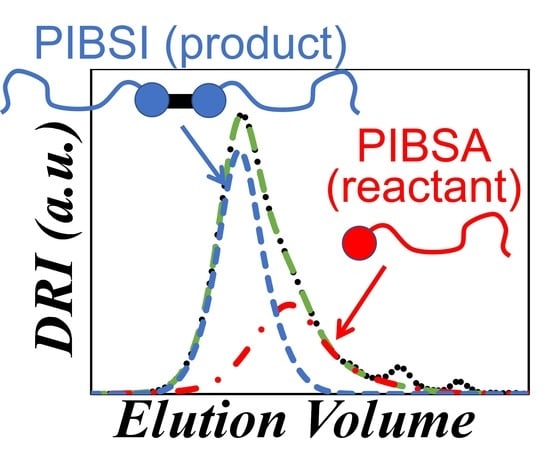
Characterization of Polyisobutylene Succinic Anhydride (PIBSA) and Its PIBSI Products from the Reaction of PIBSA with Hexamethylene Diamine
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. Analysis of the Fourier-Transform Infrared (FTIR) Spectra
3.2. Gel Permeation Chromatography (GPC) Analysis
3.3. Simulations to Predict the Composition of a PIBSA–H(X) Reaction Mixture
3.4. Predicted MWD of b-PIBSI Supports That b-PIBSI Is the Only Higher-Order PIBSI Product
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lunn, D.J.; Discekici, E.H.; Read de Alaniz, J.; Gutekunst, W.R.; Hawker, C.J. Established and Emerging Strategies for Polymer Chain-End Modification. J. Polym. Sci. A Polym. Chem. 2017, 55, 2903–2914. [Google Scholar] [CrossRef]
- Dey, A.; Haldar, U.; De, P. Block Copolymer Synthesis by the Combination of Living Cationic Polymerization and Other Polymerization Methods. Front. Chem. 2021, 9, 644547. [Google Scholar] [CrossRef]
- Hilf, S.; Kilbinger, A.F.M. Functional End Groups for Polymers Prepared Using Ring-Opening Metathesis Polymerization. Nat. Chem. 2009, 1, 537–546. [Google Scholar] [CrossRef]
- Kim, J.; Jung, H.Y.; Park, M.J. End-Group Chemistry and Junction Chemistry in Polymer Science: Past, Present, and Future. Macromolecules 2020, 53, 746–763. [Google Scholar] [CrossRef]
- Zhou, D.; Zhu, L.-W.; Wu, B.-H.; Xu, Z.-K.; Wan, L.-S. End-Functionalized Polymers by Controlled/Living Radical Polymerizations: Synthesis and Applications. Polym. Chem. 2022, 13, 300–358. [Google Scholar] [CrossRef]
- Yu, X.; Li, Y.; Dong, X.-H.; Yue, K.; Lin, Z.; Feng, X.; Huang, M.; Zhang, B.-W.; Cheng, S.Z.D. Giant Surfactants Based on Molecular Nanoparticles: Precise Synthesis and Solution Self-Assembly. J. Polym. Sci. B Polym. Phys. 2014, 52, 1309–1325. [Google Scholar] [CrossRef]
- Coudane, J.; Nottelet, B.; Mouton, J.; Garric, X.; Van Den Berghe, H. Poly(e-caprolactone)-Based Graft Copolymers: Synthesis Methods and Applications in the Biomedical Field: A Review. Molecules 2022, 27, 7339. [Google Scholar] [CrossRef] [PubMed]
- Soares, D.C.F.; Oda, C.M.R.; Monteiro, L.O.F.; Branco de Barros, A.L.; Tebaldi, M.L. Responsive Polymer Conjugates for Drug Delivery Applications: Recent Advances in Bioconjugation Methodologies. J. Drug Target. 2019, 27, 355–366. [Google Scholar] [CrossRef]
- Gauthier, M. Arborescent Polymers and Other Dendigraft Polymers: A Journey into Structural Duversity. J. Polym. Sci. A Polym. Chem. 2007, 45, 3803–3810. [Google Scholar] [CrossRef]
- Teertstra, S.; Gauthier, M. Dendrigraft Polymers: Macromolecular Engineering on a Mesoscopic Scale. Prog. Polym. Sci. 2004, 29, 277–327. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Fréchet, J.M. Discovery of Dendrimers and Dendritic Polymers: A Brief Historical Perspective. J. Polym. Sci. A Polym Chem. 2002, 40, 2719–2728. [Google Scholar] [CrossRef]
- Hadjichristidis, N.; Pitsikalis, M.; Pispas, S.; Iatrou, H. Polymers with Complex Architecture by Living Anionic Polymerization. Chem. Rev. 2001, 101, 3747–3792. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, T.; Newland, B.; Liu, W.; Wang, W.; Wang, W. Catechol Functionalized Hyperbranched Polymers as Biomedical Materials. Prog. Polym. Sci. 2018, 78, 47–55. [Google Scholar] [CrossRef]
- Blasco, E.; Pinol, M.; Oriol, L. Responsive Linear-Dendritic Block Copolymers. Macromol. Rapid Commun. 2014, 35, 1090–1115. [Google Scholar] [CrossRef]
- Lee, H.-I.; Pietrasik, J.; Sheiko, S.S.; Matyjaszewski, K. Stimuli-Responsive Molecular Brushes. Prog. Polym. Sci. 2010, 35, 24–44. [Google Scholar] [CrossRef]
- Sheiko, S.S.; Sumerlin, B.S.; Matyjaszewski, K. Cylindrical Molecular Brushes: Synthesis, Characterization, and Properties. Prog. Polym. Sci. 2008, 33, 759–785. [Google Scholar] [CrossRef]
- Pelras, T.; Mahon, C.S.; Mullner, M. Synthesis and Applications of Compartmentalized Molecular Polymer Brushes. Angew. Chem. Int. Ed. 2018, 57, 6982–6994. [Google Scholar] [CrossRef] [PubMed]
- Chernikova, E.V.; Kudryavtsev, Y.V. RAFT-Based Polymers for Click Reactions. Polymers 2022, 14, 570. [Google Scholar] [CrossRef] [PubMed]
- Frasca, F.; Duhamel, J. End Group Analysis of Polyisobutyelene Succinic Anhydride (PIBSA) Carried Out with Pyrene Excimer Fluorescence. Ind. Eng. Chem. Res. 2022, 61, 14747–14759. [Google Scholar] [CrossRef]
- Jerome, R.; Henrioulle-Grandville, M.; Boutevin, B.; Robin, J.J. Telechelic Polymers: Synthesis, Characterization and Applications. Prog. Polym. Sci. 1991, 16, 837–906. [Google Scholar] [CrossRef]
- Striegel, A.M. There’s Plenty of Gloom at the Bottom: The Many Challenges of Accurate Quantitation in Size-Based Oligomeric Separation. Anal. Bional. Chem. 2013, 405, 8959–8967. [Google Scholar] [CrossRef]
- Harrison, J.J.; Mijares, C.M.; Cheng, M.T.; Hudson, J. Negative Ion Electrospray Ionization Mass Spectrum of Polyisobutenylsuccinic anhydride: Implications for Isobutylene Polymerization. Macromolecules 2002, 35, 2494–2500. [Google Scholar] [CrossRef]
- Tessier, M.; Maréchal, E. Synthesis of Mono and Difunctional Oligoisobutylenes-III. Modification of α-Chlorooligobutylene by Reaction with Maleic Anhydride. Eur. Polym. J. 1984, 20, 269–280. [Google Scholar] [CrossRef]
- Tessier, M.; Maréchal, E. Synthesis of α-phenyl-ω-anhydride oligoisobutylene and α, ω-dianhydride oligoisobutylene. Eur. Polym. J. 1990, 26, 499–508. [Google Scholar] [CrossRef]
- Le Suer, W.M. Substituted Polyamines for Use as Additives to Lubricants, Motor Fuels, and Hydraulic Fluids. FR. Patent 1403977, 25 June 1965. [Google Scholar]
- Seddon, E.J.; Friend, C.L.; Roski, J.P. Chemistry and Technology of Lubricants; Mortier, R.M., Malcolm, F.F., Orszulik, S.T., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 213–236. [Google Scholar]
- Rizvi, S.Q.A. Fuels and Lubricants Handbook: Technology, Properties, Performance and Testing; Shah, R.J., Ed.; ASTM International: West Conshohocken, PA, USA, 2003; pp. 199–248. [Google Scholar]
- Shen, Y.; Duhamel, J. Micellization and Adsorption of a Series of Succinimide Dispersants. Langmuir 2008, 24, 10665–10673. [Google Scholar] [CrossRef]
- Mehdiabadi, S.; Lhost, O.; Vantomme, A.; Soares, J.B.P. Ethylene Polymerization Kinetics and Microstructure of Polyethylenes Made with Supported Metallocene Catalysts. Ind. Eng. Chem. Res. 2021, 60, 9739–9754. [Google Scholar] [CrossRef]
- Gaborieau, M.; Gilbert, R.G.; Gray-Weale, A.; Hernandez, J.M.; Castignolles, P. Theory of Multiple-Detection Size-Exclusion Chromatography of Complex Branched Polymers. Macromol. Theory Simul. 2007, 16, 13–28. [Google Scholar] [CrossRef]
- Laborda, F.; Bolea, E.; Géorriz, M.P.; Martín-Ruiz, M.; Ruiz-Beguería, S.; Castillo, J.R. A Speciation Methodology to Study the Contributions of Humic-Like and Fulvic-Like Acids to the Mobilization of Metals from Compost Using Size Exclusion Chromatography-Ultraviolet Absorption-Inductively Coupled Plasma Mass Spectrometry and Deconvolution Analysis. Anal. Chim. Acta 2008, 606, 1–8. [Google Scholar]
- Ogino, M.; Kameda, T.; Mutsuda, Y.; Tanaka, H.; Takahashi, J.; Okasaki, M.; Ai, M.; Ohkawa, R. Development for Internal Standard for Lipoprotein Subclass Analysis Using Dual Detection Gel-Permeation High-Performance Liquid Chromatography System. Biosci. Rep. 2022, 42, BSR20220291. [Google Scholar] [CrossRef]
- Press, W.H.; Flannery, B.P.; Teukolsky, S.A.; Vetterling, W.T. Numerical Recipes. In The Art of Scientific Computing (Fortran Version); Cambridge University Press: Cambridge, UK, 1992. [Google Scholar]
- Clay, P.L.; Gilbert, R.G. Molecular Weight Distributions in Free-Radical Polymerizations. 1. Model Development and Implications for Data Interpretation. Macromolecules 1995, 28, 552–569. [Google Scholar] [CrossRef]
- Pirouz, S.; Wang, Y.; Chong, J.M.; Duhamel, J. Characterization of the Chemical Composition of Polyisobutylene-Based Oil-Soluble Dispersants by Fluorescence. J. Phys. Chem. B 2014, 118, 3899–3911. [Google Scholar] [CrossRef]
- Walch, E.; Gaymans, R.J. Telechelic Polyisobutylene with Unsaturated End Groups and with Anhydride End Groups. Polymer 1994, 35, 1774–1778. [Google Scholar] [CrossRef]
- Rivera-Tirado, E.; Aaserud, D.J.; Wesdemiotis, C. Characterization of Polyisobutylene Succinic Anhydride Chemistries Using Mass Spectrometry. J. Appl. Polym. Sci. 2012, 124, 2682–2690. [Google Scholar] [CrossRef]
- Van der Merwe, M.M.; Landman, M.; van Rooyen, P.H.; Jordaan, J.H.L.; Otto, D.P. Structural Assignment of Commercial Polyisobutylene Succinic Anhydride-Based Surfactants. J. Surfact. Deterg. 2017, 20, 193–205. [Google Scholar] [CrossRef]
- Morales-Rivera, C.A.; Proust, N.; Burrington, J.; Mpourmpakis, G. Computational Screening of Lewis Acid Catalysts for the Ene Reaction Between Maleic Anhydride and Polyisobutyene. Ind. Eng. Chem. Res. 2021, 60, 154–161. [Google Scholar] [CrossRef]
- Morales-Rivera, C.A.; Cormack, G.; Burrington, J.; Proust, N. Understanding and Optimizing the Behavior of Al- and Ru-Based Catalysts for the Synthesis of Polyisobutenyl Succinic Anhydrides. Ind. Eng. Chem. Res. 2022, 61, 14462–14471. [Google Scholar] [CrossRef]
- Vo, M.N.; Call, M.; Kowall, C.; Johnson, J.K. Effect of Chain Length on the Dipole Moment of Polyisobutylene Succinate Anhydride. Ind. Eng. Chem. Res. 2022, 61, 2359–2365. [Google Scholar] [CrossRef]
- Streets, J.; Proust, N.; Parmar, D.; Walker, G.; Licence, P.; Woodward, S. Rate of Formation of Industrial Lubricant Additive Precursors from Maleic Anhydride and Polyisobutylene. Org. Process Res. Dev. 2022, 26, 2749–2755. [Google Scholar] [CrossRef] [PubMed]


 ) Abs(1785 cm−1)/Abs(1390 cm−1) and (
) Abs(1785 cm−1)/Abs(1390 cm−1) and ( ) Abs(1705 cm−1)/Abs(1390 cm−1) ratios as a function of the NAm/NSA ratio for the PIBSA–H(X) reaction products.
) Abs(1705 cm−1)/Abs(1390 cm−1) ratios as a function of the NAm/NSA ratio for the PIBSA–H(X) reaction products.
 ) Abs(1785 cm−1)/Abs(1390 cm−1) and (
) Abs(1785 cm−1)/Abs(1390 cm−1) and ( ) Abs(1705 cm−1)/Abs(1390 cm−1) ratios as a function of the NAm/NSA ratio for the PIBSA–H(X) reaction products.
) Abs(1705 cm−1)/Abs(1390 cm−1) ratios as a function of the NAm/NSA ratio for the PIBSA–H(X) reaction products.
 ) DRI trace and (
) DRI trace and ( ) sum of Gaussians fit of a dehydrated PIBSA sample as a function of elution volume. The contributions of the solvent peaks at 19, 20.5, and 23.5 mL were eliminated in the fitted GPC trace.
) sum of Gaussians fit of a dehydrated PIBSA sample as a function of elution volume. The contributions of the solvent peaks at 19, 20.5, and 23.5 mL were eliminated in the fitted GPC trace.
 ) DRI trace and (
) DRI trace and ( ) sum of Gaussians fit of a dehydrated PIBSA sample as a function of elution volume. The contributions of the solvent peaks at 19, 20.5, and 23.5 mL were eliminated in the fitted GPC trace.
) sum of Gaussians fit of a dehydrated PIBSA sample as a function of elution volume. The contributions of the solvent peaks at 19, 20.5, and 23.5 mL were eliminated in the fitted GPC trace.

 ) the fit with a sum of Gaussians for (
) the fit with a sum of Gaussians for ( ) the DRI trace of PIBSA–H(0.75), (
) the DRI trace of PIBSA–H(0.75), ( ) the contribution from PIBSA-like molecules, and (
) the contribution from PIBSA-like molecules, and ( ) the contribution from non-PIBSA-like molecules. (B) Fits with sums of Gaussians for the PIBSA–H(X) samples with NAm/NSA ratios of (B) (
) the contribution from non-PIBSA-like molecules. (B) Fits with sums of Gaussians for the PIBSA–H(X) samples with NAm/NSA ratios of (B) ( ) 0.19, (
) 0.19, ( ) 0.38, (
) 0.38, ( ) 0.56, (
) 0.56, ( ) 0.75, and (
) 0.75, and ( ) 0.94, and (C) (
) 0.94, and (C) ( ) 0.94, (
) 0.94, ( ) 1.13, (
) 1.13, ( ) 1.32, and (
) 1.32, and ( ) 1.51, normalized to a sum of 1. (D) Plots of the weight fraction (wPLM) of PIBSA-like molecules in the PIBSA–H(X) samples determined by fitting the DRI traces with (×) xgaussSNP and (
) 1.51, normalized to a sum of 1. (D) Plots of the weight fraction (wPLM) of PIBSA-like molecules in the PIBSA–H(X) samples determined by fitting the DRI traces with (×) xgaussSNP and ( ) linear regression.
) linear regression.
 ) the fit with a sum of Gaussians for (
) the fit with a sum of Gaussians for ( ) the DRI trace of PIBSA–H(0.75), (
) the DRI trace of PIBSA–H(0.75), ( ) the contribution from PIBSA-like molecules, and (
) the contribution from PIBSA-like molecules, and ( ) the contribution from non-PIBSA-like molecules. (B) Fits with sums of Gaussians for the PIBSA–H(X) samples with NAm/NSA ratios of (B) (
) the contribution from non-PIBSA-like molecules. (B) Fits with sums of Gaussians for the PIBSA–H(X) samples with NAm/NSA ratios of (B) ( ) 0.19, (
) 0.19, ( ) 0.38, (
) 0.38, ( ) 0.56, (
) 0.56, ( ) 0.75, and (
) 0.75, and ( ) 0.94, and (C) (
) 0.94, and (C) ( ) 0.94, (
) 0.94, ( ) 1.13, (
) 1.13, ( ) 1.32, and (
) 1.32, and ( ) 1.51, normalized to a sum of 1. (D) Plots of the weight fraction (wPLM) of PIBSA-like molecules in the PIBSA–H(X) samples determined by fitting the DRI traces with (×) xgaussSNP and (
) 1.51, normalized to a sum of 1. (D) Plots of the weight fraction (wPLM) of PIBSA-like molecules in the PIBSA–H(X) samples determined by fitting the DRI traces with (×) xgaussSNP and ( ) linear regression.
) linear regression.

 ) the xgaussSNP programs and (
) the xgaussSNP programs and ( ) the linear regression analysis of the PIBSA–H(X) samples with NAm/NSA ratios of (A) 0.19, (B) 0.38, (C) 0.56, (D) 0.75, (E) 0.94, (F) 1.13, (G) 1.32, and (H) 1.51.
) the linear regression analysis of the PIBSA–H(X) samples with NAm/NSA ratios of (A) 0.19, (B) 0.38, (C) 0.56, (D) 0.75, (E) 0.94, (F) 1.13, (G) 1.32, and (H) 1.51.
 ) the xgaussSNP programs and (
) the xgaussSNP programs and ( ) the linear regression analysis of the PIBSA–H(X) samples with NAm/NSA ratios of (A) 0.19, (B) 0.38, (C) 0.56, (D) 0.75, (E) 0.94, (F) 1.13, (G) 1.32, and (H) 1.51.
) the linear regression analysis of the PIBSA–H(X) samples with NAm/NSA ratios of (A) 0.19, (B) 0.38, (C) 0.56, (D) 0.75, (E) 0.94, (F) 1.13, (G) 1.32, and (H) 1.51.
 ) experimentally from regression of GPC traces (Figure 5D) and with (
) experimentally from regression of GPC traces (Figure 5D) and with ( ) reaction3 and (
) reaction3 and ( ) reaction4, (B) unmaleated PIB obtained with (
) reaction4, (B) unmaleated PIB obtained with ( ) reaction3 and (
) reaction3 and ( ) reaction4, and (C) PLM obtained (
) reaction4, and (C) PLM obtained ( ) experimentally from regression (Figure 5D) and with (
) experimentally from regression (Figure 5D) and with ( ) reaction3 and (
) reaction3 and ( ) reaction4 after adding a constant wPIB of 0.36.
) reaction4 after adding a constant wPIB of 0.36.
 ) experimentally from regression of GPC traces (Figure 5D) and with (
) experimentally from regression of GPC traces (Figure 5D) and with ( ) reaction3 and (
) reaction3 and ( ) reaction4, (B) unmaleated PIB obtained with (
) reaction4, (B) unmaleated PIB obtained with ( ) reaction3 and (
) reaction3 and ( ) reaction4, and (C) PLM obtained (
) reaction4, and (C) PLM obtained ( ) experimentally from regression (Figure 5D) and with (
) experimentally from regression (Figure 5D) and with ( ) reaction3 and (
) reaction3 and ( ) reaction4 after adding a constant wPIB of 0.36.
) reaction4 after adding a constant wPIB of 0.36.
 ) 1.0, (
) 1.0, ( ) 3.2 kg/mol, and (
) 3.2 kg/mol, and ( ) PIBSA and (B) simulated MWD for (
) PIBSA and (B) simulated MWD for ( , P1(M)) m-PIBSI, (
, P1(M)) m-PIBSI, ( , P2(M)) b-PIBSI, (
, P2(M)) b-PIBSI, ( , P3(M)) tri-PIBSI, (
, P3(M)) tri-PIBSI, ( , P4(M)) tetra-PIBSI, and (
, P4(M)) tetra-PIBSI, and ( ) experimental MWD for higher-order products in the PIBSA–H(0.94) reaction mixture.
) experimental MWD for higher-order products in the PIBSA–H(0.94) reaction mixture.
 ) 1.0, (
) 1.0, ( ) 3.2 kg/mol, and (
) 3.2 kg/mol, and ( ) PIBSA and (B) simulated MWD for (
) PIBSA and (B) simulated MWD for ( , P1(M)) m-PIBSI, (
, P1(M)) m-PIBSI, ( , P2(M)) b-PIBSI, (
, P2(M)) b-PIBSI, ( , P3(M)) tri-PIBSI, (
, P3(M)) tri-PIBSI, ( , P4(M)) tetra-PIBSI, and (
, P4(M)) tetra-PIBSI, and ( ) experimental MWD for higher-order products in the PIBSA–H(0.94) reaction mixture.
) experimental MWD for higher-order products in the PIBSA–H(0.94) reaction mixture.
 ) the simulated DRI traces obtained from the molar fractions obtained with reaction4, wPIB = 0.36, and the DRI traces of PIBSA and b-PIBSI given by the GPC trace obtained for the non-PLM of PIBSA–H(0.94) in Figure 6, and (
) the simulated DRI traces obtained from the molar fractions obtained with reaction4, wPIB = 0.36, and the DRI traces of PIBSA and b-PIBSI given by the GPC trace obtained for the non-PLM of PIBSA–H(0.94) in Figure 6, and ( ) the experimental* DRI traces fitted with the xgaussSNP programs of the PIBSA–H(X) products. The NAm/NSA ratios equal (A) 0.19, (B) 0.38, (C) 0.56, (D) 0.75, (E) 0.94, (F) 1.13, (G) 1.32, and (H) 1.51.
) the experimental* DRI traces fitted with the xgaussSNP programs of the PIBSA–H(X) products. The NAm/NSA ratios equal (A) 0.19, (B) 0.38, (C) 0.56, (D) 0.75, (E) 0.94, (F) 1.13, (G) 1.32, and (H) 1.51.
 ) the simulated DRI traces obtained from the molar fractions obtained with reaction4, wPIB = 0.36, and the DRI traces of PIBSA and b-PIBSI given by the GPC trace obtained for the non-PLM of PIBSA–H(0.94) in Figure 6, and (
) the simulated DRI traces obtained from the molar fractions obtained with reaction4, wPIB = 0.36, and the DRI traces of PIBSA and b-PIBSI given by the GPC trace obtained for the non-PLM of PIBSA–H(0.94) in Figure 6, and ( ) the experimental* DRI traces fitted with the xgaussSNP programs of the PIBSA–H(X) products. The NAm/NSA ratios equal (A) 0.19, (B) 0.38, (C) 0.56, (D) 0.75, (E) 0.94, (F) 1.13, (G) 1.32, and (H) 1.51.
) the experimental* DRI traces fitted with the xgaussSNP programs of the PIBSA–H(X) products. The NAm/NSA ratios equal (A) 0.19, (B) 0.38, (C) 0.56, (D) 0.75, (E) 0.94, (F) 1.13, (G) 1.32, and (H) 1.51.

 ,×) Mn and (
,×) Mn and ( ,+) Mw and (B) (
,+) Mw and (B) ( ,
, ) PDI of the PIBSA–H(X) samples as a function of the NAm/NSA ratio, (
) PDI of the PIBSA–H(X) samples as a function of the NAm/NSA ratio, ( ,
, ,
, ) simulations, and (×,+,
) simulations, and (×,+, ) experiments.
) experiments.
 ,×) Mn and (
,×) Mn and ( ,+) Mw and (B) (
,+) Mw and (B) ( ,
, ) PDI of the PIBSA–H(X) samples as a function of the NAm/NSA ratio, (
) PDI of the PIBSA–H(X) samples as a function of the NAm/NSA ratio, ( ,
, ,
, ) simulations, and (×,+,
) simulations, and (×,+, ) experiments.
) experiments.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frasca, F.; Duhamel, J. Characterization of Polyisobutylene Succinic Anhydride (PIBSA) and Its PIBSI Products from the Reaction of PIBSA with Hexamethylene Diamine. Polymers 2023, 15, 2350. https://doi.org/10.3390/polym15102350
Frasca F, Duhamel J. Characterization of Polyisobutylene Succinic Anhydride (PIBSA) and Its PIBSI Products from the Reaction of PIBSA with Hexamethylene Diamine. Polymers. 2023; 15(10):2350. https://doi.org/10.3390/polym15102350
Chicago/Turabian StyleFrasca, Franklin, and Jean Duhamel. 2023. "Characterization of Polyisobutylene Succinic Anhydride (PIBSA) and Its PIBSI Products from the Reaction of PIBSA with Hexamethylene Diamine" Polymers 15, no. 10: 2350. https://doi.org/10.3390/polym15102350
APA StyleFrasca, F., & Duhamel, J. (2023). Characterization of Polyisobutylene Succinic Anhydride (PIBSA) and Its PIBSI Products from the Reaction of PIBSA with Hexamethylene Diamine. Polymers, 15(10), 2350. https://doi.org/10.3390/polym15102350