Human Serum Albumin Nanoparticles: Synthesis, Optimization and Immobilization with Antituberculosis Drugs
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
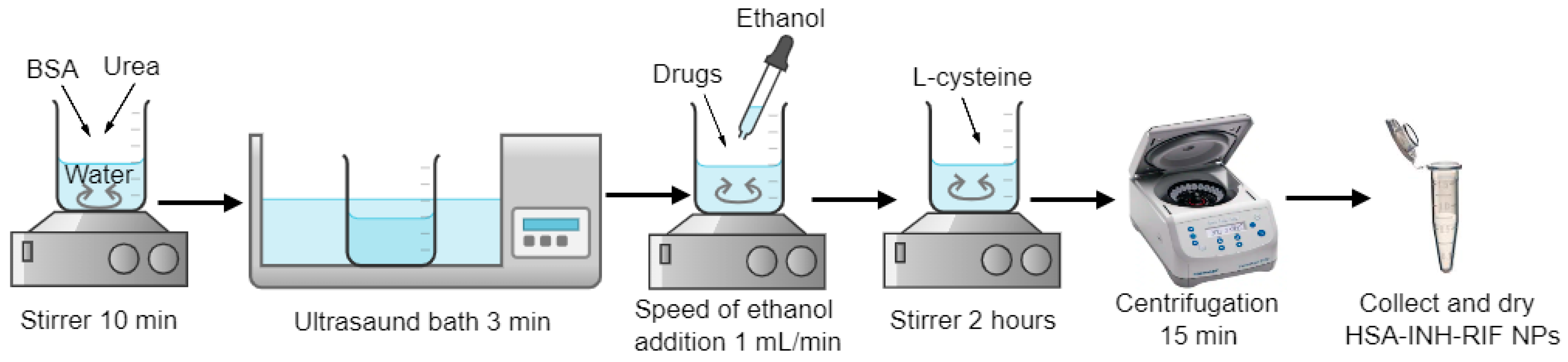
2.2. Production of Human Serum Albumin Nanoparticles Loaded with Isoniazid and Rifampicin by Desolvation
2.3. Determination of Particle Size, Polydispersity and ζ Potential
2.4. Determination of Loading Degree of Drugs and Nanoparticle Yield
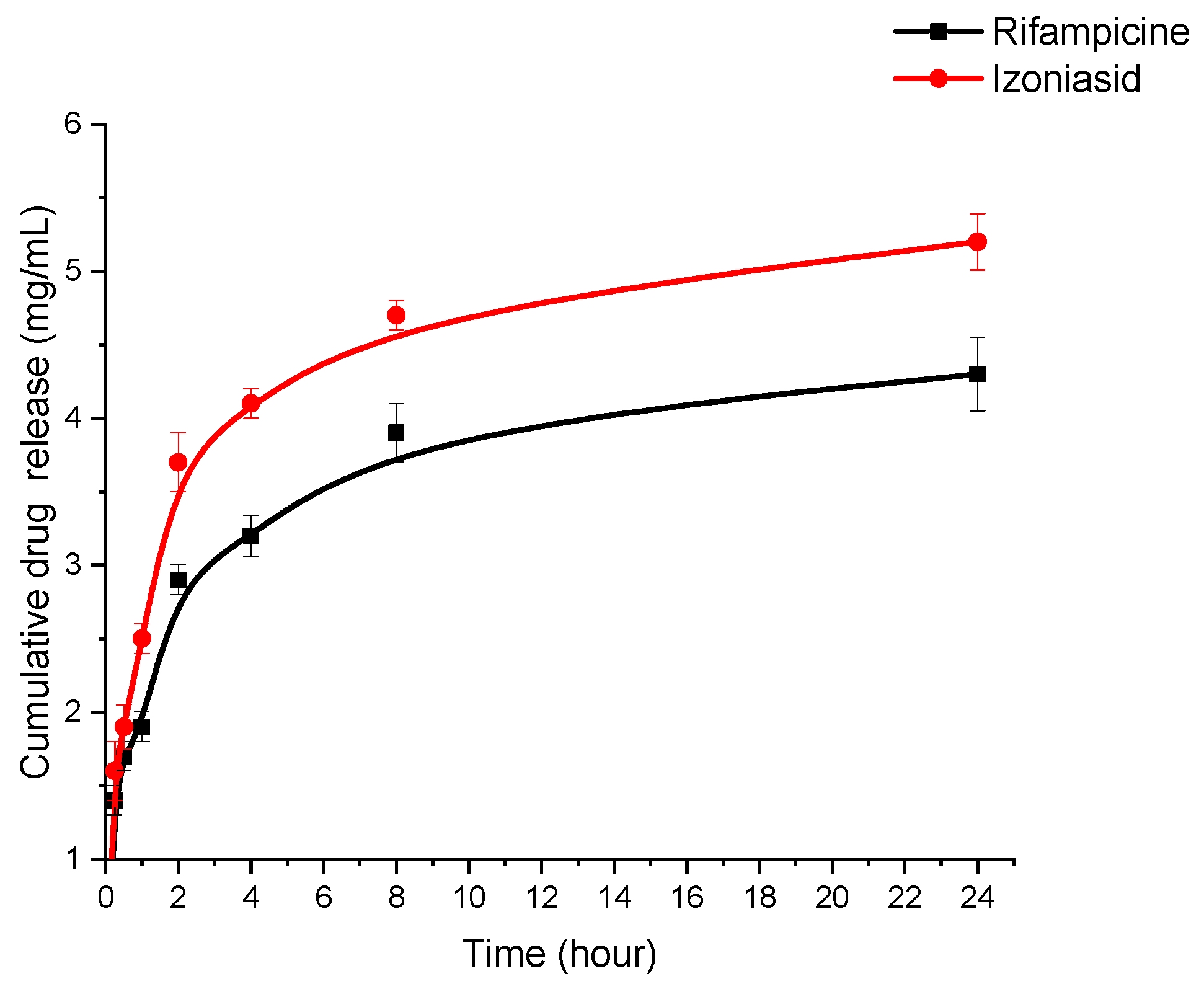
2.5. In Vitro Study of Drug Release from Polymer Nanoparticles
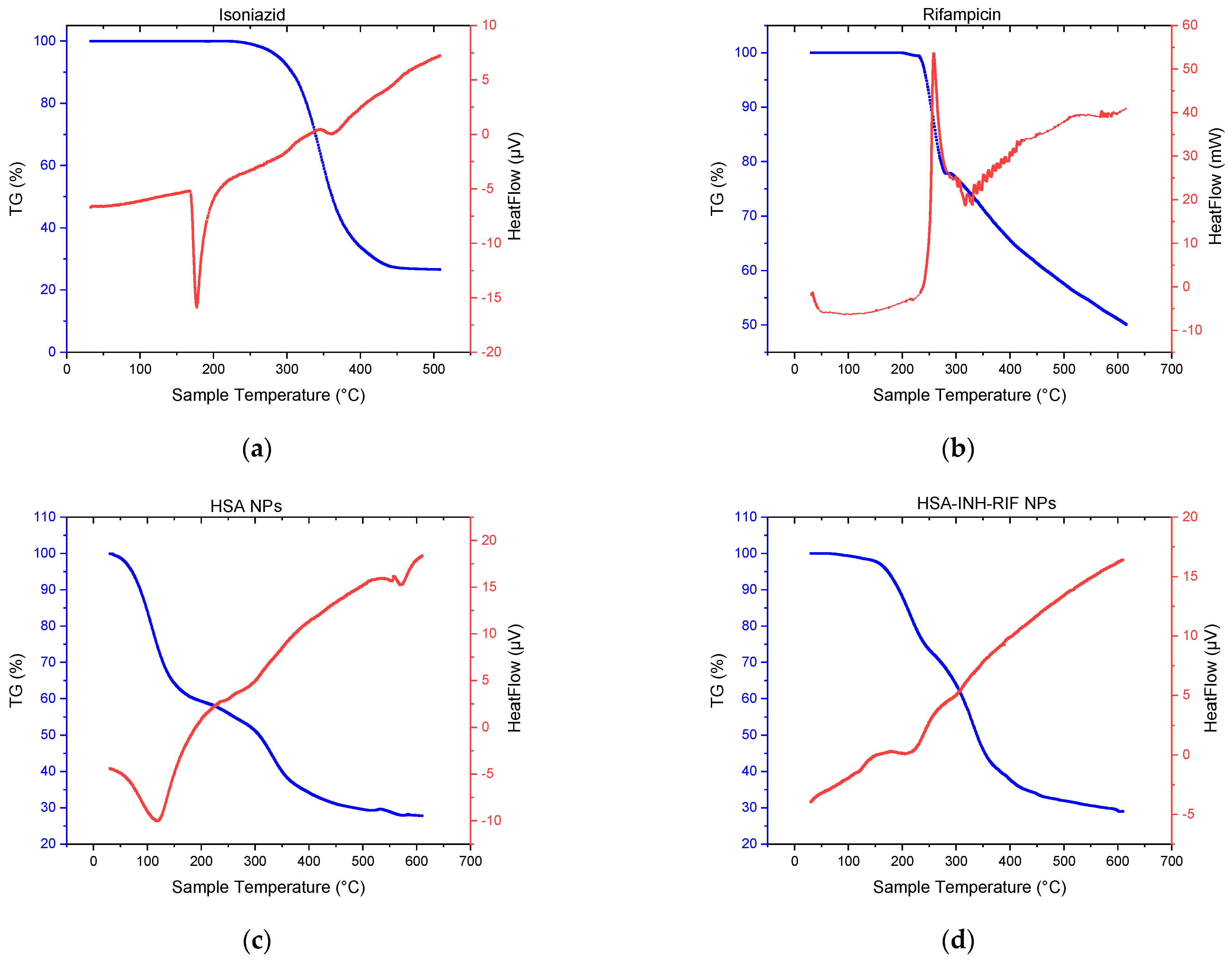
2.6. Thermogravimetric Analysis and Differential Scanning Calorimetry
2.7. Study of Prepared Nanoparticles by Infrared Spectroscopy
2.8. Experimental Design of Central Composite Design
3. Results
3.1. Optimization of Nanoparticles Preparation
3.2. Study of Physico-Chemical Parameters of the HSA-INH-RIF Nanoparticles
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2022; World Health Organization: Geneva, Switzerland, 2022; Licence: CC BY-NC-SA 3.0 IGO.
- Ge, Z.; Ma, R.; Xu, G.; Chen, Z.; Zhang, D.; Wang, Q.; Hei, L.; Ma, W. Development and In Vitro Release of Isoniazid and Rifampicin-Loaded Bovine Serum Albumin Nanoparticles. Med. Sci. Monit. 2018, 24, 473–478. [Google Scholar] [CrossRef]
- Sotgiu, G.; Centis, R.; D’ambrosio, L.; Migliori, G.B. Tuberculosis Treatment and Drug Regimens. Cold Spring Harb. Perspect. Med. 2015, 5, a017822. [Google Scholar] [CrossRef]
- Mazlan, M.K.N.; Mohd Tazizi, M.H.D.; Ahmad, R.; Noh, M.A.A.; Bakhtiar, A.; Wahab, H.A.; Mohd Gazzali, A. Antituberculosis Targeted Drug Delivery as a Potential Future Treatment Approach. Antibiotics 2021, 10, 908. [Google Scholar] [CrossRef]
- Jhun, B.W.; Koh, W.J. Treatment of Isoniazid-Resistant Pulmonary Tuberculosis. Tuberc. Respir. Dis. 2020, 83, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Somasundaram, S.; Ram, A.; Sankaranarayanan, L. Isoniazid and Rifampicin as Therapeutic Regimen in the Current Era: A Review. J. Tuberc. Res. 2014, 2, 40–51. [Google Scholar] [CrossRef]
- Kumar, P.S.; Vidya, R.; Tabassum; Jageer, M. Anti-Tuberculosis Treatment: Induced Hepatotoxicity—A Case Report. EJIFCC 2020, 31, 242–247. [Google Scholar]
- Yakar, F.; Yildiz, N.; Yakar, A.; Kılıçaslan, Z. Isoniazid- and rifampicin-induced thrombocytopenia. Multidiscip. Respir. Med. 2013, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Combrink, M.; Loots, D.T.; du Preez, I. Metabolomics describes previously unknown toxicity mechanisms of isoniazid and rifampicin. Toxicol. Lett. 2020, 322, 104–110. [Google Scholar] [CrossRef]
- Shin, H.J.; Chang, J.S.; Kim, M.S.; Koh, B.G.; Park, H.Y.; Kim, T.O.; Park, C.K.; Oh, I.J.; Kim, Y.I.; Lim, S.C.; et al. Hypersensitivity reactions to multiple anti-tuberculosis drugs. PLoS ONE 2021, 16, e0246291. [Google Scholar] [CrossRef] [PubMed]
- Nasiruddin, M.; Neyaz, M.K.; Das, S. Nanotechnology-Based Approach in Tuberculosis Treatment. Tuberc. Res. Treat. 2017, 2017, 4920209. [Google Scholar] [CrossRef]
- Nabi, B.; Rehman, S.; Aggarwal, S.; Baboota, S.; Ali, J. Nano-based anti-tubercular drug delivery: An emerging paradigm for improved therapeutic intervention. Drug Deliv. Transl. Res. 2020, 10, 1111–1121. [Google Scholar] [CrossRef]
- Borah Slater, K.; Kim, D.; Chand, P.; Xu, Y.; Shaikh, H.; Undale, V. A Current Perspective on the Potential of Nanomedicine for Anti-Tuberculosis Therapy. Trop. Med. Infect. Dis. 2023, 8, 100. [Google Scholar] [CrossRef]
- Baranyai, Z.; Soria-Carrera, H.; Alleva, M.; Millán-Placer, A.C.; Lucía, A.; Martín-Rapún, R.; Aínsa, J.A.; la Fuente, J.M. Nanotechnology-Based Targeted Drug Delivery: An Emerging Tool to Overcome Tuberculosis. Adv. Ther. 2020, 4, 2000113. [Google Scholar] [CrossRef]
- Donnellan, S.; Giardiello, M. Nanomedicines towards targeting intracellular Mtb for the treatment of tuberculosis. J. Interdiscip. Nanomed. 2019, 4, 76–85. [Google Scholar] [CrossRef]
- Tazhbayev, Y.M.; Galiyeva, A.R.; Zhumagaliyeva, T.S.; Burkeyev, M.Z.; Kazhmuratova, A.T.; Zhakupbekova, E.Z.; Zhaparova, L.Z.; Bakibayev, A.A. Synthesis and characterization of isoniazid immobilized polylactide-co-glycolide nanoparticles. Bull. Karaganda Univ. Chem. Ser. 2021, 101, 61–70. [Google Scholar] [CrossRef]
- Idrees, H.; Zaidi, S.Z.J.; Sabir, A.; Khan, R.U.; Zhang, X.; Hassan, S.U. A Review of Biodegradable Natural Polymer-Based Nanoparticles for Drug Delivery Applications. Nanomaterials 2020, 10, 1970. [Google Scholar] [CrossRef] [PubMed]
- Hornok, V. Serum Albumin Nanoparticles: Problems and Prospects. Polymers 2021, 13, 3759. [Google Scholar] [CrossRef]
- Martínez-López, A.L.; Pangua, C.; Reboredo, C.; Campión, R.; Morales-Gracia, J.; Irache, J.M. Protein-based nanoparticles for drug delivery purposes. Int. J. Pharm. 2020, 581, 119289. [Google Scholar] [CrossRef] [PubMed]
- Hassanin, I.; Elzoghby, A. Albumin-based nanoparticles: A promising strategy to overcome cancer drug resistance. Cancer Drug Resist. 2020, 3, 930–946. [Google Scholar] [CrossRef] [PubMed]
- Esim, O.; Hascicek, C. Albumin-based Nanoparticles as Promising Drug Delivery Systems for Cancer Treatment. Curr. Pharm. Anal. 2021, 17, 346–359. [Google Scholar] [CrossRef]
- Patel, A.; Redinger, N.; Richter, A.; Woods, A.; Neumann, P.R.; Keegan, G.; Childerhouse, N.; Imming, P.; Schaible, U.E.; Forbes, B.; et al. In vitro and in vivo antitubercular activity of benzothiazinone-loaded human serum albumin nanocarriers designed for inhalation. J. Control. Release 2020, 328, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Choi, D.W.; Kim, H.N.; Park, C.G.; Lee, W.; Park, H.H. Protein-Based Nanoparticles as Drug Delivery Systems. Pharmaceutics 2020, 12, 604. [Google Scholar] [CrossRef] [PubMed]
- Elzoghby, A.O.; Samy, W.M.; Elgindy, N.A. Albumin-based nanoparticles as potential controlled release drug delivery systems. J. Control. Release 2012, 157, 168–182. [Google Scholar] [CrossRef]
- Wang, G.; Uludag, H. Recent developments in nanoparticle-based drug delivery and targeting systems with emphasis on protein-based nanoparticles. Expert Opin. Drug Deliv. 2008, 25, 499–515. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Prajapati, A. Albumin and functionalized albumin nanoparticles: Production strategies, characterization, and target indications. Asian Biomed. 2020, 14, 217–242. [Google Scholar] [CrossRef]
- Tazhbayev, Y.; Galiyeva, A.; Zhumagaliyeva, T.; Burkeyev, M.; Karimova, B. Isoniazid-Loaded Albumin Nanoparticles: Taguchi Optimization Method. Polymers 2021, 13, 3808. [Google Scholar] [CrossRef]
- Shankar, K.R.; Ameta, R.K.; Singh, M. Preparation of BSA nanoparticles using aqueous urea at T = 308.15, 313.15 and 318.15 K as a function of temperature. J. Mol. Liq. 2016, 216, 808–813. [Google Scholar] [CrossRef]
- Weber, C.; Coester., C.; Kreuter., J.; Langer., K. Desolvation process and surface characterisation of protein nanoparticles. Int. J. Pharm. 2000, 194, 91–102. [Google Scholar] [CrossRef]
- Langer, K.; Balthasar, S.; Vogel, V.; Dinauer, N.; Von Briesen, H.; Schubert, D. Optimization of the preparation process for human serum albumin (HSA) nanoparticles. Int. J. Pharm. 2003, 257, 169–180. [Google Scholar] [CrossRef]
- Tazhbayev, Y.M.; Burkeyev, M.Z.; Zhaparova, L.Z.; Zhumagaliyeva, T.S.; Agdarbek, A.A. Albumin nanoparticles loaded with the antitumor drug «Hydroxycarbamide» by the incorporation method. Bull. Karaganda Univ. Chem. Ser. 2019, 93, 48–53. [Google Scholar] [CrossRef]
- Von Storp, B.; Engel, A.; Boeker, A.; Ploeger, M.; Langer, K. Albumin nanoparticles with predictable size by desolvation procedure. J. Microencapsul. 2012, 29, 138–146. [Google Scholar] [CrossRef]
- Tazhbayev, Y.; Mukashev, O.; Burkeev, M.; Kreuter, J. Hydroxyurea-Loaded Albumin Nanoparticles: Preparation, Characterization, and In Vitro Studies. Pharmaceutics 2019, 11, 410. [Google Scholar] [CrossRef] [PubMed]
- Tazhbayev, Y.; Mukashev, O.; Burkeyev, M.; Lozinsky, V.I. Synthesis and Comparative Study of Nanoparticles Derived from Bovine and Human Serum Albumins. Polymers 2020, 12, 1301. [Google Scholar] [CrossRef]
- Grigoryan, K.R. Denaturation of human serum albumin at the presence of urea–dimethylsulfoxid complexes. Proc. YSU B Chem. Biol. Sci. 2010, 44, 3–6. [Google Scholar] [CrossRef]
- Kimura, K.; Yamasaki, K.; Nakamura, H.; Haratake, M.; Taguchi, K.; Otagiri, M. Preparation and in Vitro Analysis of Human Serum Albumin Nanoparticles Loaded with Anthracycline Derivatives. Chem. Pharm. Bull. 2018, 66, 382–390. [Google Scholar] [CrossRef]
- Yang, X.; Patel, S.; Sheng, Y.; Pal, D.; Mitra, A.K. Statistical Design for Formulation Optimization of Hydrocortisone Butyrate-Loaded PLGA Nanoparticles. AAPS PharmSciTech 2014, 15, 569–587. [Google Scholar] [CrossRef] [PubMed]
- De, A.; Kuppuswamy, G.; Jaiswal, A. Implementation of two different experimental designs for screening and optimization of process parameters for metformin-loaded carboxymethyl chitosan formulation. Drug Dev. Ind. Pharm. 2019, 45, 1821–1834. [Google Scholar] [CrossRef]
- Das, P.; Das, M.K. Production and physicochemical characterization of nanocosmeceuticals. Nanocosmeceuticals 2022, 95–138. [Google Scholar] [CrossRef]
- Marsalek, R.; Kotyrba, M.; Volna, E.; Jarusek, R. Neural Network Modelling for Prediction of Zeta Potential. Mathematics 2021, 9, 3089. [Google Scholar] [CrossRef]
- Fernandes, H.P.; Cesar, C.L.; de Lourdes Barjas-Castro, M. Electrical properties of the red blood cell membrane and immunohematological investigation. Rev. Bras. Hematol. E Hemoter. 2011, 33, 297–301. [Google Scholar] [CrossRef]
- Galiyeva, A.R.; Tazhbayev, Y.M.; Zhumagaliyeva, T.S.; Sadyrbekov, D.T.; Kaikenov, D.A.; Karimova, B.N.; Shokenova, S.S. Polylactide-co-glycolide nanoparticles immobilized with isoniazid: Optimization using the experimental Taguchi method. Bull. Univ. Karaganda Chem. 2022, 105, 69–77. [Google Scholar] [CrossRef]
- Alves, R.; da Silva Reis, T.V.; da Silva, L.C.C.; Storpírtis, S.; Mercuri, L.P.; do Rosario Matos, J. Thermal behavior and decomposition kinetics of rifampicin polymorphs under isothermal and non-isothermal conditions. Braz. J. Pharm. Sci. 2010, 46, 343–351. [Google Scholar] [CrossRef]
- Gunasekaran, S.; Sailatha, E.; Seshadri, S.; Kumaresan, S. FTIR, FT Raman spectra and molecular structural confirmation of isoniazid. Indian J. Pure Appl. Phys. 2009, 47, 12–18. [Google Scholar]
- Sharma, A.; Puri, V.; Kumar, P.; Singh, I.; Huanbutta, K. Development and Evaluation of Rifampicin Loaded Alginate–Gelatin Biocomposite Microfibers. Polymers 2021, 13, 1514. [Google Scholar] [CrossRef] [PubMed]
- Ivashchenko, O.; Tomila, T.; Ulyanchich, N.; Yarmola, T.; Uvarova, I. Fourier-Transform Infrared Spectroscopy of Antibiotic Loaded Ag-Free and Ag-Doped Hydroxyapatites. Adv. Sci. Eng. Med. 2014, 6, 193–202. [Google Scholar] [CrossRef]
- HPLC Determination of Rifampicin on Newcrom a Column. Available online: https://sielc.com/hplc-determination-of-rifampicin (accessed on 14 June 2023).
- Mariappan, T.T.; Singh, B.; Singh, S. A Validated Reversed-Phase (C18) HPLC Method for Simultaneous Determination of Rifampicin, Isoniazid and Pyrazinamide in USP Dissolution Medium and Simulated Gastric Fluid. Pharm. Pharmacol. Commun. 2000, 6, 345–349. [Google Scholar] [CrossRef]
- Saraogi, G.K.; Gupta, P.; Gupta, U.D.; Jain, N.K.; Agrawal, G.P. Gelatin nanocarriers as potential vectors for effective management of tuberculosis. Int. J. Pharm. 2010, 385, 143–149. [Google Scholar] [CrossRef]






| Independent Variable | Measuring Unit | Variable Levels | ||||
|---|---|---|---|---|---|---|
| Star-Low (−1.82) | Low (−1) | Center (0) | High (1) | Star-High (1.82) | ||
| A: [HSA] | mg/mL | 10 | 20 | 40 | 80 | 100 |
| B: [Urea] | mol/L | 3 | 4 | 5 | 6 | 7 |
| C: [L-cysteine] | mg/mL | 0.1 | 0.5 | 1.25 | 2 | 2.5 |
| D: [INH] | mg/mL | 2 | 4 | 6 | 8 | 10 |
| E: [Rif] | mg/mL | 2 | 4 | 6 | 8 | 10 |
| Formulation Code | A: [HSA] (mg/mL) | B: [Urea] (mol/L) | C: [L-Cysteine] (mg/mL) | D: [INH] (mg/mL) | E: [RIF] (mg/mL) | Size (nm) | PDI | ζ Potential (mV) | Loading Degree of INH (%) | Loading Degree of RIF (%) | NP Yield (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NP1 | 80 | 4 | 2 | 8 | 4 | 304.7 ± 9.1 | 0.108 ± 0.028 | −30.0 ± 1.6 | 17 | 17 | 66 |
| NP2 | 40 | 5 | 1.25 | 6 | 10 | 351.6 ± 7.3 | 0.146 ± 0.003 | −14.9 ± 2.4 | 21 | 25 | 21 |
| NP3 | 80 | 6 | 2 | 4 | 4 | 204.7 ± 4.1 | 0.135 ± 0.037 | −31.1 ± 1.6 | 15 | 7 | 54 |
| NP4 | 40 | 5 | 1.25 | 6 | 6 | 191.4 ± 9.4 | 0.208 ± 0.009 | −19.7 ± 1.8 | 6 | 12 | 66 |
| NP5 | 40 | 5 | 2.5 | 6 | 6 | 239.6 ± 8.8 | 0.452 ± 0.039 | −14.8 ± 2.6 | 30 | 51 | 34 |
| NP6 | 100 | 5 | 1.25 | 6 | 6 | 184.2 ± 9.6 | 0.446 ± 0.004 | −18.5 ± 2.6 | 20 | 39 | 6 |
| NP7 | 40 | 5 | 1.25 | 6 | 6 | 336.6 ± 9.5 | 0.345 ± 0.048 | −20.0 ± 1.1 | 53 | 32 | 33 |
| NP8 | 40 | 5 | 0.1 | 6 | 6 | 156.1 ± 8.4 | 0.198 ± 0.043 | −24.7 ± 1.4 | 7 | 13 | 68 |
| NP9 | 80 | 6 | 0.5 | 8 | 4 | 231.4 ± 1.8 | 0.207 ± 0.049 | −34.1 ± 1.5 | 22 | 22 | 12 |
| NP10 | 40 | 5 | 1.25 | 2 | 6 | 177.5 ± 5.3 | 0.171 ± 0.039 | −22.9 ± 1.7 | 51 | 51 | 27 |
| NP11 | 40 | 5 | 1.25 | 6 | 10 | 289.5 ± 2.9 | 0.145 ± 0.004 | −24.4 ± 1.8 | 25 | 78 | 5 |
| NP12 | 20 | 4 | 2 | 8 | 8 | 233.2 ± 5.6 | 0.191 ± 0.047 | −19.6 ± 2.2 | 26 | 26 | 43 |
| NP13 | 80 | 4 | 2 | 4 | 8 | 191.6 ± 9.7 | 0.335 ± 0.016 | −35.2 ± 1.6 | 21 | 21 | 53 |
| NP14 | 20 | 6 | 2 | 8 | 4 | 232.1 ± 3.9 | 0.424 ± 0.006 | −22.7 ± 1.1 | 18 | 90 | 14 |
| NP15 | 80 | 4 | 0.5 | 8 | 8 | 215.5 ± 2.4 | 0.414 ± 0.075 | −32.1 ± 1.5 | 32 | 33 | 69 |
| NP16 | 40 | 7 | 1.25 | 6 | 6 | 136.3 ± 9.7 | 0.189 ± 0.038 | −11.2 ± 2.1 | 23 | 24 | 39 |
| NP17 | 40 | 5 | 1.25 | 6 | 6 | 216.5 ± 5.4 | 0.241 ± 0.025 | −22.6 ± 1.4 | 34 | 21 | 51 |
| NP18 | 40 | 5 | 1.25 | 6 | 6 | 134.2 ± 7.7 | 0.259 ± 0.001 | −22.6 ± 1.9 | 23 | 25 | 49 |
| NP19 | 20 | 6 | 0.5 | 8 | 8 | 243.2 ± 5.8 | 0.193 ± 0.005 | −16.0 ± 1.6 | 29 | 22 | 7 |
| NP20 | 40 | 5 | 1.25 | 6 | 6 | 134.2 ± 7.7 | 0.127 ± 0.030 | −18.3 ± 2.6 | 19 | 30 | 38 |
| NP21 | 10 | 5 | 1.25 | 6 | 6 | 253.0 ± 5.7 | 0.248 ± 0.019 | −6.2 ± 0.7 | 39 | 39 | 15 |
| NP22 | 40 | 3 | 1.25 | 6 | 6 | 221.2 ± 4.4 | 0.443 ± 0.056 | −18.4 ± 2.2 | 26 | 23 | 36 |
| NP23 | 20 | 6 | 2 | 4 | 8 | 327.3 ± 6.6 | 0.347 ± 0.002 | −22.7 ± 1.1 | 42 | 21 | 20 |
| NP24 | 40 | 5 | 1.25 | 10 | 6 | 152.8 ± 6.5 | 0.187 ± 0.016 | −11.2 ± 3.7 | 20 | 19 | 26 |
| NP25 | 20 | 4 | 0.5 | 4 | 4 | 174.9 ± 2.9 | 0.214 ± 0.007 | −22.7 ± 1.3 | 7 | 7 | 63 |
| NP26 | 80 | 6 | 0.5 | 4 | 8 | 159.6 ± 2.8 | 0.189 ± 0.032 | −19.3 ± 2.2 | 8 | 26 | 45 |
| Response | Source | Sum of Squares | Degree of Freedom | Mean Square | F-Value | p-Value | |
|---|---|---|---|---|---|---|---|
| Size | Model | 8.643 × 1012 | 21 | 4.116 × 1011 | 10.52 | 0.0171 | significant |
| Pure Error | 1.564 × 1011 | 4 | 3.911 × 1010 | ||||
| Residual | 1.564 × 1011 | 4 | 3.911 × 1010 | ||||
| Lack of Fit | 5.223 × 1011 | 1 | 5.223 × 1011 | 13.36 | 0.0217 | ||
| Cor Total | 8.800 × 1012 | 25 | |||||
| Loading degree of INH | Model | 6.619 × 108 | 20 | 3.310 × 107 | 4.70 | 0.0469 | significant |
| Pure Error | 6.019 × 106 | 4 | 1.505 × 106 | ||||
| Residual | 6.019 × 106 | 4 | 1.505 × 106 | ||||
| Lack of Fit | 2.917 × 107 | 1 | 2.917 × 107 | 19.39 | 0.0117 | ||
| Cor Total | 6.971 × 108 | 25 | |||||
| Loading degree of RIF | Model | 7.385 × 109 | 2 | 3.692 × 108 | 4.93 | 0.0426 | significant |
| Pure Error | 2.429 × 106 | 4 | 6.072 × 105 | ||||
| Residual | 2.429 × 106 | 4 | 6.072 × 105 | ||||
| Lack of Fit | 3.722 × 108 | 1 | 3.722 × 108 | 612.97 | <0.0001 | ||
| Cor Total | 7.759 × 109 | 25 |
| Size (nm) | PDI | ζ Potential (mV) | Loading Degree of Rifampicin, % | Loading Degree of Isoniazid, % | NP Yield, % | |
|---|---|---|---|---|---|---|
| Predicted | 214.2 | 0.246 | −29.6 | 43 | 29 | 44 |
| Experimental | 216.7 ± 3.7 | 0.286 ± 4.9 | −26.7 ± 1.5 | 44 | 27 | 45 |
| Error % | 1.2 | 16 | 11 | 2.3 | 6.9 | 2.3 |
| Time | Size (nm) | ζ Potential (mV) |
|---|---|---|
| Immediately after purification | 216.7 ± 3.7 | −26.7 ± 1.5 |
| After 1 h | 217.7 ± 2.4 | −25.9 ± 1.9 |
| After 2 h | 219.4 ± 2.1 | −26.1 ± 1.4 |
| After 4 h | 227.1 ± 2.7 | −26.4 ± 1.2 |
| After 8 h | 236.9 ± 3.1 | −26.5 ± 1.7 |
| After 1 day | 242.4 ± 5.7 | −27.3 ± 1.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galiyeva, A.; Daribay, A.; Zhumagaliyeva, T.; Zhaparova, L.; Sadyrbekov, D.; Tazhbayev, Y. Human Serum Albumin Nanoparticles: Synthesis, Optimization and Immobilization with Antituberculosis Drugs. Polymers 2023, 15, 2774. https://doi.org/10.3390/polym15132774
Galiyeva A, Daribay A, Zhumagaliyeva T, Zhaparova L, Sadyrbekov D, Tazhbayev Y. Human Serum Albumin Nanoparticles: Synthesis, Optimization and Immobilization with Antituberculosis Drugs. Polymers. 2023; 15(13):2774. https://doi.org/10.3390/polym15132774
Chicago/Turabian StyleGaliyeva, Aldana, Arailym Daribay, Tolkyn Zhumagaliyeva, Lyazzat Zhaparova, Daniyar Sadyrbekov, and Yerkeblan Tazhbayev. 2023. "Human Serum Albumin Nanoparticles: Synthesis, Optimization and Immobilization with Antituberculosis Drugs" Polymers 15, no. 13: 2774. https://doi.org/10.3390/polym15132774
APA StyleGaliyeva, A., Daribay, A., Zhumagaliyeva, T., Zhaparova, L., Sadyrbekov, D., & Tazhbayev, Y. (2023). Human Serum Albumin Nanoparticles: Synthesis, Optimization and Immobilization with Antituberculosis Drugs. Polymers, 15(13), 2774. https://doi.org/10.3390/polym15132774







