Fast, Multiple-Use Optical Biosensor for Point-of-Care Glucose Detection with Mobile Devices Based on Bienzyme Cascade Supported on Polyamide 6 Microparticles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Characterization Methods
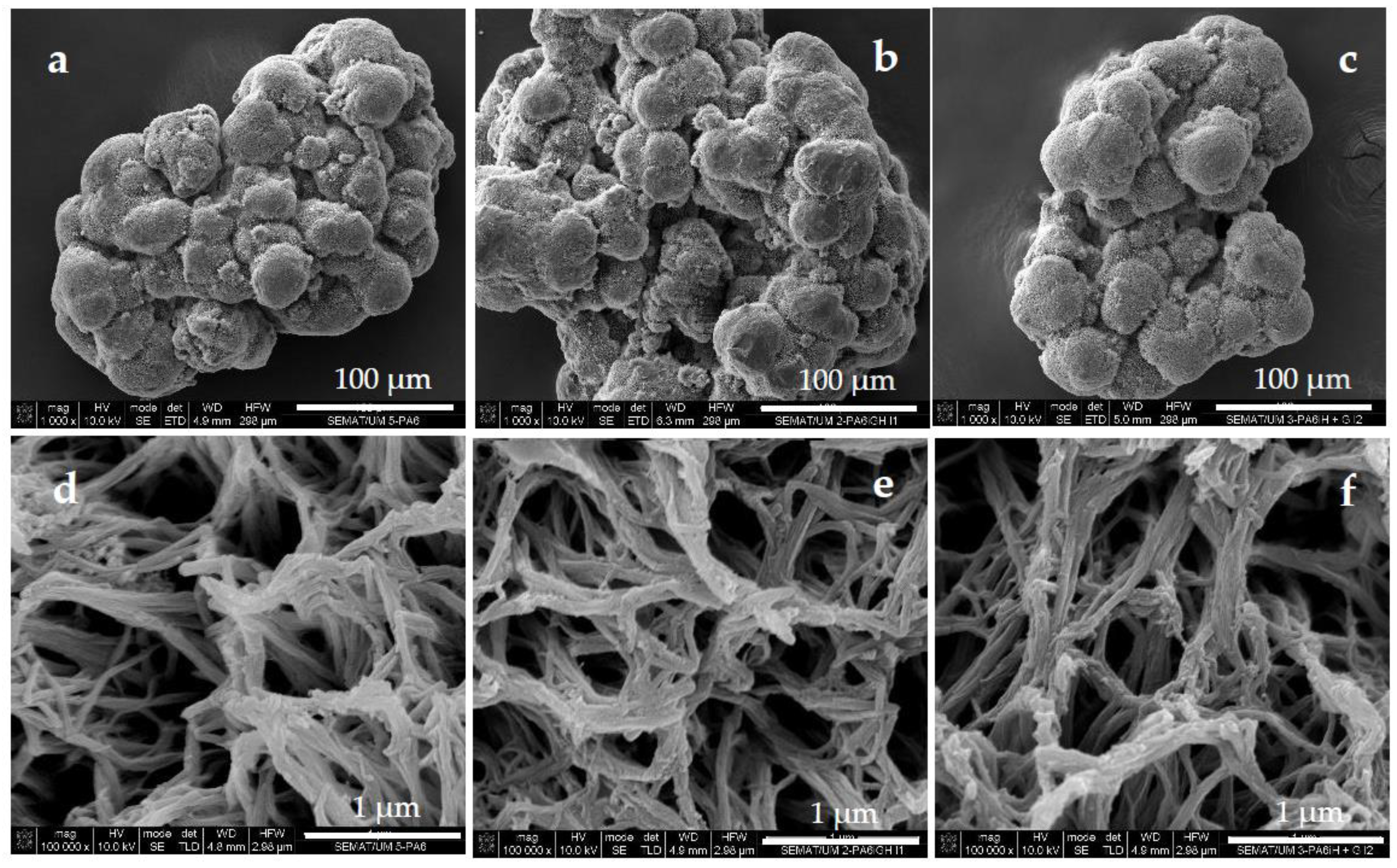
2.3. Synthesis of PA6 Microparticles
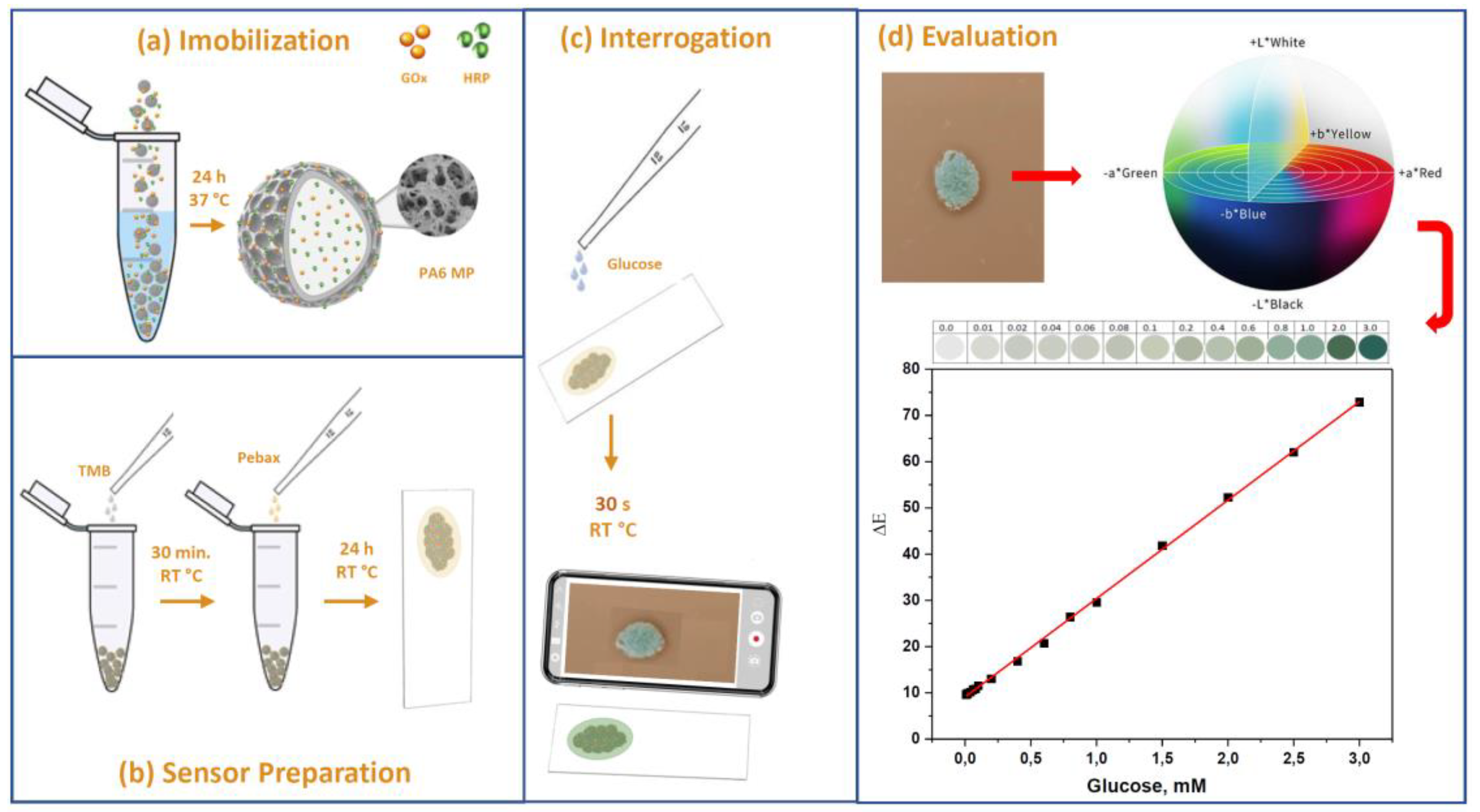
2.4. Immobilization of GOx and HRP on PA6 MPs
2.5. Determination of the Total Amount of Protein in PA6 Microparticles
2.6. Activity Measurements by UV-VIS
2.7. Biosensor Construction and Use
2.8. Detection of Glucose from Smartphone Digital Images
2.9. Detection of Glucose in Real Urine Samples
2.10. Glucose Biosensor Shelf Life and Multiple Uses
3. Results and Discussion
3.1. Morphological and Structural Characterization
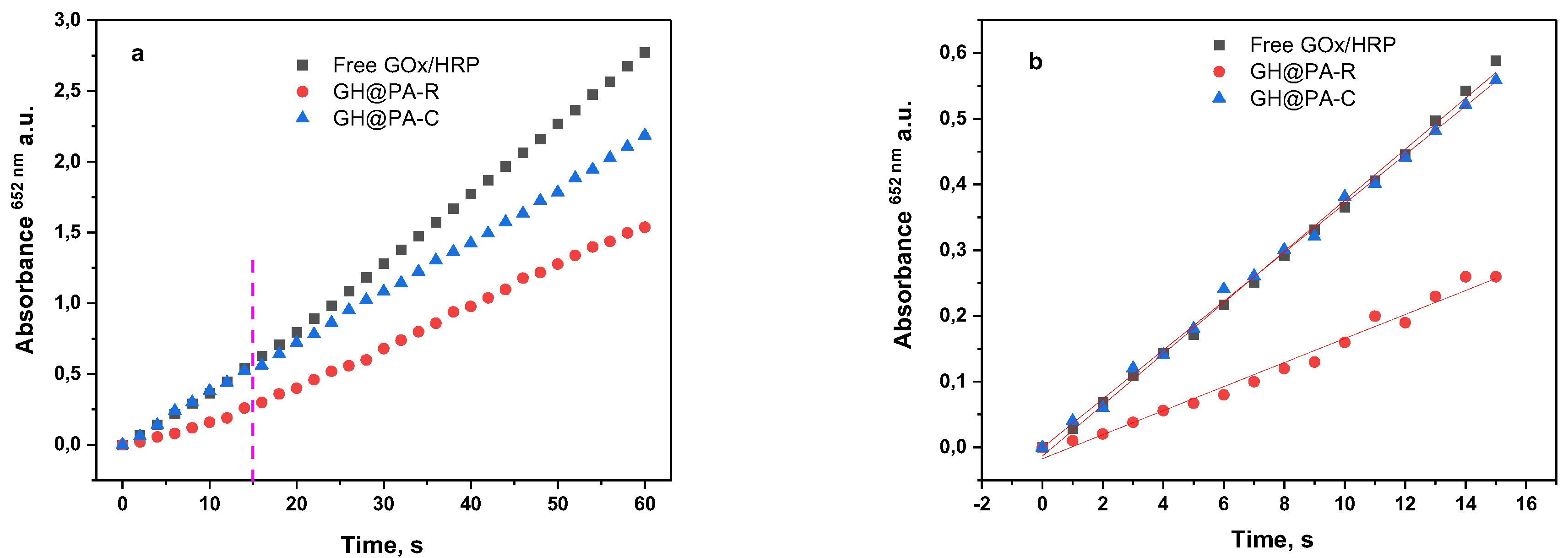
3.2. Description of the GOx-HRP Cascade Reaction
3.3. Comparative Activity Studies
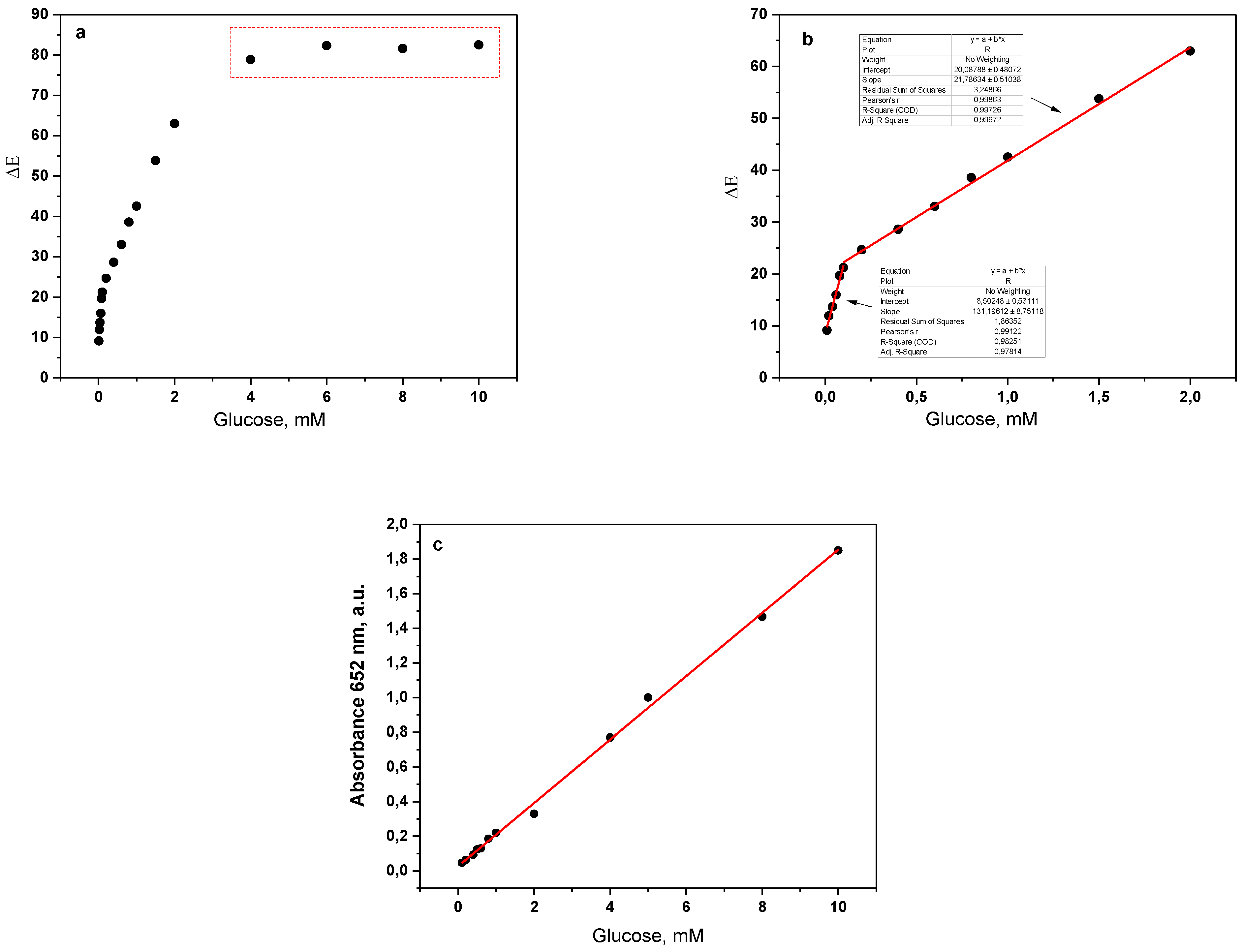
3.4. Detection of Glucose in Water Solutions
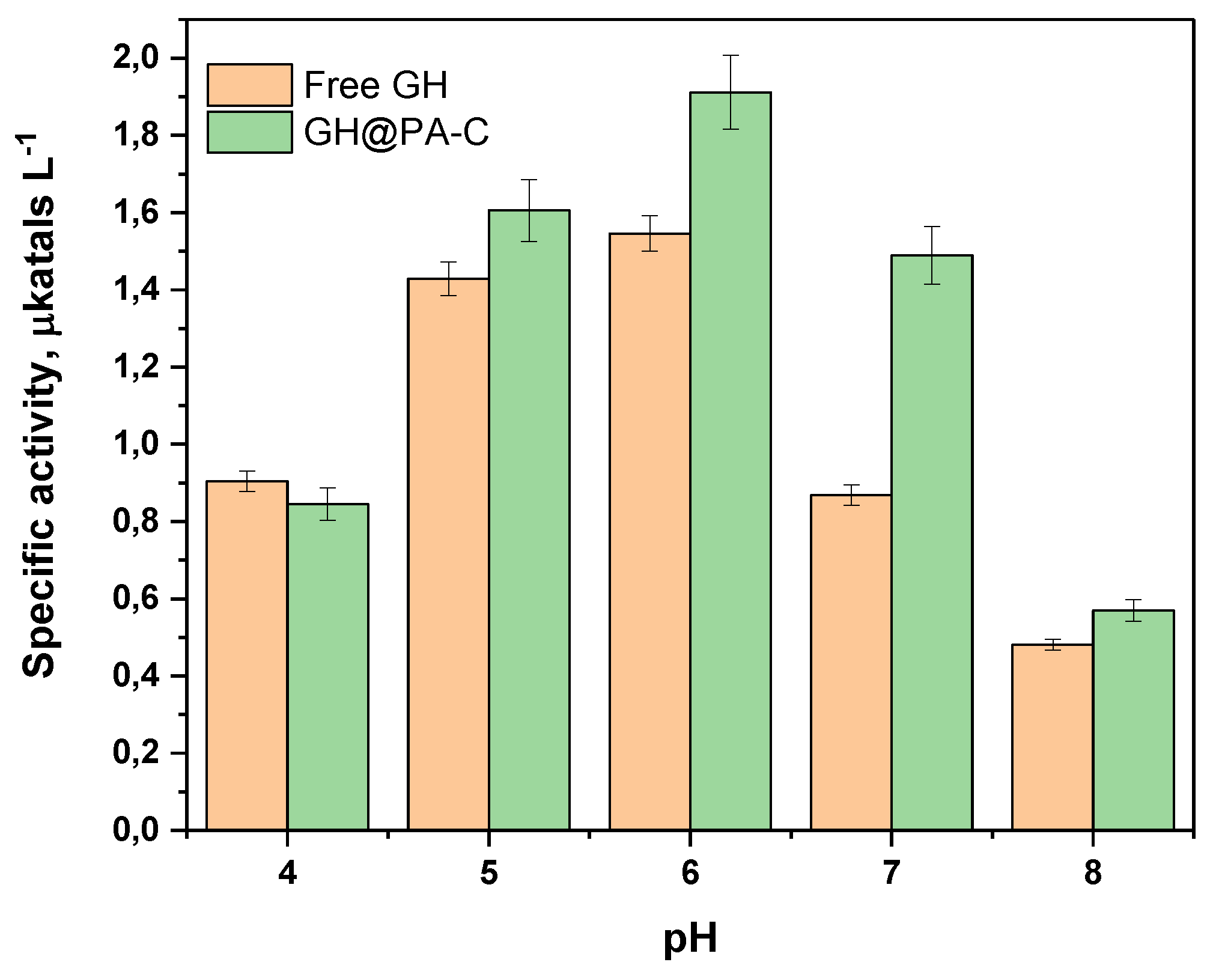
3.5. Sensor Activity at Different pH Values
3.6. Detection of Glucose in Urine
3.7. Shelf Life and Multiple Uses of the Biosensor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vyas, T.; Choudhary, S.; Kumar, N.; Joshi, A. Point-of-Care Biosensors for Glucose Sensing. In Nanobiosensors for Point-of-Care Medical Diagnostics, 1st ed.; Gogoi, M., Patra, S., Kundu, D., Eds.; Springer: Singapore, 2022; pp. 107–136. [Google Scholar]
- McMillan, J.M. Blood Glucose. In Clinical Methods: The History, Physical and Laboratory Examinations, 3rd ed.; Walker, H.K., Hall, W.D., Hurst, J.W., Eds.; Butterworths: Boston, MA, USA, 1990. Available online: https://www.ncbi.nlm.nih.gov/books/NBK248 (accessed on 9 March 2023).
- Karpova, E.V.; Shcherbacheva, E.V.; Galushin, A.A.; Vokhmyanina, D.V.; Karyakina, E.E.; Karyakin, A.A. Noninvasive Diabetes Monitoring through Continuous Analysis of Sweat Using Flow-Through Glucose Biosensor. Anal. Chem. 2019, 91, 3778–3783. [Google Scholar] [CrossRef]
- Lee, H.; Song, C.; Hong, Y.S.; Kim, M.; Cho, H.R.; Kang, T.; Shin, K.; Choi, S.H.; Hyeon, T.; Kim, D.H. Wearable/disposable sweat-based glucose monitoring device with multistage transdermal drug delivery module. Sci. Adv. 2017, 3, e1601314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.F.; Sann, E.E.; Lou, X.; Liu, R.; Dai, J.; Zuo, X.; Xia, F.; Jiang, L. Naked-eye point-of-care testing platform based on a pH-responsive superwetting surface: Toward the non-invasive detection of glucose. NPG Asia Mater. 2018, 10, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Jang, S.; Xu, C. Review of emerging approaches in non- or minimally invasive glucose monitoring and their application to physiological human body fluids. Int. J. Biosen. Bioelectron. 2018, 4, 5–9. [Google Scholar] [CrossRef]
- Bruen, D.; Delaney, C.; Florea, L.; Diamond, D. Glucose Sensing for Diabetes Monitoring: Recent Developments. Sensors 2017, 17, 1866. [Google Scholar] [CrossRef] [Green Version]
- Haxha, S.; Jhoja, J. Optical based non-invasive glucose monitoring sensor prototype. IEEE Photon. J. 2016, 8, 6805911. [Google Scholar] [CrossRef] [Green Version]
- Nery, E.W.; Kundys, M.; Jeleń, P.S.; Jönsson-Niedziółka, M. Electrochemical Glucose Sensing: Is There Still Room for Improvement? Anal. Chem. 2016, 88, 11271–11282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corrie, S.R.; Coffey, J.W.; Islam, J.; Markey, K.A.; Kendall, M.A.F. Blood, sweat, and tears: Developing clinically relevant protein biosensors for integrated body fluid analysis. Analyst 2015, 140, 4350–4364. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.J.; Feng, D.-Q.; Chen, M.; Chen, Z.D.; Zhu, R.; Fang, H.L.; Wang, W. Bienzyme colorimetric detection of glucose with self-calibration based on tree-shaped paper strip. Sens. Actuators B Chem. 2014, 190, 414–418. [Google Scholar] [CrossRef]
- Xu, K.; Chen, X.; Zheng, R.; Zheng, Y. Immobilization of multienzymes on support materials for efficient biocatalysis. Front. Bioeng. Biotechnol. 2020, 8, 660. [Google Scholar] [CrossRef]
- Ge, J.; Lei, J.; Zare, R.N. Protein–inorganic hybrid nanoflowers. Nat. Nanotechnol. 2012, 7, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Braz, J.F.; Dencheva, N.V.; Malfois, M.; Denchev, Z.Z. Synthesis of Novel Polymer-Assisted Organic-Inorganic Hybrid Nanoflowers and Their Application in Cascade Biocatalysis. Molecules 2023, 28, 839. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Ge, J.; Liu, W.; Lan, M.; Zhang, H.; Wang, P.; Wang, Y.; Niu, Z. Multi-enzyme co-embedded organic–inorganic hybrid nanoflowers: Synthesis and application as a colorimetric sensor. Nanoscale 2014, 6, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Ariza-Avidad, M.; Salinas-Castillo, A.; Capitán-Vallvey, L.F. A 3D mPAD based on a multi-enzyme organic–inorganic hybrid nanoflower reactor. Biosens. Bioelectron. 2016, 77, 51–55. [Google Scholar] [CrossRef]
- Luo, X.; Xia, J.; Jiang, X.; Yang, M.; Liu, S. Cellulose-Based Strips Designed Based on a Sensitive Enzyme Colorimetric Assay for the Low Concentration of Glucose Detection. Anal. Chem. 2019, 91, 15461–15468. [Google Scholar] [CrossRef] [PubMed]
- Hosu, O.; Lettieri, M.; Papara, N.; Ravalli, A.; Sandulescu, R.; Cristea, C.; Marrazza, G. Colorimetric multienzymatic smart sensors for hydrogen peroxide, glucose and catechol screening analysis. Talanta 2019, 204, 525–532. [Google Scholar] [CrossRef]
- Luo, M.; Li, M.; Lu, Y.; Xia, M.; Zhao, Q.; Wang, D. In-situ growth of multienzyme-inorganic hybrid nanoflowers on PVA-co-PE nanofibrous strip for colorimetric biosensor. Colloids Surf. A Physicochem. Eng. Asp. 2022, 640, 128419. [Google Scholar] [CrossRef]
- Sun, X.; Li, Y.; Yang, Q.; Xiao, Y.; Zeng, Y.; Gong, J.; Wang, Z.; Tan, X.; Li, H. Self-assembled all-inclusive organic-inorganic nanoparticles enable cascade reaction for the detection of glucose. Chin. Chem. Lett. 2021, 32, 1780–1784. [Google Scholar] [CrossRef]
- Tang, Z.; Li, X.; Tong, L.; Yang, H.; Wu, J.; Zhang, X.; Song, T.; Huang, S.; Zhu, F.; Chen, G.; et al. A Biocatalytic Cascade in an Ultrastable Mesoporous Hydrogen-Bonded Organic Framework for Point-of-Care Biosensing. Angew. Chem. Int. Ed. 2021, 60, 23608–23613. [Google Scholar] [CrossRef]
- Hosu, O.; Ravalli, A.; Piccolo, G.M.L.; Cristea, C.; Sandulescu, R.; Marrazza, G. Smartphone-based immuno-sensor for CA125 detection. Talanta 2017, 166, 234–240. [Google Scholar] [CrossRef]
- Dencheva, N.; Denchev, Z.; Lanceros-Méndez, S.; Ezquerra, T.E. One-step in-situ synthesis of polyamide microcapsules with inorganic payload and their transformation into responsive thermoplastic composite materials. Macromol. Mater. Eng. 2016, 301, 119–124. [Google Scholar] [CrossRef]
- Dencheva, N.; Braz, J.; Nunes, T.G.; Oliveira, F.D.; Denchev, Z. One-pot low temperature synthesis and characterization of hybrid poly(2-pyrrolidone) microparticles suitable for protein immobilization. Polymer 2018, 145, 402–415. [Google Scholar] [CrossRef]
- Cano-Raya, C.; Dencheva, N.V.; Braz, J.F.; Malfois, M.; Denchev, Z.Z. Optical biosensor for catechol determination based on laccase-immobilized anionic polyamide 6 microparticles. J. Appl. Polym. Sci. 2020, 137, e49131. [Google Scholar] [CrossRef]
- Ashiotis, G.; Deschildre, A.; Nawaz, Z.; Wright, J.P.; Karkoulis, D.; Picca, F.E.; Kieffer, J. The fast azimuthal integration Python library: pyFAI. J. Appl. Crystallogr. 2015, 48, 510–519. [Google Scholar] [CrossRef]
- Bartczak, Z.; Galeski, A.; Argon, A.S.; Cohen, R.E. On the plastic deformation of the amorphous component in semicrystalline polymers. Polymer 1996, 37, 2113–2123. [Google Scholar] [CrossRef]
- Josephy, P.D.; Eling, T.; Mason, R.P. The horseradish peroxidase-catalyzed oxidation of 3,5,3′,5′-tetramethylbenzidine. Free radical and charge-transfer complex intermediates. J. Biol. Chem. 1982, 257, 3669–3675. [Google Scholar] [CrossRef]
- Misono, Y.; Ohkata, Y.; Morikawa, T.; Itoh, K. Resonance raman and absorption spectroscopic studies on the electrochemical oxidation processes of 3,3′,5,5′-tetramethylbenzidine. J. Electroanal. Chem. 1997, 436, 203–212. [Google Scholar] [CrossRef]
- Lin, J.L.; Palomec, L.; Wheeldon, I. Design and analysis of enhanced catalysis in scaffolded multienzyme cascade reactions. ACS Catal. 2014, 4, 505–511. [Google Scholar] [CrossRef]
- Luo, A.M.; Shao, Y.; Zhang, K.J.; Wang, Y.W.; Peng, Y. Syntheses of three terbium complexes as fluorescent probes and their application on the pH detection of routine urine test. Chin. Chem. Lett. 2017, 28, 2009–2013. [Google Scholar] [CrossRef]


| Sample | Total Protein, mg | Slope, Abs s−1 (×104) | Specific Activity a, mol L−1 s−1 (×107) | Specific Activity, µkatals.L−1 | Specific Activity, U.L−1 | Relative Activity, % |
|---|---|---|---|---|---|---|
| GH Free | 0.01750 | 389 ± 5 | 9.97692 | 0.99769 | 59.861 | 100.0 |
| GH@PA-R | 0.00501 | 183 ± 6 | 4.68974 | 0.46897 | 28.138 | 47.0 |
| GH@PA-C | 0.00499 | 372 ± 5 | 9.52564 | 0.95256 | 57.154 | 95.5 |
| Quantification | LOD | Linear Range | Response Time | Analyte | Ref. |
|---|---|---|---|---|---|
| UV/VIS | 0.2 µM | 0.5–50 µM | - | Urine | [15] |
| Digital image | 15.6 µM | 0.5–300 µM | 15 min | Urine | [16] |
| UV/VIS Digital image | 0.45 mM | 1–11 mM | 5 min | Urine, water | [17] |
| Digital image UV/VIS | 49 µM 28 µM | 25–200 µM | - | Water | [18] |
| Digital image | 0.14 mM | 0.25–10 µM | - | Water | [19] |
| UV/VIS Thermal | 0.012 mM 0.049 mM | 0.05–10 mM | - | Water | [20] |
| Digital image UV/VIS | 30.7 µM 64.9 µM | 0.01–3.0 mM | 20 s | Urine | This study |
| Sample | No. of Uses | Glucose Added mM | Detection Mean ± SDD mM * | RSD ** n = 3 % | 95% Confidence Interval, mM *** | Recovery, n = 3 % |
|---|---|---|---|---|---|---|
| 1 | 2.00 | 1.9033 ± 0.0308 | 1.616 | 1.9033 ± 0.0348 | 95.163 | |
| 2 | 1st | 0.40 | 0.4076 ± 0.0076 | 1.863 | 0.4076 ± 0.0086 | 101.898 |
| 3 | 0.04 | 0.0458 ± 0.0006 | 1.417 | 0.0458 ± 0.0007 | 114.584 | |
| 4 | 2.00 | 2.0812 ± 0.0194 | 0.930 | 2.0812 ± 0.0219 | 104.060 | |
| 5 | 1st | 0.40 | 0.4002 ± 0.0117 | 2.920 | 0.4002 ± 0.0133 | 100.042 |
| 6 | 0.04 | 0.0402 ± 0.0041 | 10.200 | 0.0402 ± 0.0046 | 100.576 | |
| 4 | 2.00 | 2.0438 ± 0.0221 | 1.083 | 2.0438 ± 0.0251 | 102.193 | |
| 5 | 2nd | 0.40 | 0.4089 ± 0.0262 | 6.427 | 0.4089 ± 0.0297 | 102.219 |
| 6 | 0.04 | 0.0402 ± 0.0008 | 21.000 | 0.0402 ± 0.0100 | 100.579 | |
| 4 | 2.00 | 2.0928 ± 0.0579 | 2.766 | 2.0928 ± 0.0655 | 104.642 | |
| 5 | 3rd | 0.40 | 0.4138 ± 0.0251 | 6.068 | 0.4138 ± 0.0284 | 103.451 |
| 6 | 0.04 | 0.04276 ± 0.012 | 29.114 | 0.0428 ± 0.0141 | 106.898 | |
| 4 | 2.00 | 2.1028 ± 0.0461 | 2.1923 | 2.1028 ± 0.0522 | 105.141 | |
| 5 | 4th | 0.40 | 0.4290 ± 0.0237 | 5.525 | 0.4290 ± 0.0268 | 107.256 |
| 6 | 0.04 | 0.0442 ± 0.099 | 22.418 | 0.0442 ± 0.0112 | 110.553 | |
| 4 | 2.00 | 2.1428 ± 0.0807 | 3.766 | 2.1428 ± 0.0913 | 107.138 | |
| 5 | 5th | 0.40 | 0.4174 ± 0.0138 | 3.299 | 0.4174 ± 0.0156 | 104.351 |
| 6 | 0.04 | 0.0427 ± 0.0022 | 5.222 | 0.0427 ± 0.0025 | 106.811 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Braz, J.F.; Dencheva, N.V.; Tohidi, S.D.; Denchev, Z.Z. Fast, Multiple-Use Optical Biosensor for Point-of-Care Glucose Detection with Mobile Devices Based on Bienzyme Cascade Supported on Polyamide 6 Microparticles. Polymers 2023, 15, 2802. https://doi.org/10.3390/polym15132802
Braz JF, Dencheva NV, Tohidi SD, Denchev ZZ. Fast, Multiple-Use Optical Biosensor for Point-of-Care Glucose Detection with Mobile Devices Based on Bienzyme Cascade Supported on Polyamide 6 Microparticles. Polymers. 2023; 15(13):2802. https://doi.org/10.3390/polym15132802
Chicago/Turabian StyleBraz, Joana F., Nadya V. Dencheva, Shafagh D. Tohidi, and Zlatan Z. Denchev. 2023. "Fast, Multiple-Use Optical Biosensor for Point-of-Care Glucose Detection with Mobile Devices Based on Bienzyme Cascade Supported on Polyamide 6 Microparticles" Polymers 15, no. 13: 2802. https://doi.org/10.3390/polym15132802
APA StyleBraz, J. F., Dencheva, N. V., Tohidi, S. D., & Denchev, Z. Z. (2023). Fast, Multiple-Use Optical Biosensor for Point-of-Care Glucose Detection with Mobile Devices Based on Bienzyme Cascade Supported on Polyamide 6 Microparticles. Polymers, 15(13), 2802. https://doi.org/10.3390/polym15132802










