Efficiency Assessment between Entrapment and Covalent Bond Immobilization of Mutant β-Xylosidase onto Chitosan Support
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Mutagenesis, Overexpression, and Partial Purification of XynB2Y509E
2.3. Preparation of Chitosan Spheres and Immobilization
2.4. Enzymatic Assays
2.5. Optimization of Immobilization Processes
2.6. Biochemical Characterization of Immobilized XynB2Y509E
3. Results and Discussion
3.1. Immobilization of XynB2Y509E in Chitosan Spheres by Entrapment
3.2. Imobilization of XynB2Y509E on Chitosan Spheres by Covalent Bond Formation
3.3. Effect of Temperature and Thermostability
3.4. Effect of pH on Activity and Stability
3.5. Effect of Immobilization on Kinetic Parameters
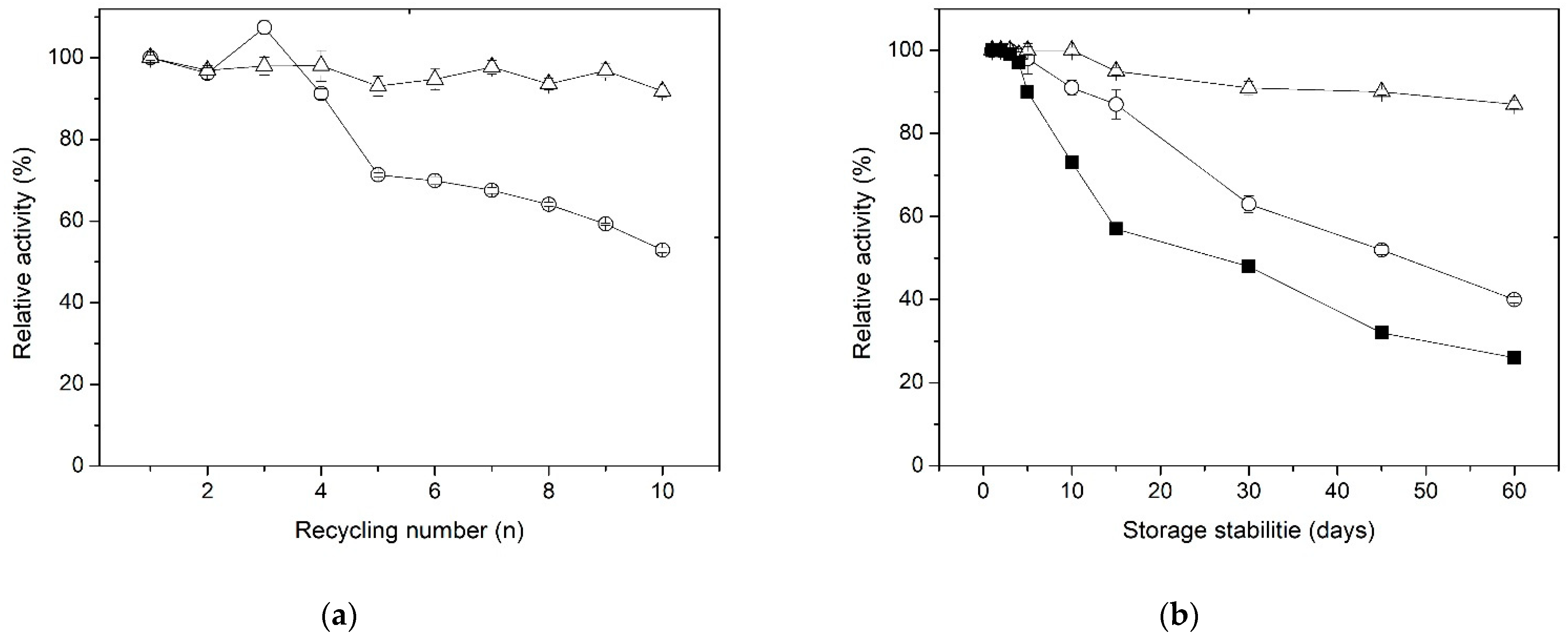
3.6. Operational and Storage Stability
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Boudrant, J.; Woodley, J.M.; Fernandez-Lafuente, R. Parameters necessary to define an immobilized enzyme preparation. Process Biochem. 2020, 90, 66–80. [Google Scholar] [CrossRef]
- Mohamad, N.R.; Marzuki, N.H.C.; Buang, N.A.; Huyop, F.; Wahab, R.A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A. Engineering a more sustainable world through catalysis and green chemistry. J. R. Soc. Interface 2016, 13, 20160087. [Google Scholar] [CrossRef] [PubMed]
- Meldal, M.; Schoffelen, S. Recent advances in covalent, site-specific protein immobilization. F1000Research 2016, 5, 2303. [Google Scholar] [CrossRef]
- Arana-Peña, S.; Carballares, D.; Morellon-Sterlling, R.; Berenguer-Murcia, Á.; Alcántara, A.R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Enzyme co-immobilization: Always the biocatalyst designers’ choice…or not? Biotechnol. Adv. 2020, 51, 107584. [Google Scholar] [CrossRef] [PubMed]
- Guisan, J.M. (Ed.) New opportunities for immobilization of enzymes. In Methods in Molecular Biology; Springer: Clifton, NJ, USA, 2013; pp. 1–13. [Google Scholar] [CrossRef]
- Sheldon, R.; van Pelt, S. Enzyme immobilization in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223. [Google Scholar] [CrossRef]
- Rehm, F.B.H.; Chen, S.; Rehm, B.H.A. Enzyme engineering for in situ immobilization. Molecules 2016, 21, 1370. [Google Scholar] [CrossRef] [PubMed]
- Brena, B.; González-Pombo, P.; Batista-Viera, F. Immobilization of enzymes: A literature survey. Methods Mol. Biol. 2013, 1051, 15–31. [Google Scholar] [CrossRef]
- Ali, S.; Zafar, W.; Shafiq, S.; Manzoor, M. Enzymes Immobilization: An overview of techniques, support materials and its applications. Int. J. Sci. Res. 2017, 6, 4–72. [Google Scholar]
- Graebin, N.; Schöffer, J.; de Andrades, D.; Hertz, P.; Ayub, M.; Rodrigues, R. Immobilization of glycoside hydrolase familie GH1, GH13, and GH70: State of the art and perspectives. Molecules 2016, 21, 1074. [Google Scholar] [CrossRef]
- Sadaqat, B.; Sha, C.; Dar, M.A.; Dhanavade, M.J.; Sonawane, K.D.; Mohamed, H.; Shao, W.; Song, Y. Modifying Thermostability and Reusability of Hyperthermophilic Mannanase by Immobilization on Glutaraldehyde Cross-Linked Chitosan Beads. Biomolecules 2022, 12, 999. [Google Scholar] [CrossRef] [PubMed]
- Nunes, Y.L.; de Menezes, F.L.; de Sousa, I.G.; Cavalcante, A.L.G.; Cavalcante, F.T.T.; da Silva Moreira, K.; de Oliveira, A.L.B.; Mota, G.F.; Souza, J.E.D.S.; Falcão, I.R.D.A.; et al. Chemical and physical Chitosan modification for designing enzymatic industrial biocatalysts: How to choose the best strategy? Int. J. Biol. Macromol. 2021, 181, 1124–1170. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, P.S.; Selvakumar, D.; Kadirvelu, K.; Kumar, N.S. Chitosan as an environment friendly biomaterial—A review on recent modifications and applications. Int. J. Biol. Macromol. 2020, 150, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Sulej, J.; Osińska-Jaroszuk, M.; Jaszek, M.; Olszewska, A.; Belcarz, A.; Piątek-Gołda, W. Chitosan as a Promising Support of a CDH Activity Preservation System for Biomedical and Industrial Applications. Int. J. Mol. Sci. 2023, 24, 4535. [Google Scholar] [CrossRef]
- Verma, M.L.; Kumar, S.; Das, A.; Randhawa, J.; Chamundeeswar, M. Chitin and chitosan-based support materials for enzyme immobilization and biotechnological applications. Environ. Chem. Lett. 2020, 18, 315–323. [Google Scholar] [CrossRef]
- Kaczmarek, M.B.; Struszczyk-Swita, K.; Li, X.; Szczęsna-Antczak, M.; Daroch, M. Enzymatic modifications of chitin, chitosan, and chitooligosaccharides. Front. Bioeng. Biotechnol. 2019, 7, 243. [Google Scholar] [CrossRef]
- Wu, Z.; Zhou, W.; Deng, W.; Xu, C.; Cai, Y.; Wang, X. Antibacterial and hemostatic thiol-modified chitosan-immobilized AgNPs composite sponges. ACS Appl. Mater. Interfaces 2020, 12, 20307–20320. [Google Scholar] [CrossRef]
- Lawrie, G.; Keen, I.; Drew, B.; Chandler-Temple, A.; Rintoul, L.; Fredericks, P.; Grøndahl, L. Interactions between alginate and chitosan biopolymers characterized using FTIR and XPS. Biomacromolecules 2007, 8, 2533–2541. [Google Scholar] [CrossRef]
- Mathew, S.A.; Arumainathan, S. Crosslinked Chitosan-Gelatin Biocompatible Nanocomposite as a Neuro Drug Carrier. ACS Omega 2022, 7, 18732–18744. [Google Scholar] [CrossRef]
- Rana, M.; Kumari, A.; Chauhan, G.S.; Chauhan, K. Modified chitosan microspheres in non-aggregated amylase immobilization. Int. J. Biol. Macromol. 2014, 66, 46–51. [Google Scholar] [CrossRef]
- Corradini, F.A.; Milessi, T.S.; Gonçalves, V.M.; Ruller, R.; Sargo, C.R.; Lopes, L.A.; Zangirolami, T.C.; Tardioli, P.W.; Giordano, R.C.; Giordano, R.L. High stabilization and hyperactivation of a Recombinant β-xylosidase through Immobilization Strategies. Enzym. Microb. Technol. 2021, 145, 109725. [Google Scholar] [CrossRef]
- Canio, D.; Bari, D.; Patrizi, R. Latest Frontiers in the Biotechnologies for Ethanol Production from Lignocellulosic Biomass. In Biofuel Production-Recent Developments and Prospects; InTech: London, UK, 2011. [Google Scholar] [CrossRef]
- Bravman, T.; Zolotnitsky, G.; Shulami, S.; Belakhov, V.; Solomon, D.; Baasov, T.; Shoham, G.; Shoham, Y. Stereochemistry of family 52 glycosyl hydrolases: A beta-xylosidase from Bacillus stearothermophilus T-6 is a retaining enzyme. FEBS Lett. 2001, 495, 39–43. [Google Scholar] [CrossRef]
- Contreras, L.M.; Gómez, J.; Prieto, J.; Clemente-Jiménez, J.M.; Las Heras-Vázquez, F.J.; Rodríguez-Vico, F.; Blanco, F.J.; Neira, J.L. The family 52 β-xylosidase from Geobacillus stearothermophilus is a dimer: Structural and biophysical characterization of a glycoside hydrolase. Biochim. Biophys. Acta-Proteins Proteom. 2008, 1784, 1924–1934. [Google Scholar] [CrossRef] [PubMed]
- Bravman, T.; Belakhov, V.; Solomon, D.; Shoham, G.; Henrissat, B.; Baasov, T.; Shoham, Y. Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus stearothermophilus T-6. J. Biol. Chem. 2003, 278, 26742–26749. [Google Scholar] [CrossRef]
- Bravman, T.; Zolotnitsky, G.; Belakhov, V.; Shoham, G.; Henrissat, B.; Baasov, T.; Shoham, Y. Detailed kinetic analysis of a family 52 glycoside hydrolase: A β-xylosidase from Geobacillus stearothermophilus. Biochemistry 2003, 42, 10528–10536. [Google Scholar] [CrossRef]
- Kurz, L.; García, V.; Wilkesman, J.; Contreras, L.M. Enzymatic characterization of the recombinant beta-xylosidase XynB2. J. Med. Biol. Sci. 2014, 1, 14–19. [Google Scholar]
- Ben-David, A.; Bravman, T.; Balazs, Y.S.; Czjzek, M.; Schomburg, D.; Shoham, G.; Shoham, Y. Glycosynthase activity of Geobacillus stearothermophilus GH52 β-xylosidase: Efficient synthesis of xylooligosaccharides from a D-xylopyranosyl fluoride through a conjugated reaction. Chembiochem 2007, 8, 2145–2151. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Liu, X.; Zhang, S.; Liu, Z. GH52 xylosidase from Geobacillus stearothermophilus: Characterization and introduction of xylanase activity by site-directed mutagenesis of Tyr509. J. Ind. Microbiol. Biotechnol. 2014, 41, 65–74. [Google Scholar] [CrossRef]
- Hong, S.; Kyung, M.; Jo, I.; Kim, Y.R.; Ha, N.C. Structure-based protein engineering of bacterial β-xylosidase to increase the production yield of xylobiose from xylose. Biochem. Biophys. Res. Commun. 2018, 501, 703–710. [Google Scholar] [CrossRef]
- Romero, G.; Contreras, L.M.; Aguirre, C.; Wilkesman, J.; Clemente-Jiménez, J.M.; Rodríguez-Vico, F.; Rodríguez-Vico, F.; Heras-Vázquez, F.J.L. Characterization of Cross-Linked Enzyme Aggregates of the Y509E Mutant of a Glycoside Hydrolase Family 52 β-xylosidase from G. stearothermophilus. Molecules 2021, 26, 451. [Google Scholar] [CrossRef]
- Kamburov, M.; Lalov, I. Preparation of chitosan beads for trypsin immobilization. Biotechnol. Biotechnol. Equip. 2012, 26 (Suppl. 1), 156–163. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Moehlenbrock, M.J.; Minteer, S.D. Introduction to the field of enzyme immobilization and stabilization. Methods Mol. Biol. 2010, 679, 1–7. [Google Scholar] [CrossRef]
- Imam, H.T.; Marr, P.C.; Marr, A.C. Enzyme entrapment, biocatalyst immobilization without covalent attachment. Green Chem. 2021, 23, 4980–5005. [Google Scholar] [CrossRef]
- Tanaka, H.; Matsumura, M.; Veliky, I.A. Diffusion characteristics of substrates in Ca-alginate gel beads. Biotechnol. Bioeng. 1984, 26, 53–58. [Google Scholar] [CrossRef]
- Sun, S.F.; Zhang, Y. A novel process to prepare chitosan macrospheres without shrinkage and its application to immobilize β-galactosidase. J. Chem. 2009, 6, 1211–1220. [Google Scholar] [CrossRef]
- Bilal, M.; Jing, Z.; Zhao, Y.; Iqbal, H.M.N. Immobilization of fungal laccase on glutaraldehyde cross-linked chitosan beads and its bio-catalytic potential to degrade bisphenol A. Biocatal. Agric. Biotechnol. 2019, 19, 101174. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in bio-catalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef]
- Migneault, I.; Dartiguenave, C.; Bertrand, M.J.; Waldron, K.C. Glutaraldehyde: Behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques 2004, 37, 790–802. [Google Scholar] [CrossRef]
- Betancor, L.; López-Gallego, F.; Hidalgo, A.; Alonso-Morales, N.; Mateo, G.D.O.C.; Fernández-Lafuente, R.; Guisán, J.M. Different mechanisms of protein immobilization on glutaraldehyde activated supports: Effect of support activation and immobilization conditions. Enzym. Microb. Technol. 2006, 39, 877–882. [Google Scholar] [CrossRef]
- Chitosan; Matexcel: Shirley, NY, USA, 2023; Available online: https://www.matexcel.com/category/products/natural-materials/naturally-extracted-materials/chitosan/#:~:text=Chitosan%20is%20produced%20commercially%20from,can%20be%20converted%20to%20chitosan (accessed on 19 June 2023).
- Chitosan-Low Molecular Weight. Product Specification; Sigma-Aldrich: Saint Louis, MO, USA, 2023; Available online: https://www.sigmaaldrich.com/specification-sheets/462/398/448869-BULK_______ALDRICH__.pdf (accessed on 19 June 2023).
- Zaak, H.; Siar, E.H.; Kornecki, J.F.; Fernandez-Lopez, L.; Pedrero, S.G.; Virgen-Ortíz, J.J.; Fernandez-Lafuente, R. Effect of immobilization rate and enzyme crowding on enzyme stability under different conditions. The case of lipase from Thermomyces lanuginosus immobilized on octyl agarose beads. Process Biochem. 2017, 56, 117–123. [Google Scholar] [CrossRef]
- Alsarra, I.A.; Betigeri, S.S.; Zhang, H.; Evans, B.A.; Neau, S.H. Molecular weight and degree of deacetylation effects on lipase-loaded chitosan bead characteristics. Biomaterials 2002, 23, 3637–3644. [Google Scholar] [CrossRef]
- Karami, F.; Ghorbani, M.; Mahoonak, A.S.; Khodarahmi, R. Fast, inexpensive purification of β-glucosidase from Aspergillus niger and improved catalytic/physicochemical properties upon the enzyme immobilization: Possible broad prospects for industrial applications. Food Sci. Technol. 2020, 118, 108770. [Google Scholar] [CrossRef]
- Singh, A.N.; Singh, S.; Suthar, N.; Dubey, V.K. Glutaraldehyde-activated chitosan matrix for immobilization of a novel cysteine protease, procerain B. J. Agric. Food Chem. 2011, 59, 6256–6262. [Google Scholar] [CrossRef]
- Munjal, N.; Sawhney, S.K. Stability and properties of mushroom tyrosinase entrapped in alginate, polyacrylamide and gelatin gels. Enzym. Microb. Technol. 2002, 30, 613–619. [Google Scholar] [CrossRef]
- Asgher, M.; Noreen, S.; Bilal, M. Enhancement of catalytic, reusability, and long-term stability features of Trametes versicolor IBL-04 laccase immobilized on different polymers. Int. J. Biol. Macromol. 2017, 95, 54–62. [Google Scholar] [CrossRef]
- Roy, S.K.; Raha, S.K.; Dey, S.K.; Chakrabarty, S.L. Immobilization of β-glucosidase from Myceliophthora thermophila D-14. Enzym. Microb. Technol. 1989, 11, 431–435. [Google Scholar] [CrossRef]
- Abdel-Naby, M.A. Immobilization of Aspergillus niger NRC 107 xylanase and b-xylosidase, and properties of the immobilized enzymes. Appl. Biochem. Biotechnol. 1993, 38, 69–81. [Google Scholar] [CrossRef]
- Figueira, J.D.A.; Dias, F.F.G.; Sato, H.H.; Fernandes, P. Screening of Supports for the Immobilization of-Glucosidase. Enzym. Res. 2011, 2011, 642460. [Google Scholar] [CrossRef]
- Asgher, M.; Noreen, S.; Bilal, M. Enhancing catalytic functionality of Trametes versicolor IBL-04 laccase by immobilization on chitosan microspheres. Chem. Eng. Res. Des. 2017, 119, 1–11. [Google Scholar] [CrossRef]
- Aslam, S.; Asgher, M.; Khan, N.A.; Bilal, M. Immobilization of Pleurotus nebrodensis WC 850 laccase on glutaraldehyde cross-linked chitosan beads for enhanced biocatalytic degradation of textile dyes. J. Water Process Eng. 2021, 40, 101971. [Google Scholar] [CrossRef]
- Blagodatskikh, I.V.; Kulikov, S.N.; Vyshivannaya, O.V.; Bezrodnykh, E.A.; Tikhonov, V.E. N-reacetylated oligochitosan: pH dependence of self-assembly properties and antibacterial activity. Biomacromolecules 2017, 18, 1491–1498. [Google Scholar] [CrossRef]
- Jaiswal, N.; Pandey, V.P.; Dwivedi, U.N. Immobilization of papaya laccase in chitosan led to improved multipronged stability and dye discoloration. Int. J. Biol. Macromol. 2016, 86, 288–295. [Google Scholar] [CrossRef]
- Chen, H.; Liu, L.; Lv, S.; Liu, X.; Wang, M.; Song, A.; Jia, X. Immobilization of Aspergillus niger xylanase on chitosan using dialdehyde starch as a coupling agent. Appl. Biochem. Biotechnol. 2010, 162, 24–32. [Google Scholar] [CrossRef]
- Jiang, Y.; Wu, Y.; Li, H. Immobilization of Thermomyces lanuginosus Xylanase on Aluminum Hydroxide Particles Through Adsorption: Characterization of Immobilized Enzyme. J. Microbiol. Biotechnol. 2015, 25, 2016–2023. [Google Scholar] [CrossRef]
- Işık, M. High Stability of Immobilized Acetylcholinesterase on Chitosan Beads. ChemistrySelect 2020, 5, 4623–4627. [Google Scholar] [CrossRef]
- Monier, M.; Ayad, D.M.; Wei, Y.; Sarhan, A.A. Immobilization of horseradish peroxidase on modified chitosan beads. Int. J. Biol. Macromol. 2010, 46, 324–330. [Google Scholar] [CrossRef]
- Pal, A.; Khanum, F. Covalent immobilization of xylanase on glutaraldehyde activated alginate beads using response surface methodology: Characterization of the immobilized enzyme. Process Biochem. 2011, 46, 1315–1322. [Google Scholar] [CrossRef]
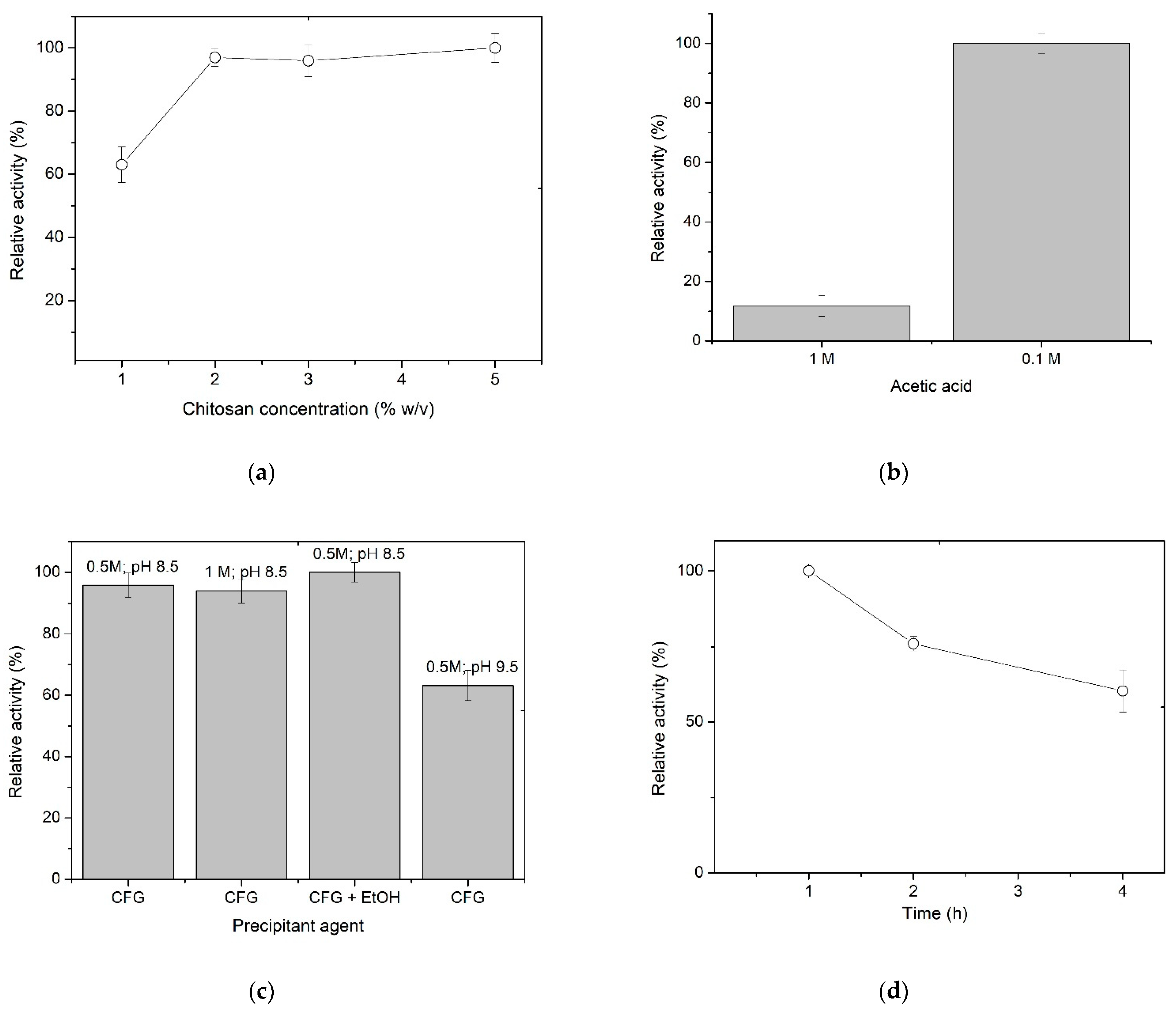
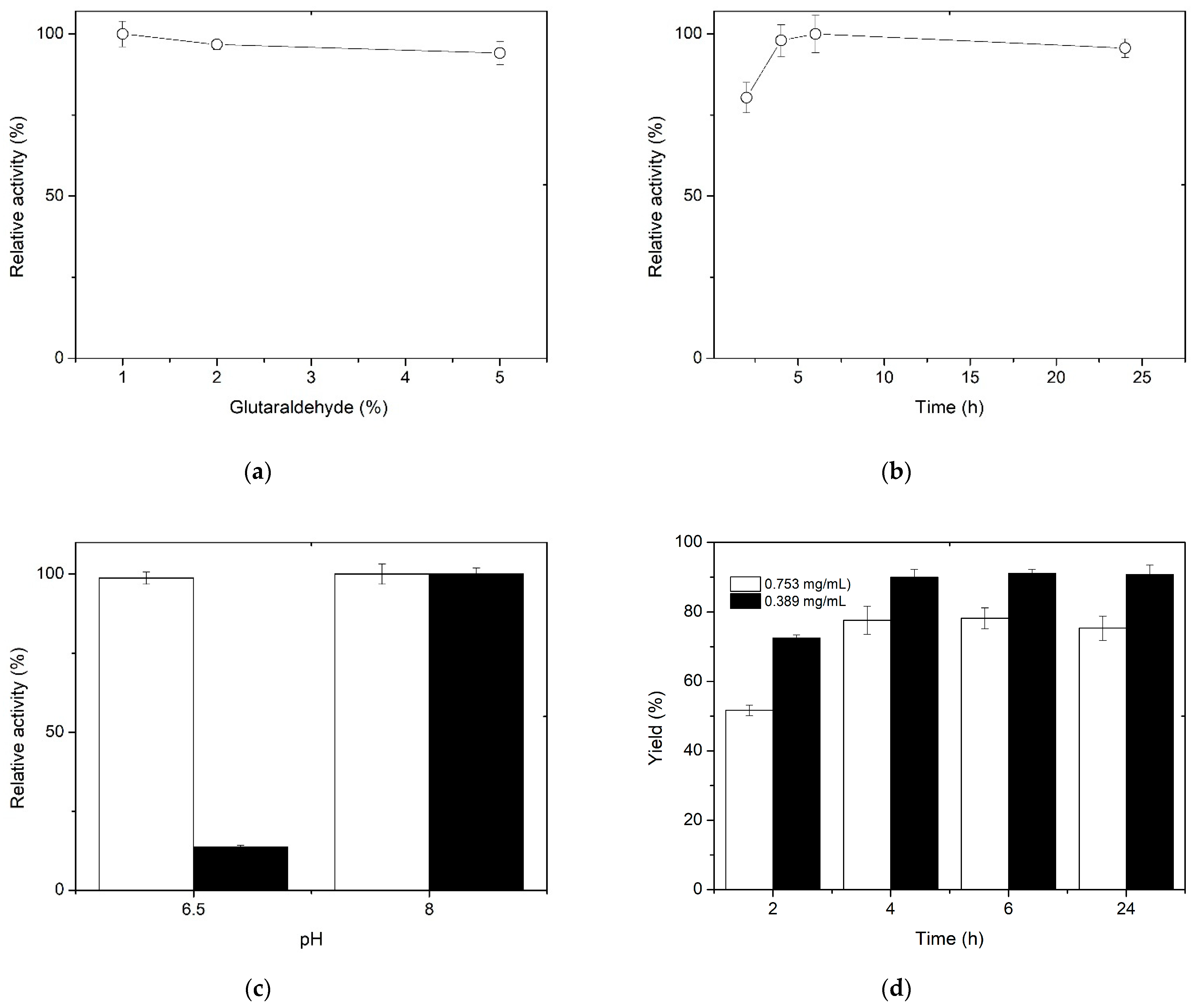
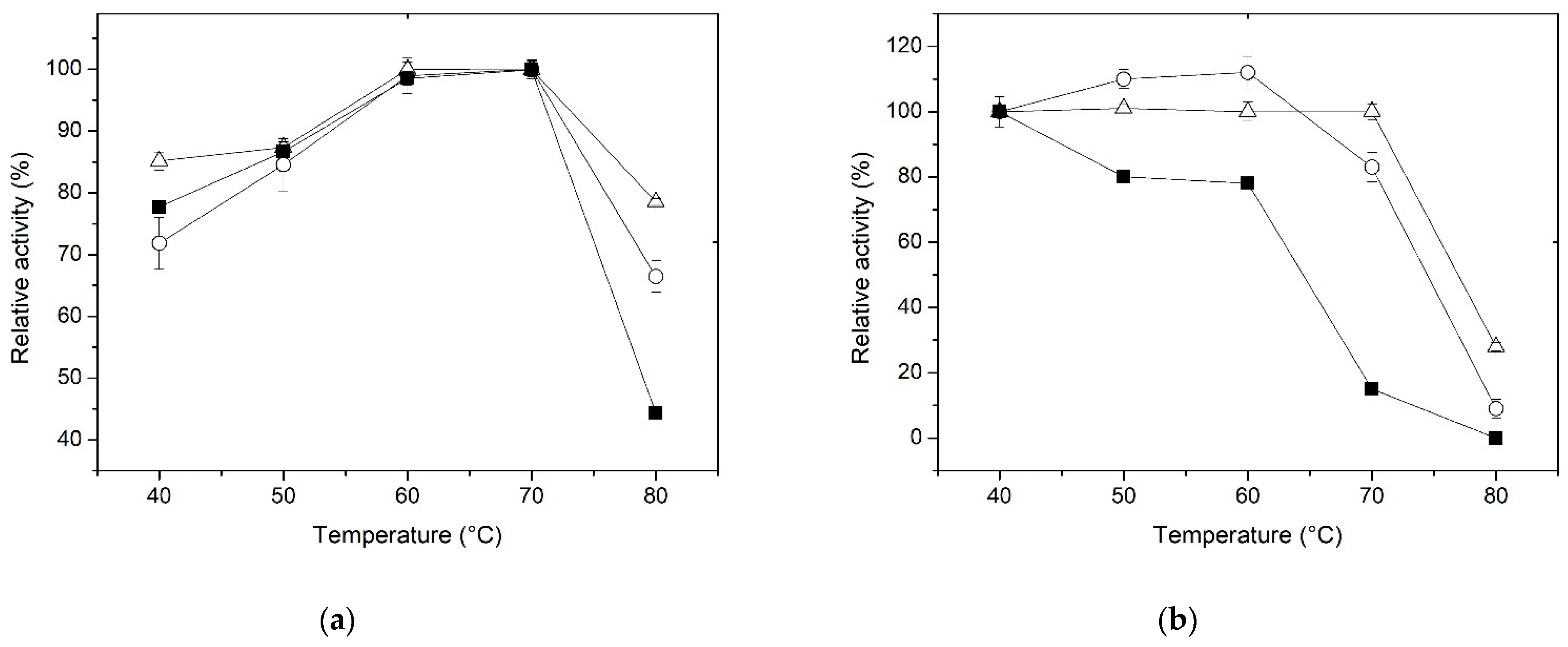
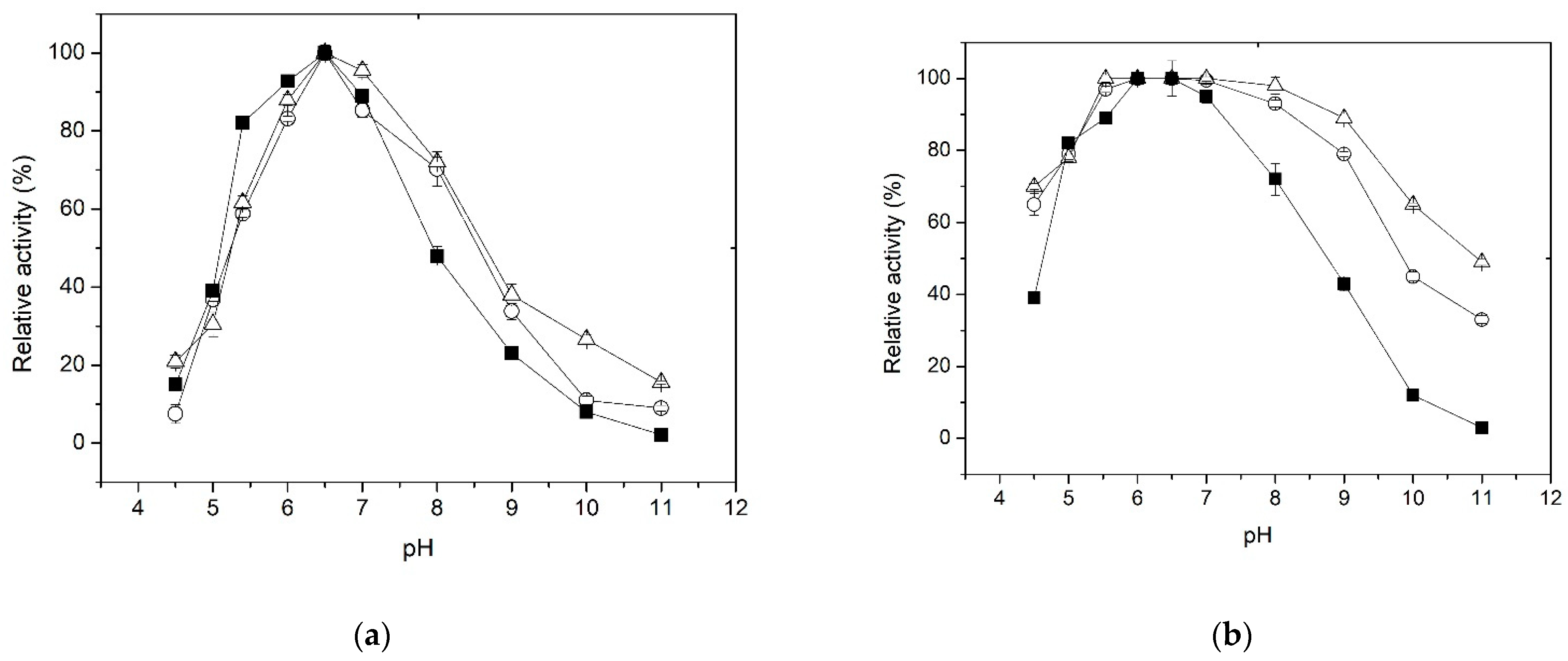
| Immobilization Parameter | Method | |
|---|---|---|
| Entrapment | Covalent | |
| PIY (%) a | 68 | 90 |
| EY (%) b | 7.9 | 30 |
| EAY (%) c | 38 | 67 |
| AEI (IU.g−1 support) d | 99.4 ± 0.5 | 122.3 ± 1.3 |
| XynB2Y509E | Km (mM) | Vmax (nmol/min) | kcat (s−1) | kcat/Km (s−1 M−1) |
|---|---|---|---|---|
| Free | 0.9 ±0.1 | 1.1 ± 0.2 | 3.8 | 4.2 |
| Covalent bond immobilization | 0.9 ±0.1 | 1.0 ±0.3 | 0.1 | 0.1 |
| Entrapment immobilization | 0.7 ± 0.1 | 0.3 ± 0.1 | 0.01 | 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romero, G.; Contreras, L.M.; Aguirre Céspedes, C.; Wilkesman, J.; Clemente-Jiménez, J.M.; Rodríguez-Vico, F.; Las Heras-Vázquez, F.J. Efficiency Assessment between Entrapment and Covalent Bond Immobilization of Mutant β-Xylosidase onto Chitosan Support. Polymers 2023, 15, 3170. https://doi.org/10.3390/polym15153170
Romero G, Contreras LM, Aguirre Céspedes C, Wilkesman J, Clemente-Jiménez JM, Rodríguez-Vico F, Las Heras-Vázquez FJ. Efficiency Assessment between Entrapment and Covalent Bond Immobilization of Mutant β-Xylosidase onto Chitosan Support. Polymers. 2023; 15(15):3170. https://doi.org/10.3390/polym15153170
Chicago/Turabian StyleRomero, Gabriela, Lellys M. Contreras, Carolina Aguirre Céspedes, Jeff Wilkesman, Josefa María Clemente-Jiménez, Felipe Rodríguez-Vico, and Francisco Javier Las Heras-Vázquez. 2023. "Efficiency Assessment between Entrapment and Covalent Bond Immobilization of Mutant β-Xylosidase onto Chitosan Support" Polymers 15, no. 15: 3170. https://doi.org/10.3390/polym15153170
APA StyleRomero, G., Contreras, L. M., Aguirre Céspedes, C., Wilkesman, J., Clemente-Jiménez, J. M., Rodríguez-Vico, F., & Las Heras-Vázquez, F. J. (2023). Efficiency Assessment between Entrapment and Covalent Bond Immobilization of Mutant β-Xylosidase onto Chitosan Support. Polymers, 15(15), 3170. https://doi.org/10.3390/polym15153170









