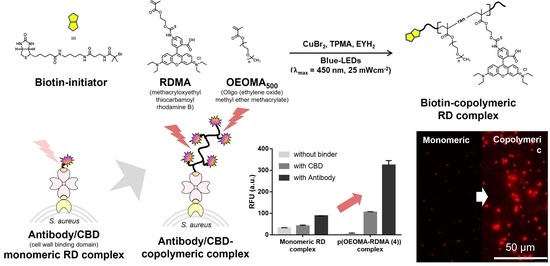
Highly Sensitive Detection of Bacteria by Binder-Coupled Multifunctional Polymeric Dyes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of Biotinylated Dye Copolymers
2.3. Characterization of Biotinylated Copolymeric Rhodamine B by 1H NMR Spectroscopy and Size Exclusion Chromatography with Multi-Angle Light Scattering (SEC–MALS) Detectors
2.4. Kinetics of Photoinduced ATRP
2.5. Photostability Assessment of Biotinylated Copolymeric Rhodamine B
2.6. Preparation of Cell-Wall Binding Domain (CBD)
2.7. Construction and Characterization of Antibody/CBD-Copolymeric Rhodamine B Complex
2.8. Confocal Laser Scanning Microscopy (CLSM)
3. Results and Discussion
3.1. Synthesis and Characterization of Biotinylated Copolymeric Rhodamine B
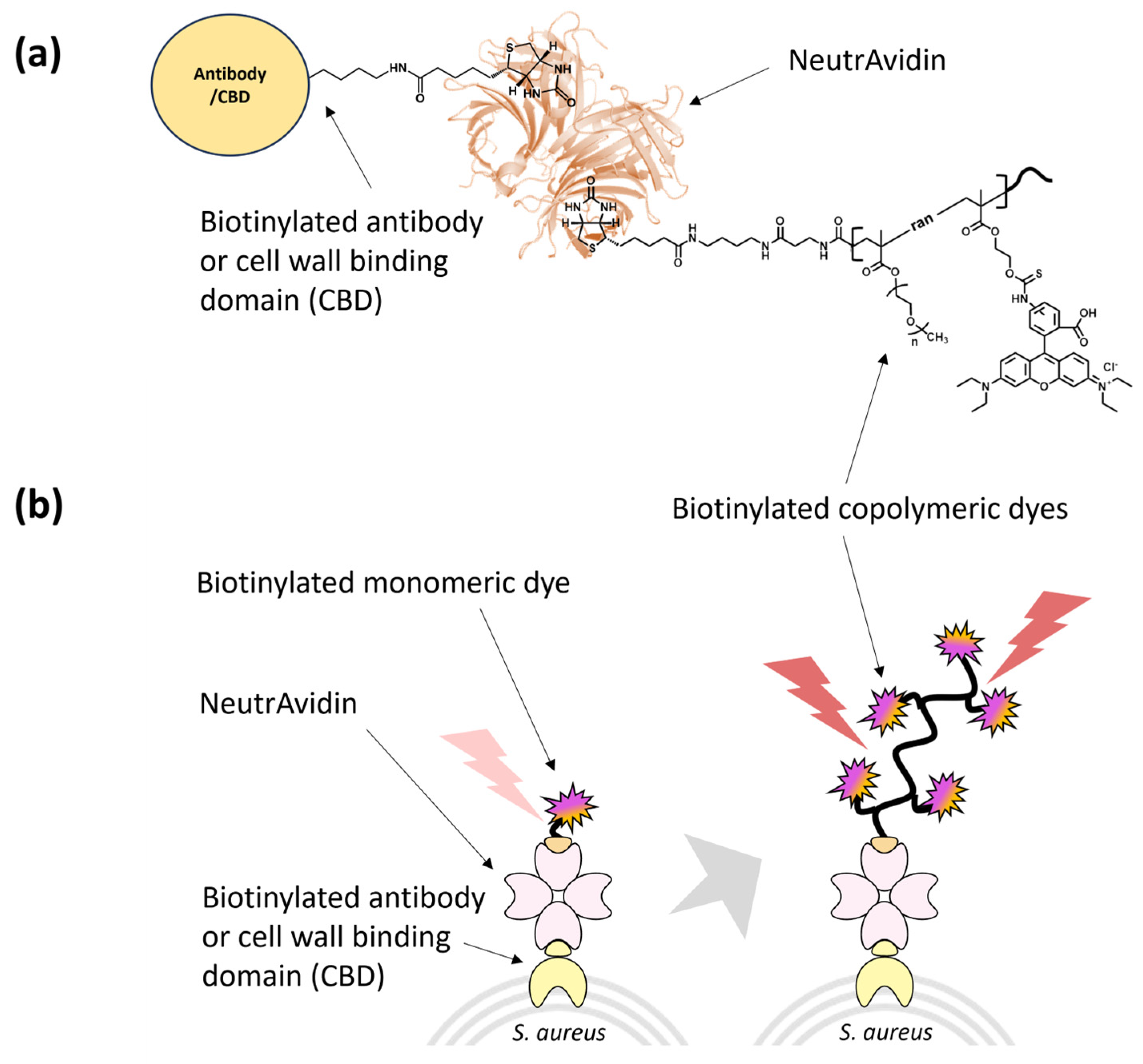
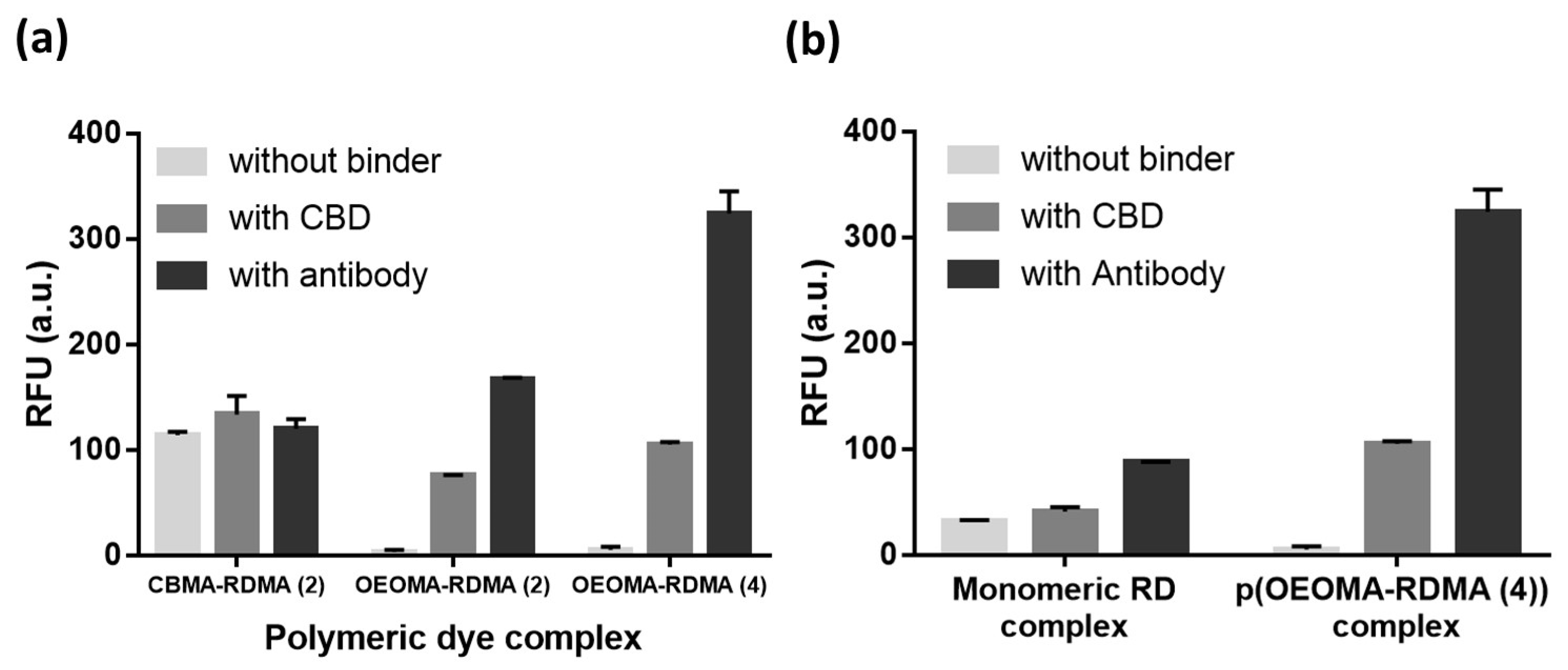
3.2. Characterization of Biotinylated Polymeric Dyes and Their Complexes with Selective Binders
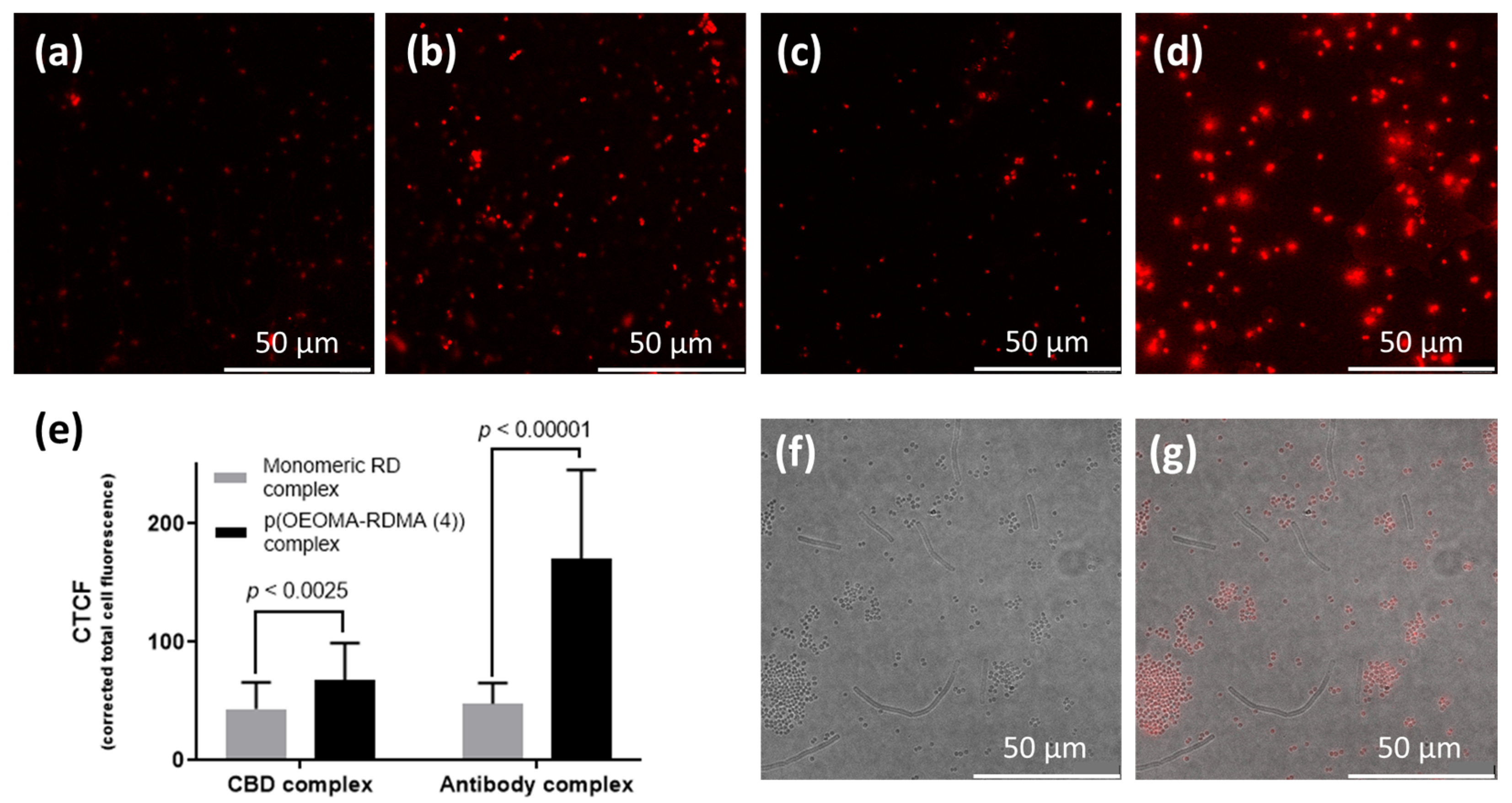
3.3. Antibody/CBD-Polymeric Dyes Complex for Bioimaging Application
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holmes, K.K.; Bertozzi, S.; Bloom, B.R.; Jha, P.; Gelband, H.; DeMaria, L.M.; Horton, S. Major Infectious Diseases: Key Messages from Disease Control Priorities, Third Edition. In Disease Control Priorities, Third Edition (Volume 6): Major Infectious Diseases; World Bank: Washington, DC, USA, 2017; pp. 1–27. [Google Scholar]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.A.; Park, S.Y.; Cha, Y.J.; Gopala, L.; Lee, M.H. Strategies of Detecting Bacteria Using Fluorescence-Based Dyes. Front. Chem. 2021, 9, 743923. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Rothman, R.E. PCR-based diagnostics for infectious diseases: Uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 2004, 4, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Váradi, L.; Luo, J.L.; Hibbs, D.E.; Perry, J.D.; Anderson, R.J.; Orenga, S.; Groundwater, P.W. Methods for the detection and identification of pathogenic bacteria: Past, present, and future. Chem. Soc. Rev. 2017, 46, 4818–4832. [Google Scholar] [CrossRef] [PubMed]
- Hameed, S.; Xie, L.; Ying, Y. Conventional and emerging detection techniques for pathogenic bacteria in food science: A review. Trends Food Sci. Technol. 2018, 81, 61–73. [Google Scholar] [CrossRef]
- Franco-Duarte, R.; Černáková, L.; Kadam, S.; Kaushik, K.S.; Salehi, B.; Bevilacqua, A.; Corbo, M.R.; Antolak, H.; Dybka-Stępień, K.; Leszczewicz, M.; et al. Advances in Chemical and Biological Methods to Identify Microorganisms—From Past to Present. Microorganisms 2019, 7, 130. [Google Scholar] [CrossRef] [Green Version]
- Yao, Z.; Carballido-López, R. Fluorescence Imaging for Bacterial Cell Biology: From Localization to Dynamics, From Ensembles to Single Molecules. Annu. Rev. Microbiol. 2014, 68, 459–476. [Google Scholar] [CrossRef]
- Guo, Z.; Zeng, J.; Liu, W.; Chen, Y.; Jiang, H.; Weizmann, Y.; Wang, X. Formation of bio-responsive nanocomposites for targeted bacterial bioimaging and disinfection. Chem. Eng. J. 2021, 426, 130726. [Google Scholar] [CrossRef]
- Si, Y.; Grazon, C.; Clavier, G.; Rieger, J.; Tian, Y.Y.; Audibert, J.F.; Sclavi, B.; Meallet-Renault, R. Fluorescent Copolymers for Bacterial Bioimaging and Viability Detection. ACS Sens. 2020, 5, 2843–2851. [Google Scholar] [CrossRef]
- Lu, L.L.; Suscovich, T.J.; Fortune, S.M.; Alter, G. Beyond binding: Antibody effector functions in infectious diseases. Nat. Rev. Immunol. 2018, 18, 46–61. [Google Scholar] [CrossRef]
- Dammes, N.; Peer, D. Monoclonal antibody-based molecular imaging strategies and theranostic opportunities. Theranostics 2020, 10, 938–955. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Liu, X.; Zu, Y. Oligonucleotide aptamers for pathogen detection and infectious disease control. Theranostics 2021, 11, 9133–9161, Review. [Google Scholar] [CrossRef]
- Davydova, A.; Vorobjeva, M.; Pyshnyi, D.; Altman, S.; Vlassov, V.; Venyaminova, A. Aptamers against pathogenic microorganisms. Crit. Rev. Microbiol. 2016, 42, 847–865. [Google Scholar] [CrossRef] [Green Version]
- Fischetti, V.A. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 2008, 11, 393–400. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Kwon, S.-J.; Kim, J.; Kane, R.S.; Dordick, J.S. Biocatalytic Nanocomposites for Combating Bacterial Pathogens. Annu. Rev. Chem. Biomol. Eng. 2017, 8, 87–113. [Google Scholar] [CrossRef]
- Bhagwat, A.; Mixon, M.; Collins, C.H.; Dordick, J.S. Opportunities for broadening the application of cell wall lytic enzymes. Appl. Microbiol. Biotechnol. 2020, 104, 9019–9040. [Google Scholar] [CrossRef] [PubMed]
- Li, B.H.; Lu, L.F.; Zhao, M.Y.; Lei, Z.H.; Zhang, F. An Efficient 1064 nm NIR-II Excitation Fluorescent Molecular Dye for Deep-Tissue High-Resolution Dynamic Bioimaging. Angew. Chem. -Int. Ed. 2018, 57, 7483–7487. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Li, M.F.; Ma, H.L.; Hu, Z.B.; Wang, X.Y.; Ma, R.; Jiang, Y.Y.; Sun, H.T.; Zhu, S.J.; Liang, Y.Y. Furan Donor for NIR-II Molecular Fluorophores with Enhanced Bioimaging Performance. Research 2023, 2023, 0039. [Google Scholar] [CrossRef]
- Chang, P.; Han, C.M.; Xu, H. Research progress of near infrared organic small-molecule electroluminescent materials. Chin. J. Liq. Cryst. Disp. 2021, 36, 62–77. [Google Scholar] [CrossRef]
- Hama, H.; Kurokawa, H.; Kawano, H.; Ando, R.; Shimogori, T.; Noda, H.; Fukami, K.; Sakaue-Sawano, A.; Miyawaki, A. Scale: A chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 2011, 14, 1481–1488. [Google Scholar] [CrossRef]
- Romei, M.G.; Boxer, S.G. Split Green Fluorescent Proteins: Scope, Limitations, and Outlook. Annu. Rev. Biophys. 2019, 48, 19–44. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, M. Green fluorescent protein (GFP): Applications, structure, and related photophysical behavior. Chem. Rev. 2002, 102, 759–781. [Google Scholar] [CrossRef] [PubMed]
- Vetschera, P.; Mishra, K.; Fuenzalida-Werner, J.P.; Chmyrov, A.; Ntziachristos, V.; Stiel, A.C. Characterization of Reversibly Switchable Fluorescent Proteins in Optoacoustic Imaging. Anal. Chem. 2018, 90, 10527–10535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ban, Q.F.; Li, Y.; Wu, S. Self-fluorescent polymers for bioimaging. View 2022, 3, 20200135. [Google Scholar] [CrossRef]
- Bentolila, A.; Totre, J.; Zozulia, I.; Levin-Elad, M.; Domb, A.J. Fluorescent Cyanoacrylate Monomers and Polymers for Fingermark Development. Macromolecules 2013, 46, 4822–4828. [Google Scholar] [CrossRef]
- Deng, H.P.; Su, Y.; Hu, M.X.; Jin, X.; He, L.; Pang, Y.; Dong, R.J.; Zhu, X.Y. Multicolor Fluorescent Polymers Inspired from Green Fluorescent Protein. Macromolecules 2015, 48, 5969–5979. [Google Scholar] [CrossRef]
- Adjili, S.; Favier, A.; Massin, J.; Bretonniere, Y.; Lacour, W.; Lin, Y.C.; Chatre, E.; Place, C.; Favard, C.; Muriaux, D.; et al. Synthesis of multifunctional lipid-polymer conjugates: Application to the elaboration of bright far-red fluorescent lipid probes. RSC Adv. 2014, 4, 15569–15578. [Google Scholar] [CrossRef]
- Duret, D.; Haftek-Terreau, Z.; Carretier, M.; Berki, T.; Ladaviere, C.; Monier, K.; Bouvet, P.; Marvel, J.; Leverrier, Y.; Charreyre, M.T.; et al. Labeling of native proteins with fluorescent RAFT polymer probes: Application to the detection of a cell surface protein using flow cytometry. Polym. Chem. 2018, 9, 1857–1868. [Google Scholar] [CrossRef]
- Jiang, R.M.; Huang, L.; Liu, M.Y.; Deng, F.J.; Huang, H.Y.; Tian, J.W.; Wen, Y.Q.; Cao, Q.Y.; Zhang, X.Y.; Wei, Y. Ultrafast microwave-assisted multicomponent tandem polymerization for rapid fabrication of AIE-active fluorescent polymeric nanoparticles and their potential utilization for biological imaging. Mater. Sci. Eng. C-Mater. Biol. Appl. 2018, 83, 115–120. [Google Scholar] [CrossRef]
- Huang, H.Y.; Jiang, R.M.; Ma, H.J.; Li, Y.S.; Zeng, Y.; Zhou, N.G.; Liu, L.J.; Zhang, X.Y.; Wei, Y. Fabrication of claviform fluorescent polymeric nanomaterials containing disulfide bond through an efficient and facile four-component Ugi reaction. Mater. Sci. Eng. C-Mater. Biol. Appl. 2021, 118, 111437. [Google Scholar] [CrossRef]
- Dong, J.D.; Jiang, R.M.; Huang, H.Y.; Chen, J.Y.; Tian, J.W.; Deng, F.J.; Dai, Y.F.; Wen, Y.Q.; Zhang, X.Y.; Wei, Y. Facile preparation of fluorescent nanodiamond based polymer nanoparticles via ring-opening polymerization and their biological imaging. Mater. Sci. Eng. C-Mater. Biol. Appl. 2020, 106, 110297. [Google Scholar] [CrossRef]
- Reisch, A.; Klymchenko, A.S. Fluorescent Polymer Nanoparticles Based on Dyes: Seeking Brighter Tools for Bioimaging. Small 2016, 12, 1968–1992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thapaliya, E.K.; Zhang, Y.; Dhakal, P.; Brown, A.S.; Wilson, J.N.; Collins, K.M.; Raymo, F.M. Bioimaging with Macromolecular Probes Incorporating Multiple BODIPY Fluorophores. Bioconjugate Chem. 2017, 28, 1519–1528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kloepfer, J.A.; Mielke, R.E.; Wong, M.S.; Nealson, K.H.; Stucky, G.; Nadeau, J.L. Quantum dots as strain- and metabolism-specific microbiological labels. Appl. Environ. Microbiol. 2003, 69, 4205–4213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalmers, N.I.; Palmer, R.J.; Du-Thumm, L.; Sullivan, R.; Shi, W.Y.; Kolenbrander, P.E. Use of quantum dot luminescent probes to achieve single-cell resolution of human oral bacteria in biofilms. Appl. Environ. Microbiol. 2007, 73, 630–636. [Google Scholar] [CrossRef] [Green Version]
- Gazouli, M.; Liandris, E.; Andreadou, M.; Sechi, L.A.; Masala, S.; Paccagnini, D.; Ikonomopoulos, J. Specific Detection of Unamplified Mycobacterial DNA by Use of Fluorescent Semiconductor Quantum Dots and Magnetic Beads. J. Clin. Microbiol. 2010, 48, 2830–2835. [Google Scholar] [CrossRef] [Green Version]
- Xue, X.H.; Pan, J.; Xie, H.M.; Wang, J.H.; Zhang, S. Fluorescence detection of total count of Escherichia coli and Staphylococcus aureus on water-soluble CdSe quantum dots coupled with bacteria. Talanta 2009, 77, 1808–1813. [Google Scholar] [CrossRef] [PubMed]
- Resch-Genger, U.; Grabolle, M.; Cavaliere-Jaricot, S.; Nitschke, R.; Nann, T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 2008, 5, 763–775. [Google Scholar] [CrossRef]
- Wolfbeis, O.S. An overview of nanoparticles commonly used in fluorescent bioimaging. Chem. Soc. Rev. 2015, 44, 4743–4768. [Google Scholar] [CrossRef] [Green Version]
- Bou, S.; Klymchenko, A.S.; Collot, M. Fluorescent labeling of biocompatible block copolymers: Synthetic strategies and applications in bioimaging. Mater. Adv. 2021, 2, 3213–3233. [Google Scholar] [CrossRef]
- Zhang, L.E.; Zhang, Z.K.; Liu, C.R.; Zhang, X.K.; Fan, Q.L.; Wu, W.; Jiangsu, X.Q. NIR-II Dye-Labeled Cylindrical Polymer Brushes for in Vivo Imaging. Acs Macro Lett. 2019, 8, 1623–1628. [Google Scholar] [CrossRef] [PubMed]
- Trofymchuk, K.; Valanciunaite, J.; Andreiuk, B.; Reisch, A.; Collot, M.; Klymchenko, A.S. BODIPY-loaded polymer nanoparticles: Chemical structure of cargo defines leakage from nanocarrier in living cells. J. Mater. Chem. B 2019, 7, 5199–5210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Zhu, X.L.; Cheng, Z.P.; Zhang, W.; Sun, B. Synthesis of poly(methyl methacrylate) labeled with fluorescein moieties via atom transfer radical polymerization. J. Macromol. Sci. Part A-Pure Appl. Chem. 2008, 45, 495–501. [Google Scholar] [CrossRef]
- Chaney, E.J.; Tang, L.; Tong, R.; Cheng, J.J.; Boppart, S.A. Lymphatic Biodistribution of Polylactide Nanoparticles. Mol. Imaging 2010, 9, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.J.; Li, X.; Zhang, J.B.; Chen, J.L.; Xu, B.; Fu, X.Q.; Tian, W.J. Folic acid-functionalized AIE Pdots based on amphiphilic PCL-b-PEG for targeted cell imaging. Polym. Chem. 2014, 5, 3824–3830. [Google Scholar] [CrossRef]
- Wang, K.; Luo, Y.M.; Huang, S.; Yang, H.B.; Liu, B.; Wang, M.F. Highly Fluorescent Polycaprolactones Decorated with Di(thiophene-2-yl)-diketopyrrolopyrrole: A Covalent Strategy of Tuning Fluorescence Properties in Solid States. J. Polym. Sci. Part A-Polym. Chem. 2015, 53, 1032–1042. [Google Scholar] [CrossRef]
- Lu, X.J.; Zhang, L.F.; Meng, L.Z.; Liu, Y.H. Synthesis of poly(N-isopropylacrylamide) by ATRP using a fluorescein-based initiator. Polym. Bull. 2007, 59, 195–206. [Google Scholar] [CrossRef]
- Breul, A.M.; Hager, M.D.; Schubert, U.S. Fluorescent monomers as building blocks for dye labeled polymers: Synthesis and application in energy conversion, biolabeling and sensors. Chem. Soc. Rev. 2013, 42, 5366–5407. [Google Scholar] [CrossRef]
- Wan, Q.; Liu, M.Y.; Mao, L.C.; Jiang, R.M.; Xu, D.Z.; Huang, H.Y.; Dai, Y.F.; Deng, F.J.; Zhang, X.Y.; Wei, Y. Preparation of PEGylated polymeric nanoprobes with aggregation-induced emission feature through the combination of chain transfer free radical polymerization and multicomponent reaction: Self-assembly, characterization and biological imaging applications. Mater. Sci. Eng. C-Mater. Biol. Appl. 2017, 72, 352–358. [Google Scholar] [CrossRef] [Green Version]
- Li, F.J.; Zhu, A.P.; Song, X.L.; Ji, L.J.; Wang, J. The internalization of fluorescence-labeled PLA nanoparticles by macrophages. Int. J. Pharm. 2013, 453, 506–513. [Google Scholar] [CrossRef]
- Guan, X.L.; Lai, S.J.; Su, Z.X. Facile Preparation and Potential Application of Water-Soluble Polymeric Temperature/pH Probes Bearing Fluorescein. J. Appl. Polym. Sci. 2011, 122, 1968–1975. [Google Scholar] [CrossRef]
- Kim, C.; Wallace, J.U.; Chen, S.H.; Merkel, P.B. Effects of dilution, polarization ratio, and energy transfer on photoalignment of liquid crystals using coumarin-containing polymer films. Macromolecules 2008, 41, 3075–3080. [Google Scholar] [CrossRef]
- Manickasundaram, S.; Kannan, P.; Kumaran, R.; Velu, R.; Ramamurthy, P.; Hassan, Q.M.A.; Palanisamy, P.K.; Senthil, S.; Narayanan, S.S. Holographic grating studies in pendant xanthene dyes containing poly(alkyloxymethacrylate)s. J. Mater. Sci. -Mater. Electron. 2011, 22, 25–34. [Google Scholar] [CrossRef]
- Berger, S.; Synytska, A.; Ionov, L.; Eichhorn, K.J.; Stamm, M. Stimuli-Responsive Bicomponent Polymer Janus Particles by “Grafting from”/“Grafting to” Approaches. Macromolecules 2008, 41, 9669–9676. [Google Scholar] [CrossRef]
- Li, G.; Bai, L.P.; Tao, F.R.; Deng, A.X.; Wang, L.P. A dual chemosensor for Cu2+ and Hg2+ based on a rhodamine-terminated water-soluble polymer in 100% aqueous solution. Analyst 2018, 143, 5395–5403. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.M.; Dai, L.; Liu, S.Y. Analyte-Reactive Amphiphilic Thermoresponsive Diblock Copolymer Micelles-Based Multifunctional Ratiometric Fluorescent Chemosensors. Macromolecules 2011, 44, 4699–4710. [Google Scholar] [CrossRef]
- Hu, J.M.; Zhang, X.Z.; Wang, D.; Hu, X.L.; Liu, T.; Zhang, G.Y.; Liu, S.Y. Ultrasensitive ratiometric fluorescent pH and temperature probes constructed from dye-labeled thermoresponsive double hydrophilic block copolymers. J. Mater. Chem. 2011, 21, 19030–19038. [Google Scholar] [CrossRef]
- Ma, C.P.; Xie, G.Y.; Zhang, X.Q.; Yang, L.T.; Li, Y.; Liu, H.L.; Wang, K.; Wei, Y. Biocompatible fluorescent polymers from PEGylation of an aggregation-induced emission dye. Dye. Pigment. 2017, 139, 672–680. [Google Scholar] [CrossRef] [Green Version]
- Nicolas, J.; San Miguel, V.; Mantovani, G.; Haddleton, D.M. Fluorescently tagged polymer bioconjugates from protein derived macroinitiators. Chem. Commun. 2006, 45, 4697–4699. [Google Scholar] [CrossRef] [Green Version]
- Madsen, J.; Canton, I.; Warren, N.J.; Themistou, E.; Blanazs, A.; Ustbas, B.; Tian, X.H.; Pearson, R.; Battaglia, G.; Lewis, A.L.; et al. Nile Blue-Based Nanosized pH Sensors for Simultaneous Far-Red and Near-Infrared Live Bioimaging. J. Am. Chem. Soc. 2013, 135, 14863–14870. [Google Scholar] [CrossRef]
- Truong, N.P.; Jones, G.R.; Bradford, K.G.E.; Konkolewicz, D.; Anastasaki, A. A comparison of RAFT and ATRP methods for controlled radical polymerization. Nat. Rev. Chem. 2021, 5, 859–869. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Yoon, J.A.; Kim, J.; Huang, C.F.; Matyjaszewski, K.; Kim, E. Excimer Emission from Self-Assembly of Fluorescent Diblock Copolymer Prepared by Atom Transfer Radical Polymerization. Chem. Mater. 2010, 22, 4426–4434. [Google Scholar] [CrossRef]
- Neugebauer, D.; Charasim, D.; Swinarew, A.; Stolarzewicz, A.; Krompiec, M.; Janeczek, H.; Simokaitiene, J.; Grazulevicius, J.V. Polymethacrylates with anthryl and carbazolyl groups prepared by atom transfer radical polymerization. Polym. J. 2011, 43, 448–454. [Google Scholar] [CrossRef] [Green Version]
- Spiniello, M.; Blencowe, A.; Qiao, G.G. Synthesis and characterization of fluorescently labeled core cross-linked star polymers. J. Polym. Sci. Part A-Polym. Chem. 2008, 46, 2422–2432. [Google Scholar] [CrossRef]
- Madsen, J.; Warren, N.J.; Armes, S.P.; Lewis, A.L. Synthesis of Rhodamine 6G-Based Compounds for the ATRP Synthesis of Fluorescently Labeled Biocompatible Polymers. Biomacromolecules 2011, 12, 2225–2234. [Google Scholar] [CrossRef]
- Yang, Q.A.; Jin, H.; Xu, Y.D.; Shen, Z.H.; Fan, X.H.; Zou, D.C.; Zhou, Q.F. Electroluminescent Block Copolymers Containing Oxadiazole and Thiophene via ATRP. J. Polym. Sci. Part A-Polym. Chem. 2010, 48, 5670–5678. [Google Scholar] [CrossRef]
- Yang, Q.; Xu, Y.D.; Jin, H.; Shen, Z.H.; Chen, X.F.; Zou, D.C.; Fan, X.H.; Zhou, Q.F. A Novel Mesogen-Jacketed Liquid Crystalline Electroluminescent Polymer with Both Thiophene and Oxadiazole in Conjugated Side Chain. J. Polym. Sci. Part A-Polym. Chem. 2010, 48, 1502–1515. [Google Scholar] [CrossRef]
- Trzebicka, B.; Szweda, R.; Kosowski, D.; Szweda, D.; Otulakowski, L.; Haladjova, E.; Dworak, A. Thermoresponsive polymer-peptide/protein conjugates. Prog. Polym. Sci. 2017, 68, 35–76. [Google Scholar] [CrossRef]
- Dimitrov, I.; Trzebicka, B.; Muller, A.H.E.; Dworak, A.; Tsvetanov, C.B. Thermosensitive water-soluble copolymers with doubly responsive reversibly interacting entities. Prog. Polym. Sci. 2007, 32, 1275–1343. [Google Scholar] [CrossRef]
- Yamamoto, S.; Pietrasik, J.; Matyjaszewski, K. Temperature- and pH-responsive dense copolymer brushes prepared by ATRP. Macromolecules 2008, 41, 7013–7020. [Google Scholar] [CrossRef]
- Nese, A.; Lebedeva, N.V.; Sherwood, G.; Averick, S.; Li, Y.C.; Gao, H.F.; Peteanu, L.; Sheiko, S.S.; Matyjaszewski, K. pH-Responsive Fluorescent Molecular Bottlebrushes Prepared by Atom Transfer Radical Polymerization. Macromolecules 2011, 44, 5905–5910. [Google Scholar] [CrossRef]
- Mielanczyk, A.; Skonieczna, M.; Bernaczek, K.; Neugebauer, D. Fluorescein nanocarriers based on cationic star copolymers with acetal linked sugar cores. Synthesis and biochemical characterization. Rsc Adv. 2014, 4, 31904–31913. [Google Scholar] [CrossRef]
- Zhao, K.; Cheng, Z.P.; Zhang, Z.B.; Zhu, J.; Zhu, X.L. Synthesis of fluorescent poly(methyl methacrylate) via AGET ATRP. Polym. Bull. 2009, 63, 355–364. [Google Scholar] [CrossRef]
- Li, Y.; Bai, Y.G.; Zheng, N.; Liu, Y.; Vincil, G.A.; Pedretti, B.J.; Cheng, J.J.; Zimmerman, S.C. Crosslinked dendronized polyols as a general approach to brighter and more stable fluorophores. Chem. Commun. 2016, 52, 3781–3784. [Google Scholar] [CrossRef] [PubMed]
- Chien, M.P.; Carlini, A.S.; Hu, D.H.; Barback, C.V.; Rush, A.M.; Hall, D.J.; Orr, G.; Gianneschi, N.C. Enzyme-Directed Assembly of Nanoparticles in Tumors Monitored by in Vivo Whole Animal Imaging and ex Vivo Super-Resolution Fluorescence Imaging. J. Am. Chem. Soc. 2013, 135, 18710–18713. [Google Scholar] [CrossRef] [Green Version]
- Yu, K.K.; Li, K.; Hou, J.T.; Yang, J.; Xie, Y.M.; Yu, X.Q. Rhodamine based pH-sensitive “intelligent” polymers as lysosome targeting probes and their imaging applications in vivo. Polym. Chem. 2014, 5, 5804–5812. [Google Scholar] [CrossRef]
- Hench, L.L.; Polak, J.M. Third-generation biomedical materials. Science 2002, 295, 1014–1017. [Google Scholar] [CrossRef] [Green Version]
- Anderson, D.G.; Burdick, J.A.; Langer, R. Materials science—Smart biomaterials. Science 2004, 305, 1923–1924. [Google Scholar] [CrossRef]
- Wang, J.S.; Matyjaszewski, K. Controlled/“Living” Radical Polymerization. Halogen Atom Transfer Radical Polymerization Promoted by a Cu(I)/Cu(II) Redox Process. Macromolecules 1995, 28, 7901–7910. [Google Scholar] [CrossRef]
- Patten, T.E.; Xia, J.; Abernathy, T.; Matyjaszewski, K. Polymers with very low polydispersities from atom transfer radical polymerization. Science 1996, 272, 866–868. [Google Scholar] [CrossRef]
- Matyjaszewski, K.; Xia, J.H. Atom transfer radical polymerization. Chem. Rev. 2001, 101, 2921–2990. [Google Scholar] [CrossRef]
- Corrigan, N.; Jung, K.; Moad, G.; Hawker, C.J.; Matyjaszewski, K.; Boyer, C. Reversible-deactivation radical polymerization (Controlled/living radical polymerization): From discovery to materials design and applications. Prog. Polym. Sci. 2020, 111, 101311. [Google Scholar] [CrossRef]
- Chmielarz, P.; Fantin, M.; Park, S.; Isse, A.A.; Gennaro, A.; Magenau, A.J.D.; Sobkowiak, A.; Matyjaszewski, K. Electrochemically mediated atom transfer radical polymerization (eATRP). Prog. Polym. Sci. 2017, 69, 47–78. [Google Scholar] [CrossRef]
- Pan, X.C.; Tasdelen, M.A.; Laun, J.; Junkers, T.; Yagci, Y.; Matyjaszewski, K. Photomediated controlled radical polymerization. Prog. Polym. Sci. 2016, 62, 73–125. [Google Scholar] [CrossRef] [Green Version]
- Matyjaszewski, K.; Tsarevsky, N.V. Macromolecular Engineering by Atom Transfer Radical Polymerization. J. Am. Chem. Soc. 2014, 136, 6513–6533. [Google Scholar] [CrossRef] [PubMed]
- Matyjaszewski, K.; Tsarevsky, N.V. Nanostructured functional materials prepared by atom transfer radical polymerization. Nat. Chem. 2009, 1, 276–288. [Google Scholar] [CrossRef]
- Ribelli, T.G.; Lorandi, F.; Fantin, M.; Matyjaszewski, K. Atom Transfer Radical Polymerization: Billion Times More Active Catalysts and New Initiation Systems. Macromol. Rapid Commun. 2019, 40, 1800616. [Google Scholar] [CrossRef]
- Enciso, A.E.; Fu, L.; Russell, A.J.; Matyjaszewski, K. A Breathing Atom-Transfer Radical Polymerization: Fully Oxygen-Tolerant Polymerization Inspired by Aerobic Respiration of Cells. Angew. Chem. Int. Ed. 2018, 57, 933–936. [Google Scholar] [CrossRef]
- Oh, J.K.; Min, K.; Matyjaszewski, K. Preparation of poly(oligo(ethylene glycol) monomethyl ether methacrylate) by homogeneous aqueous AGET ATRP. Macromolecules 2006, 39, 3161–3167. [Google Scholar] [CrossRef]
- Baker, S.L.; Kaupbayeva, B.; Lathwal, S.; Das, S.R.; Russell, A.J.; Matyjaszewski, K. Atom Transfer Radical Polymerization for Biorelated Hybrid Materials. Biomacromolecules 2019, 20, 4272–4298. [Google Scholar] [CrossRef]
- Jakubowski, W.; Min, K.; Matyjaszewski, K. Activators Regenerated by Electron Transfer for Atom Transfer Radical Polymerization of Styrene. Macromolecules 2006, 39, 39–45. [Google Scholar] [CrossRef]
- Magenau, A.J.D.; Strandwitz, N.C.; Gennaro, A.; Matyjaszewski, K. Electrochemically Mediated Atom Transfer Radical Polymerization. Science 2011, 332, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Matyjaszewski, K. Atom Transfer Radical Polymerization (ATRP): Current status and future perspectives. Macromolecules 2012, 45, 4015–4039. [Google Scholar] [CrossRef]
- Fung, A.K.K.; Coote, M.L. A mechanistic perspective on atom transfer radical polymerization. Polym. Int. 2021, 70, 918–926. [Google Scholar] [CrossRef]
- Lorandi, F.; Fantin, M.; Matyjaszewski, K. Atom Transfer Radical Polymerization: A Mechanistic Perspective. J. Am. Chem. Soc. 2022, 144, 15413–15430. [Google Scholar] [CrossRef] [PubMed]
- Jin-Shan Wang and Krzysztof, M. Controlled/”living” radical polymerization. atom transfer radical polymerization in the presence of transition-metal complexes. J. Am. Chem. Soc. 1995, 117, 5614–5615. [Google Scholar]
- Tsarevsky, N.V.; Pintauer, T.; Matyjaszewski, K. The rate of deactivation in atom transfer radical polymerization in protic and aqueous media. Polym. Prepr. (Am. Chem. Soc. Div. Polym. Chem.) 2004, 45, 1067–1068. [Google Scholar]
- Simakova, A.; Averick, S.E.; Konkolewicz, D.; Matyjaszewski, K. Aqueous ARGET ATRP. Macromolecules 2012, 45, 6371–6379. [Google Scholar] [CrossRef]
- Tsarevsky, N.V.; Matyjaszewski, K. “Green” atom transfer radical polymerization: From process design to preparation of well-defined environmentally friendly polymeric materials. Chem. Rev. 2007, 107, 2270–2299. [Google Scholar] [CrossRef]
- Ouchi, M.; Terashima, T.; Sawamoto, M. Transition Metal-Catalyzed Living Radical Polymerization: Toward Perfection in Catalysis and Precision Polymer Synthesis. Chem. Rev. 2009, 109, 4963–5050. [Google Scholar] [CrossRef]
- Pintauer, T.; Matyjaszewski, K. Atom transfer radical addition and polymerization reactions catalyzed by ppm amounts of copper complexes. Chem. Soc. Rev. 2008, 37, 1087–1097. [Google Scholar] [CrossRef] [PubMed]
- Konkolewicz, D.; Magenau, A.J.D.; Averick, S.E.; Simakova, A.; He, H.; Matyjaszewski, K. ICAR ATRP with ppm Cu catalyst in water. Macromolecules 2012, 45, 4461–4468. [Google Scholar] [CrossRef]
- Szczepaniak, G.; Fu, L.Y.; Jafari, H.; Kapil, K.; Matyjaszewski, K. Making ATRP More Practical: Oxygen Tolerance. Acc. Chem. Res. 2021, 54, 1779–1790. [Google Scholar] [CrossRef]
- Fu, L.; Wang, Z.; Lathwal, S.; Enciso, A.E.; Simakova, A.; Das, S.R.; Russell, A.J.; Matyjaszewski, K. Synthesis of Polymer Bioconjugates via Photoinduced Atom Transfer Radical Polymerization under Blue Light Irradiation. ACS Macro Lett. 2018, 7, 1248–1253. [Google Scholar] [CrossRef] [PubMed]
- Szczepaniak, G.; Łagodzińska, M.; Dadashi-Silab, S.; Gorczyński, A.; Matyjaszewski, K. Fully oxygen-tolerant atom transfer radical polymerization triggered by sodium pyruvate. Chem. Sci. 2020, 11, 8809–8816. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.K.; Szczepaniak, G.; Dadashi-Silab, S.; Lin, T.C.; Kowalewski, T.; Matyjaszewski, K. Cu-Catalyzed Atom Transfer Radical Polymerization: The Effect of Cocatalysts. Macromol. Chem. Phys. 2023, 224, 2200347. [Google Scholar] [CrossRef]
- Szczepaniak, G.; Jeong, J.; Kapil, K.; Dadashi-Silab, S.; Yerneni, S.S.; Ratajczyk, P.; Lathwal, S.; Schild, D.J.; Das, S.R.; Matyjaszewski, K. Open-air green-light-driven ATRP enabled by dual photoredox/copper catalysis. Chem. Sci. 2022, 13, 11540–11550. [Google Scholar] [CrossRef]
- Kapil, K.; Jazani, A.M.; Szczepaniak, G.; Murata, H.; Olszewski, M.; Matyjaszewski, K. Fully Oxygen-Tolerant Visible-Light-Induced ATRP of Acrylates in Water: Toward Synthesis of Protein-Polymer Hybrids. Macromolecules 2023, 56, 2017–2026. [Google Scholar] [CrossRef]
- Kapil, K.; Szczepaniak, G.; Martinez, M.R.; Murata, H.; Jazani, A.M.; Jeong, J.; Das, S.R.; Matyjaszewski, K. Visible-Light-Mediated Controlled Radical Branching Polymerization in Water. Angew. Chem. Int. Ed. 2023, 62, e202217658. [Google Scholar] [CrossRef]
- Cummings, C.; Murata, H.; Koepsel, R.; Russell, A.J. Tailoring enzyme activity and stability using polymer-based protein engineering. Biomaterials 2013, 34, 7437–7443. [Google Scholar] [CrossRef]
- Pan, X.; Lathwal, S.; Mack, S.; Yan, J.; Das, S.R.; Matyjaszewski, K. Automated Synthesis of Well-Defined Polymers and Biohybrids by Atom Transfer Radical Polymerization Using a DNA Synthesizer. Angew. Chem. Int. Ed. 2017, 56, 2740–2743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Cao, M.; Feng, Y.; Liang, R.; Fu, X.; Zhong, M. Site-Specifically Initiated Controlled/Living Branching Radical Polymerization: A Synthetic Route toward Hierarchically Branched Architectures. J. Am. Chem. Soc. 2019, 141, 794–799. [Google Scholar] [CrossRef] [PubMed]
- Kaupbayeva, B.; Murata, H.; Lucas, A.; Matyjaszewski, K.; Minden, J.S.; Russell, A.J. Molecular Sieving on the Surface of a Nano-Armored Protein. Biomacromolecules 2019, 20, 1235–1245. [Google Scholar] [CrossRef] [PubMed]
- Kaupbayeva, B.; Boye, S.; Munasinghe, A.; Murata, H.; Matyjaszewski, K.; Lederer, A.; Colina, C.M.; Russell, A.J. Molecular Dynamics-Guided Design of a Functional Protein-ATRP Conjugate That Eliminates Protein-Protein Interactions. Bioconjugate Chem. 2021, 32, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Averick, S.E.; Dey, S.K.; Grahacharya, D.; Matyjaszewski, K.; Das, S.R. Solid-phase incorporation of an ATRP initiator for polymer-DNA biohybrids. Angew. Chem. Int. Ed. 2014, 53, 2739–2744. [Google Scholar] [CrossRef] [PubMed]
- Lathwal, S.; Yerneni, S.S.; Boye, S.; Muza, U.L.; Takahashi, S.; Sugimoto, N.; Lederer, A.; Das, S.R.; Campbell, P.G.; Matyjaszewski, K. Engineering exosome polymer hybrids by atom transfer radical polymerization. Proc. Natl. Acad. Sci. USA 2021, 118, e2020241118. [Google Scholar] [CrossRef]
- Bateman, A.; Rawlings, N.D. The CHAP domain: A large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 2003, 28, 234–237. [Google Scholar] [CrossRef]
- Kim, D.; Kwon, S.-J.; Sauve, J.; Fraser, K.; Kemp, L.; Lee, I.; Nam, J.; Kim, J.; Dordick, J.S. Modular Assembly of Unique Chimeric Lytic Enzymes on a Protein Scaffold Possessing Anti-Staphylococcal Activity. Biomacromolecules 2019, 20, 4035–4043. [Google Scholar] [CrossRef]
- Szczepaniak, G.; Piatkowski, J.; Nogas, W.; Lorandi, F.; Yerneni, S.S.; Fantin, M.; Ruszczynska, A.; Enciso, A.E.; Bulska, E.; Grela, K.; et al. An isocyanide ligand for the rapid quenching and efficient removal of copper residues after Cu/TEMPO-catalyzed aerobic alcohol oxidation and atom transfer radical polymerization. Chem. Sci. 2020, 11, 4251–4262. [Google Scholar] [CrossRef] [Green Version]
- Erfani, A.; Seaberg, J.; Aichele, C.P.; Ramsey, J.D. Interactions between Biomolecules and Zwitterionic Moieties: A Review. Biomacromolecules 2020, 21, 2557–2573. [Google Scholar] [CrossRef]
- Brown, S.; Maria, J.P.S.; Walker, S. Wall Teichoic Acids of Gram-Positive Bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Entry | Sample Name | [M]/[Dye- M]/[I] | αM [b] (%) | Mn,th [c] | Mn, abs [d] | Ð |
|---|---|---|---|---|---|---|
| Biotin- | ||||||
| 1 | p(CBMA- | 200/2/1 | 80% | 36 600 | 40 200 | 1.25 |
| RDMA (2)) | ||||||
| Biotin- | ||||||
| 2 | p(OEOMA500- | 400/2/1 | 84% | 168 000 | 150 850 | 1.17 |
| RDMA (2)) | ||||||
| Biotin- | ||||||
| 3 | p(OEOMA500- | 400/4/1 | 78% | 158 600 | 149 200 | 1.23 |
| RDMA (4)) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kapil, K.; Xu, S.; Lee, I.; Murata, H.; Kwon, S.-J.; Dordick, J.S.; Matyjaszewski, K. Highly Sensitive Detection of Bacteria by Binder-Coupled Multifunctional Polymeric Dyes. Polymers 2023, 15, 2723. https://doi.org/10.3390/polym15122723
Kapil K, Xu S, Lee I, Murata H, Kwon S-J, Dordick JS, Matyjaszewski K. Highly Sensitive Detection of Bacteria by Binder-Coupled Multifunctional Polymeric Dyes. Polymers. 2023; 15(12):2723. https://doi.org/10.3390/polym15122723
Chicago/Turabian StyleKapil, Kriti, Shirley Xu, Inseon Lee, Hironobu Murata, Seok-Joon Kwon, Jonathan S. Dordick, and Krzysztof Matyjaszewski. 2023. "Highly Sensitive Detection of Bacteria by Binder-Coupled Multifunctional Polymeric Dyes" Polymers 15, no. 12: 2723. https://doi.org/10.3390/polym15122723
APA StyleKapil, K., Xu, S., Lee, I., Murata, H., Kwon, S.-J., Dordick, J. S., & Matyjaszewski, K. (2023). Highly Sensitive Detection of Bacteria by Binder-Coupled Multifunctional Polymeric Dyes. Polymers, 15(12), 2723. https://doi.org/10.3390/polym15122723